The Role of dinB in the Appearing of Antibiotics Resistance in E.coli
2016-02-13XIYunHUANGJingTAOLingZHAOPengCHENYoumingFUYongmeiSUNHengbiaoYOUXuXIAOGang
XI Yun,HUANG Jing,TAO Ling,ZHAO Peng,CHEN You-ming,FU Yong-mei,SUN Heng-biao,YOU Xu,XIAO Gang*
(1.Dept.of Lab.,3rd Affil.Hosp.of Zhongshan Uni.;2.Dept.of Lab.,3rd Affil.Hosp.of S.Med.Uni.; 3.Dept.of Parm,3rd Affil.Hosp.of Zhongshan Uni;4.Dept.of Emergency,3rd Affil.Hosp.of Zhongshan Uni.,Guangzhou 510630)
The Role of dinB in the Appearing of Antibiotics Resistance in E.coli
XI Yun1,HUANG Jing2,TAO Ling3,ZHAO Peng2,CHEN You-ming2,FU Yong-mei4,SUN Heng-biao2,YOU Xu2,XIAO Gang2*
(1.Dept.of Lab.,3rd Affil.Hosp.of Zhongshan Uni.;2.Dept.of Lab.,3rd Affil.Hosp.of S.Med.Uni.; 3.Dept.of Parm,3rd Affil.Hosp.of Zhongshan Uni;4.Dept.of Emergency,3rd Affil.Hosp.of Zhongshan Uni.,Guangzhou 510630)
The role of dinB gene in the appearing of antibiotics resistance was studied.Plasmid containing multi-copy dinB gene was transfected into E.coli to create an overexpression.The strains carrying multi-copies of dinB gene demonstrate a significant survival advantage over the wild strain.In vitro experiment,the dinB-overexpressed strain evolved resistance within 8 hours,while wild strain could not.In vivo experiment with mice model infected with dinB-overexpressed strain,resistant clones emerged significantly earlier and demonstrated significant higher level of resistance than those infected with the wild control strain.The results showed that dinB gene made a contribution in the appearing of the antibiotics resistance and has a potential as a target for prevention from the appearing of antibiotic resistance.
dinB gene;Escherichia coli;overexpression
Bacteria resistant to antibiotics have become a major concern globally because they may kill,spread and impose huge costs to society.Resistant bacteria drastically prevent colonization of the normal microbial inhabitants and thus perturb digestion and uptake of essential nutrients.Major causes of appearing of antibiotic resistance are selective pressure of antibiotics to which bacteria are exposed to.Within the classical paradigm,mutations are detrimental.However,accumulated evidence suggests that mutations play a key role in the appearing of drug resistance in bacteria[1-2].In the prolonged exposure of bacteria to sublethal levels of antibiotics,a small population of them experiences a brief state of high mutation rate,in which they relieve the selective pressure[3].SOS response is a protective response in bacteria,activated under stress conditions,such as exposure to antibiotics.dinB encoding Pol IV is one of more than 30 genes of the SOS system.
DNA polymerase Pol IV was identified in E.coli in 1999,which is encoded by dinB gene.In E.coli,the expression of dinB is elevated during the DNA damage-induced SOS response.The polymerase can replicate past DNA lesions but at the cost of frequent mutation.Under adverse conditions,cells may use this error-prone polymerase to produce variants that allow their descendents to survive[4].The enzyme has the potential to mediate template-directed DNA replication without 3'-5'exonuclease(proofreading)activity.It can easily extend the misaligned primer/template mechanism and is thus considered to be error-prone DNA polymerase[5].
Error-prone replication by various low-fidelity polymerases is reported to contribute to the generation of bacterial resistance to antibiotics because of increased mutations[6].To further understand the mechanisms under the appearing of bacteria resistance to antibiotics,in this study,the biological significance of dinB gene using overexpression approach was investigated.
1 Materials and Methods
1.1 Animals and mouse infection model
The institute guidelines for animal use and care were followed in all animal studies.Female C57BL/ 6 mice,6-8 weeks of age,were purchased from Medical Experimental Animal Center of Guangdong (Guangzhou,China).A mouse infection model was established as described by Vogelman[7].Briefly,specific-pathogen-free female C57BL/6 mice were rendered neutropenia by cyclophosphamide(Jiang Su HengRui Medicine,China).Neutropenia was defined as neutrophil counts less than 108/L.150 mg/kg cyclophosphamide was injected intraperitoneally 4 days before infection and 100 mg/kg cyclophosphamide 24 hours before infection.The neutropenia produced via this approach lasted for 5 days in previous studies[8].
Freshly plated bacteria were inoculated into Mueller-Hinton(MH)broth and grown to logarithmic phase at OD600of approximately 0.3,and then diluted 110 in MH broth.100 μL of the diluted broth culture with approximately 106cfu was injected intothethighofthepentobarbital-anesthetized mouse.Starting from 2 hours after the infection,which was defined as time 0,mouse was injected subcutaneously with 40 mg/kg ceftazidime every 12 hours for 3 days.At hour 24,48 and 72,mouse was sacrificed and both thighs were collected and homogenized.Serial dilutions of homogenates of each thigh were plated on MH agar and incubated at 37℃.After 24 hours,visible colonies were counted.
1.2 dinB transcription assay
dinB transcription was tested with a disk plate assay as described previously[9].Briefly,E.coli strain GW1030[dinB1::Mud I(Ap lac)],harboring a dinB::lacZ operon fusion[10],was cultured overnight.100 μL of the culture was plated onto LB plates containing 50 μg 5-bromo-4-chloro-3-indolylβ-D-galactopyranoside(X-Gal).Antibiotic disks were put onto the seeded bacteria.After incubation at 30℃for 24 hours,the induction of the transcrip-tion of the dinB::lacZ fusion can be deduced from a blue band in the border of the inhibition halo.For measurement of dinB transcription level,E.coli strains were exposed to different ceftazidime(CAZ) concentrations around the MIC(which was determined to be 2 μg/mL in the preliminary study).The β-galactosidase levels were measured after different exposure time.MICs were determined using micro-dilutionmethodsdescribedinM07-A09 (2013)from the Clinical and Laboratory Standards Institute(CLSI).
1.3 Tool enzymes and other reagents
Restriction enzyme Sal I,EcoR I,BamH I,Hind III were bought from TaKaRa;T4 DNA ligase were bought from New England Biolab;Pfu enzyme and Taq Plus enzyme were bought from Shanghai ShengGong;Plasmid extraction kit and DNA gel recovery kit were products of QIAGEN.
1.4 Construction of recombinant plasmid pUC-dinB-rrnb
The transcription terminator rrnb region was amplified from the pBV220 template using polymerase Pfu.The primers used were as follows:forward primer:5'-TAAGTCGACCTGTTTTGGCGGATGAGAG-3',reverse primer:5'-CCCAAGCTTAAACAAAAGAGTTTGTAGAA-3'.The PCR product,a 450 bp fragment, was digested with Hin d III and Sal I and inserted in puc19 at the same digestion site,and then transformed into Top 10 E.coli strain to construct pUC-rrnb.The sequencing results showed that amplification and insertion of rrnb fragment were correct.
dinB gene including promoter sequence was amplified from E.coli AB1157 genome with Taq Plus polymerase.The PCR product,a 1 kb fragment,was digested with EcoR and Sal I and inserted in pUC-rrnb at the same digestion site,and then transformed into Top 10 E.coli strain to construct pUC-dinB-rrnb.The sequencing results showed that amplification and insertion were correct.
2 Results
2.1 Ceftazidime induced dinB transcription
As dinB is a component of SOS system,whether the dinB transcription level will be elevated whenE.coli is exposed was tested to CAZ.dinB transcription was tested with a disk plate assay.As expected,CAZ-mediated dinB transcription induction was observedinthedinB-harboringstrain (GW1030)but not in the wild AB1157 strain.CAZ induces dinB transcription in a dose-dependent fashion(Fig.1).These results are similar to those reported by Tatiana[9].
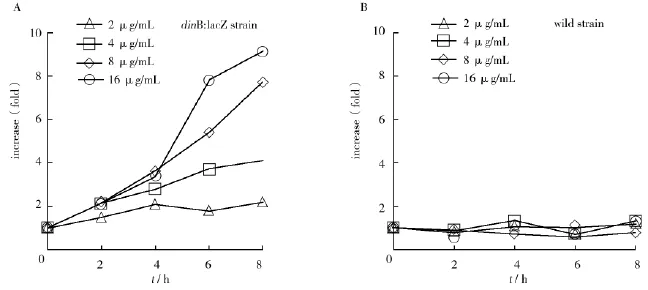
Fig.1 Ceftazidime mediated induction of transcription of dinBWild(B)and dinB:lacZ(A)E.coli strains were exposed to different CAZ concentrations around the MIC.The transcription increase levels were calculated by assigning a value of 1 to the transcription level of the nontreated culture at each time point.The presented is the mean values of results from three independent experiments
2.2 dinB overexpression approach conferred drug resistance in vitro
The fact that dinB transcription induction can be mediated by CAZ as described above was prompted to consider whether dinB overexpression would affect the drug resistance of the strains.The growth curve of dinB-overexpressed and wild E.coli strains in the presence of CAZ is shown in Fig.2.As expected,the dinB-overexpressed strain demonstrated significantly higher growth rate.
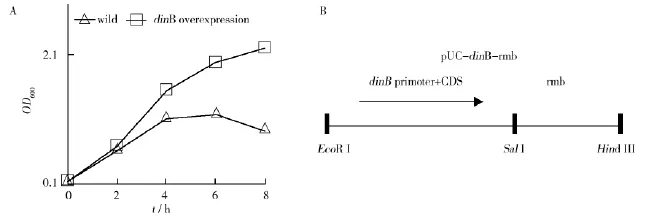
Fig.2 Effects of dinB overproduction on CAZ resistance in E.coli in vitroA:The panel shows in vitro growth curve of wild(AB1157)and dinB-overexpressed(pUC-dinB-rrnb/AB1157)strains with CAZ(2 mg/L)in the media;B:The diagram shows inserted segment in recombinant plasmids pUC-dinB-rrnb
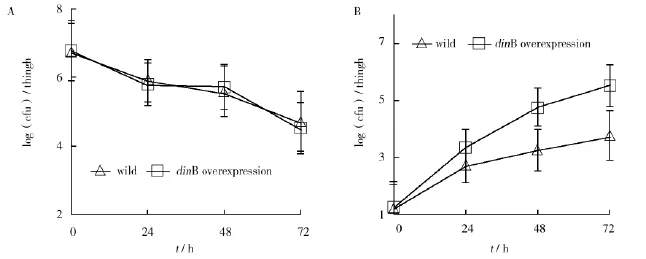
Fig.3 Survival of E.coli strains in vivo after starting ceftazidime therapyWild(AB1157)and dinB-overexpressed(pUC-dinB-rrnb/AB1157)E.coli strains were infected in thighs of neutropenic mice and treated with cefazidime.The thigh homogenates were prepared at 24 h intervals after starting ceftazidime therapy,and plated on media with(A)or without(B)ceftazidime for cfu counting
2.3 dinB overexpression promotes the evolution of resistance to ceftazidime in vivo
TotestwhetherdinBoverexpressionhave effects on the evolution of drug resistance in vivo,a neutropenic murine model[7]with E.coli infection (described in the method section)was used.The cyclophosphamide-treated mice were infected with 106cfu of either wild or dinB-overexpressed strain.After infection,the mice were treated with 40 mg/kg ceftazidime every 12 hours for 3 days.At hour 24, 48 and 72,mice were sacrificed and their thighs were collected and homogenized.Serial dilutions of homogenates were plated on MH agar with and without ceftazidime and incubated for cfu counting(Fig.3).Therapy with ceftazidime had the same bactericidal efficacy on both strains.The total cfu/thigh was reduced approximately 200 fold during the 72 hours of treatment.Resistant strains emerged in the mice infected with dinB-overexpressed strain by hour 7 2,amounting to approximately 5%of the entirepopulation,significantly higher than that of wild strain.Nine clones of either strains were analyzed for minimum inhibitory concentration(MIC)and individual clones were found to have MICs as high as 64 μg/mL,significantly different from the 2 μg/mL of MIC for wild strains.
3 Discussion
It was previously reported that some antibiotics in clinical use,such as quinolones,induces the activation of SOS system and,consequently significantly enhances the mutation rate[11].
While stable transmission of hereditary information is very important for the survival and passage of an organism,mutation is usually detrimental.However,mutation provides selective opportunities for evolution.Several studies have shown that dinB gene causedelevatedmutationrateswhenoverexpressed[12].Under stressful conditions of antibiotics,DNA synthesis may become more erroneous because of the induction of error-prone DNA polymerases,such as dinB-encoding Pol IV[13].Two possible mechanisms might be responsible for the induction of drug resistance.First,high levels of dinB expression may help the cell survive certain classes of DNA damage encountered in stressful conditions resulted from antibiotics.Second,by increasing the errors in DNA synthesis,high levels of dinB production may produce more genetic variants,from which drug resistance emerges.
In this study,an overexpression approach by putting dinB gene into multi-copy of plasmid in E.coli was used.The strains carrying multi-copies of dinB gene demonstrate a significant survival advantage over the wild strain.In the in vitro experiment,the dinB-overexpressedstrainevolvedresistance within 8 hours,while wild strain did not(Fig.2).To further confirm the efficacy of dinB-overexpression,the in vivo appearing of resistance using a neutropenic murine model was studied.In mice infected withdinB-overexpressedstrain,resistantclones emerged significantly earlier and demonstrated significant higher level of resistance than those infected with the wild control strain.In summary,dinB-overexpression enhances the mutation rate,and thus the ability of bacteria to appear resistance to ceftazidime.In the conventional paradigm,antibiotics which prevent bacterial DNA replication can increase mutation frequency via the activity of Pol V[11].In the study,ceftazidime,which is known to inhibit cell wall synthesis,other than DNA replication,is able to induce dinB transcription.And overexpression of dinB increases the level of resistance to CAZ.These results suggest that direct impact on DNA replication is dispensable for the activation of the SOS genes,while any situation which produces stress can induce the activation of the SOS system and thus increase mutation frequency.
The findings indicated that dinB gene plays a role in the appearing of antibiotics resistance and could serve as a novel therapeutic target for prevent of evolution of antibiotic resistance.
[1]B.Yeiser,E.D.Pepper,M.F.Goodman,et al.SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness[J].Proc Natl Acad Sci U S A,2002,99(13): 8737-8741.
[2]B.Tippin,P.Pham,M.F.Goodman.Error-prone replication for better or worse[J].Trends Microbiol,2004,12(6):288-295.
[3]Dalhoff A.Global fluoroquinolone resistance epidemiology and implictions for clinical use[J].Interdiscip Perspect Infect Dis,2012:1-37.
[4]J.C.Layton,P.L.Foster.Error-prone DNA polymerase IV is controlled by the stress-response sigma factor,RpoS,in Escherichia coli[J].Mol Microbiol,2003,50(2):549-561.
[5]K.A.Bunting,S.M.Roe,L.H.Pearl.Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp[J].EMBO J,2003,22(21):5883-5892.
[6]B.Michel.After 30 years of study,the bacterial SOS response still surprises us[J].PLoS Biol,2005,3(7):e255.
[7]B.Vogelman,S.Gudmundsson,J.Leggett,et al.Correlation of antimicrobial pharmacokinetic parameters with therapeutic efficacy in an animal model[J].J Infect Dis,1988,158(4): 831-847.
[8]D.Andes,W.A.Craig.In vivo activities of amoxicillin and amoxicillin-clavulanate against Streptococcus pneumoniae:application to breakpoint determinations[J].Antimicrob AgentsChemother,1998,42(9):2375-2379.
[9]T.Perez-Capilla,M.R.Baquero,J.M.Gomez-Gomez,et al.SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli[J].J Bacteriol,2005,187(4):1515-1518.
[10]C.J.Kenyon,G.C.Walker.DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli[J].Proc Natl Acad Sci U S A,1980,77(5):2819-2823.
[11]P.Ysern,B.Clerch,M.Castano,et al.Induction of SOS genes in Escherichia coli and mutagenesis in Salmonella typhimurium by fluoroquinolones[J].Mutagenesis,1990,5(1):63-66.
[12]S.R.Kim,G.Maenhaut-Michel,M.Yamada,et al.Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA[J].Proc Natl Acad Sci U S A,1997,94(25): 13792-13797.
[13]M.Kivisaar.Stationary phase mutagenesis:mechanisms that accelerate adaptation of microbial populations under environmental stress[J].Environ Microbiol,2003,5(10):814-827.
dinB基因在大肠埃希菌抗生素耐性产生中的作用
席云1,黄敬2,陶玲3,赵鹏2,陈佑明2,付永玫4,孙恒彪2,尤旭2,肖刚2*
(1.中山大学第三附属医院检验科,广东广州510630;2.南方医科大学第三附属医院检验科,广东广州510630; 3.中山大学第三附属医院药剂科,广东广州510630;4.中山大学第三附属医院急诊科,广东广州510630)
本研究探讨dinB基因在抗生素耐性产生中的作用。多拷贝含dinB基因质粒被转染到大肠埃希菌造成超表达。携带多拷贝dinB基因的菌株比野生菌更有生长优势。在体外实验中,dinB超表达株在8 h内即显示出耐药性,而野生株不能。在鼠模型体内实验中,与野生株相比,dinB超表达株出现耐药株时间更短,耐药水平更高。结果显示dinB基因在抗生素耐性产生中有贡献并有作为防止抗生素耐性靶点的潜力。
dinB基因;大肠埃希菌;超表达
Q939.93;R378.2+1
A
1005-7021(2016)04-0047-06
广东省科技计划社会发展项目(2009B030803035);广州市科技计划一般项目(2006Z1-E0141);北京市科技计划一般项目(D07050201170701)
席云女,硕士,副主任技师,硕士生导师。从事临床微生物学检验及研究。Tel:020-85252331,E-mail:xiyun1993@163.com
10.3969/j.issn.1005-7021.2016.04.008
Funds Projecd:This work was supported by Guangdong province science and technology plan(2009B030803035);Guangzhou city science and technology plan(2006Z1-E0141);Beijing city science and technology plan(D07050201170701)
Author profile:XI Yun,female,with a master’s degree in clinical laboratory medicine,is a clinical microbiologist and a deputy Chief technologist.Tel:020-85252331,E-mail:xiyun 1993@163.com
Dr.XIAO Gang,male,is a Chief technologist and an investigator in the field of clinical immunology.
Tel:020-62784787,E-mail:xiaogang2993@yeah.net
Recived:2015-07-20
*通讯作者。男,博士,主任技师,硕士生导师。主要研究方向为临床免疫学。Tel:020-62784787,E-mail:xiaogang2993@yeah.net