退火解旋酶SMARCAL1在维持基因组稳定中的作用与机制
2019-12-24文雅蕾吕柯孬徐小康张欣丁良潘学峰
文雅蕾,吕柯孬,徐小康,张欣,丁良,潘学峰,,3
退火解旋酶SMARCAL1在维持基因组稳定中的作用与机制
文雅蕾1,吕柯孬2,徐小康1,张欣1,丁良1,潘学峰1,2,3
1. 河北大学医学院药理室,保定 071000 2. 北京理工大学生命学院,北京 100081 3. 河北大学化学与环境学院,保定 071000
SMARCAL1是属于SWI/SNF (SWItch/Sucrose Non-Fermentable)相关、基质相关和激动蛋白依赖的染色质调节因子家族成员ATP驱动的DNA退火解旋酶。SMARCAL1在体外和体内能催化单链结合蛋白RPA结合的DNA单链与其互补链退火成双链DNA。人基因的突变与Schimke免疫骨性发育不良(Schimke immuno-osseous dysplasia, SIOD)所能表现出的临床症状呈高度相关。本文对SMARCAL1在DNA损伤部位DNA复制叉的重塑、在DNA双链断裂(double-stranded DNA, dsDNA)处参与经典的非同源末端连接(non-homologous end joining, NHEJ)修复,以及在人染色体端粒完整性维护等方面的作用与机制进行了梳理,对基因突变类型与SIOD症状之间的对应关系进行了更新,并对SMARCAL1在三核苷酸重复序列扩增关联的神经–肌肉退行性病变过程中的可能作用进行了分析和讨论,旨在更好地理解该退火解旋酶在维持基因组稳定中的作用和机制。
SMARCAL1;RPA;DNA复制叉;SIOD;三核苷酸重复序列扩增
基因组不稳定常见于人类遗传疾病、癌症及神经–肌肉退行性疾病的病理过程[1]。常见的基因组不稳定主要包括基因的点突变、插入/缺失突变、DNA链断裂(单链断裂和双链断裂)、DNA链交联、基因组的倍性改变等类型[1,2]。引发基因组不稳定的原因可依据DNA是否受损分为两类:一类源于细胞内源性或细胞外源性因素造成DNA损伤;一类则起因于基因组内特定DNA序列在DNA复制、转录或重组过程中发生了错误折叠[1,2]。据统计,无论是原核细胞还是真核细胞,均需要130种以上的蛋白组份参与基因组稳定维护[1]。其中,退火解旋酶SMARCAL1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1),因广泛参与DNA损伤部位DNA复制叉的重塑、DNA双链断裂部位的非同源末端连接(non-homologous end joining, NHEJ)修复、以及染色体端粒的维护而受到高度重视[3~7]。
SMARCAL1普遍存在于真核生物细胞中,属于一类依赖ATP的染色质“重塑因子”SNF2 (sucrose non-fermenting 2, SNF2)家族的一员[4]。SNF2家族的染色质重塑因子需要利用ATP水解供能,广泛参与细胞周期调控、基因转录、DNA重组、DNA损伤修复和DNA甲基化修饰等过程[5]。除SMARCAL1之外,通过影响DNA复制过程维持基因组稳定性的ZRANB3和HLTF也是SNF2家族的重要成员[6]。这些蛋白均含有一个由7个保守基序(motif)组成的类似“解旋酶”(与常见的DNA和RNA解旋酶类似)的ATPase结构域[7]。尽管SMARCAL1具有DNA“退火解旋酶”(annealing helicase)活性,但目前并未被归类于已有的6个DNA解旋酶超家族(DNA helicase superfamily)中[8]。
人基因定位于2q34-q36区段,含17个外显子,编码一个由954个氨基酸残基组成的蛋白质[7]。在人()和小鼠()的所有组织中均见基因表达,如在人免疫系统中,在单核细胞、B淋巴细胞、CD4+T细胞、CD8+T细胞、NK细胞中的表达量分别为1.259‰、1.584‰、1.259‰、1.995‰和0.100‰;在内分泌系统的胰腺细胞、前列腺细胞中表达量约为0.100‰和0.016‰;在睾丸细胞中表达量则为0.158‰[9~16]。基因突变与Schimke免疫骨性发育不良(Schimke immuno-osseous dysplasia, SIOD)密切相关[17,18]。SIOD是一种累及多系统、进行性加重的罕见常染色体遗传病,主要表现为T细胞免疫缺陷、局灶节段性肾小球硬化、脑发育受损、肾功能衰竭和骨骼发育不良造成的生长迟缓等[8,12,19~21]。除此之外,部分SIOD患者还表现有甲状腺功能减退、骨髓衰竭、头发稀薄、角膜混浊、动脉粥样硬化、中风和偏头痛等[8,12,21]。
本文将对SMARCAL1在DNA损伤部位借退火解旋酶活性重塑DNA复制叉,在DNA双链断裂部位参与非同源末端连接(NHEJ)修复,以及在端粒完整性维护过程等方面的作用与机制进行梳理。同时,对基因突变型与SIOD症状的相关性的最新进展进行了更新,对其在累及多达40余种人神经–肌肉退行性疾病中的三核苷酸重复序列扩增性不稳定和脆性不稳定发生过程中的可能作用进行了分析。
1 SMARCAL1的结构与功能
SMARCAL1含有复制蛋白A(replication protein A , RPA)结合基序、退火解旋酶活性结构域、ATP酶结构域等(图1)[22]。其中,与RPA作用的基序位于N-末端(28个高度保守的氨基酸残基序列);与“退火解旋活性”有关的结构域位于239~307和331~ 4002区间的“HARP”结构域(每个“HARP”各含60个氨基酸残基)[23]。C-末端是解旋酶结构域,具有ATP酶催化活性(由115个氨基酸残基组成的“RecA样”结构域,DEXDc和HELICc)和SWI/SNF“核小体重塑蛋白”结构域(图1)[24]。
SMARCAL1的ATP依赖DNA退火解旋酶含有2个HARP结构域组成的ATPase[25~27]。当DNA出现损伤时,与单链DNA结合的单链DNA结合蛋白RPA32识别位于SMARCAL1的N端RPA作用结构域(图1),并招募SMARCAL1至dsDNA-ssDNA的单链DNA一侧,这一反应可见于S期细胞周期关卡通路激活、停滞的复制叉重塑、端粒DNA完整性维护以及利用NHEJ机制修复DNA双链断裂损伤等过程[3,24,25,28~31]。
1.1 SMARCAL1与DNA复制叉的维护
在人体细胞中,缺失常影响细胞对DNA复制逆境(replication stress)应答能力[4]。缺陷的细胞内可见DNA双链断裂,且对多种影响DNA复制的物质敏感,表明SMARCAL1与DNA复制忠实性的监管有关[3,24,28]。研究发现,SMARCAL1可通过与RPA作用参与停滞的DNA复制叉的“重塑”[29],否则停滞的DNA复制叉有可能衍生为DNA双链断裂从而使DNA突变和染色体重排的风险增高[32]。真核生物的RPA是一个由RPA70、RPA32、RPA14亚基组成的异源三聚体,可借4个DNA结合结构域(DNA-binding domain, DBD)与单链DNA结合[33]。RPA异三聚体参与DNA损伤部位修复蛋白的募集和组装,同时规避单链DNA错误折叠出非B型DNA构象[33,34]。SMARCAL1借其N-末端与RPA32的N端结合,从而被快速高效地定位到复制叉的单链DNA部位[3,35~37]。
活性氧自由基、紫外辐射及一些化学物质(如放线菌素等)会损伤DNA模板,影响细胞周期S时相的DNA复制[3]。为此,细胞需针对相应的DNA损伤类型做出应答,包括利用ATR对单链空缺(single stranded gap, SSG)损伤应答,利用ATM、DNA-PK等激酶对DNA DSBs应答等[1]。这些激酶对SMARCAL1蛋白的磷酸化修饰有助于有序启动DNA损伤修复[38]。比如,SMARCAL1第652位丝氨酸(S652)残基被ATR磷酸化修饰,可抑制SMARCAL1的ATP酶活性,从而降低其DNA复制叉重塑能力,避免DNA复制叉崩解[39]。
体外研究表明,SMARCAL1的退火解旋酶活性的发挥需要与DNA结合,并由ATP水解供能[22]。在这个过程中,SMARCAL1借RPA结合蛋白与单链DNA结合,并利用ATP水解提供的能量催化彼此互补的单链DNA重新形成双链DNA,并同时解除RPA与单链DNA的结合(图2,A和B)[24]。现已发现,上述反应可以发生在DNA复制叉部位,帮助DNA复制叉内的单链DNA发生“分支迁移”和“分支重塑”[4,22,40],使停滞的DNA复制叉“重塑”出“鸡爪”状四臂交叉中间体结构[41,42](图2C)。图2C中的“鸡爪”结构可以稳定停滞的DNA复制叉,又能使带伤的单链DNA重新“退回”双链DNA状态,可以赢得修复时间,或利用“模板转换”(template switching)使新生DNA链的3 ʹ生长端“绕过”带伤DNA模板后重启DNA复制[6,27,29,33,43](图2,B和C)。

图1 退火解旋酶SMARCAL1的功能结构域分布
SMARCAL1在其N端区域包含与RPA相互作用基序和2个退火活性所需的HARP结构域;在其C末端含有解旋酶结构域,为DEXDc和HELICc基序[23,25]。
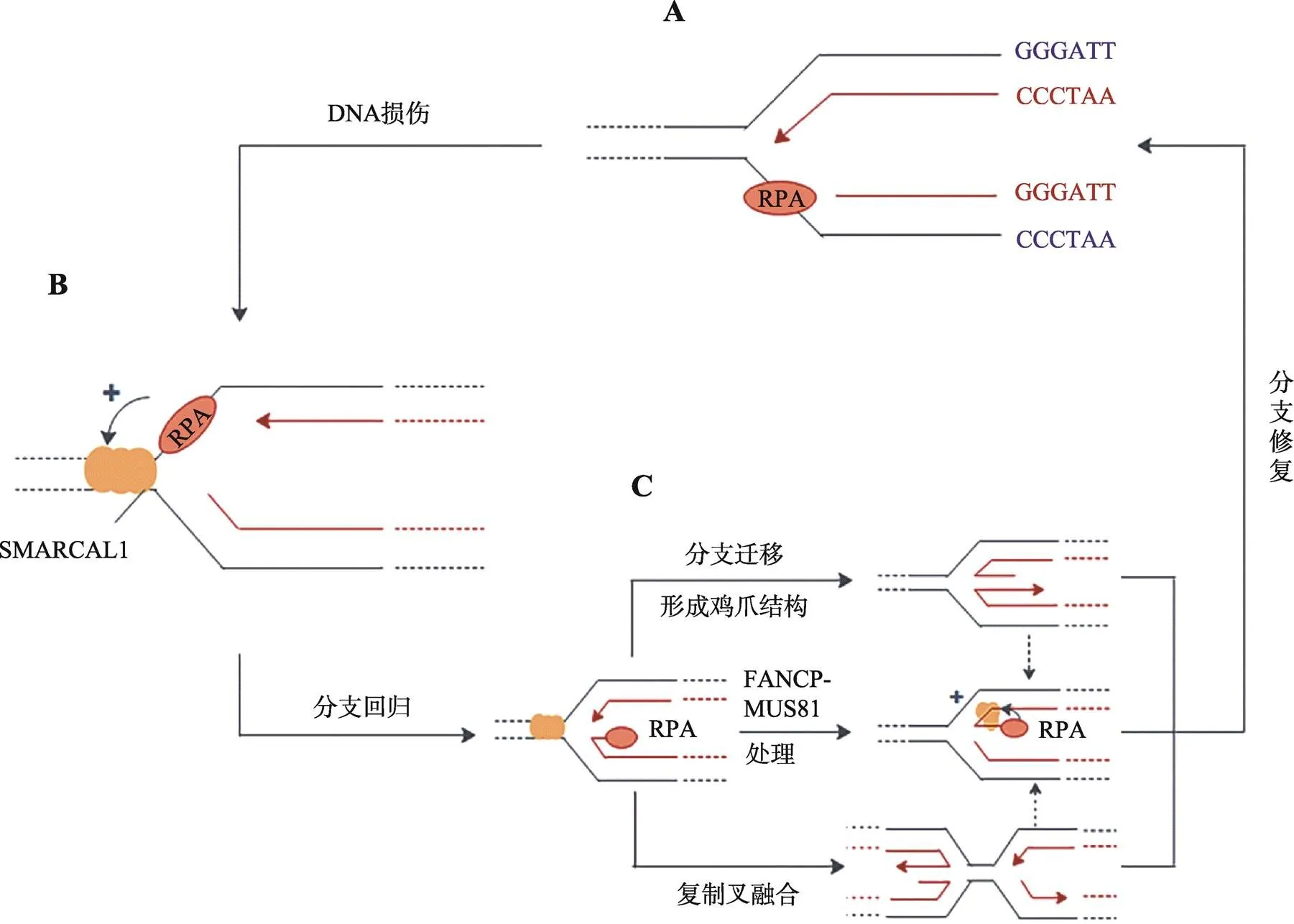
图2 SMARCAL1修复受损DNA复制叉的机制
A:DNA链复制遇阻,复制型DNA聚合酶和所“偶联”的DNA解旋酶脱离,导致前导链模板上产生ssDNA空缺;B:在停滞的复制叉上,RPA与单链DNA结合后行成RPA-ssDNA,并招募SMARCAL1,启动复制叉回转;C:SMARCAL1在复制叉回转后以3种可能的方式催化复制叉的修复,包括持续分支迁移产生“鸡爪”状的Holliday结构;与相邻的重新建立的DNA复制叉发生“融合”获得拯救;以及用FANCP-MUS81处理后产生对应于新生前导链的ssDNA链,然后生成单末端DNA双链断裂(DSB)。RPA协助SMARCAL1用互补的模板链与新生的ssDNA前导链“退火”,重构出可正常复制的DNA复制叉[25,39,40]。
需要特别指出的是SMARCAL1催化的上述DNA复制叉重塑在体外也可以由大肠杆菌的DNA解旋酶RecG催化[29]。大肠杆菌RecG是一个3ʹ→5ʹDNA解旋酶,具有催化Holliday中间体(同源重组过程中出现的交叉结构)发生分支迁移能力[29]。在催化DNA复制叉回转时,RecG类似SMARCAL1,需要借DNA单链结合蛋白与DNA复制叉结合。在这个过程中DNA单链结合蛋白还可以进一步稳定RecG与DNA复制叉的结合,有利于RecG催化DNA复制叉回转(图2)[44]。因此,虽然SMARCAL1在结构上与大肠杆菌RNA聚合酶结合蛋白HepA相似(被称为HepA相关蛋白,HARP),但在催化DNA底物结构重塑时则更类似于大肠杆菌的DNA解旋酶RecG[9~11]。
1.2 SMARCAL1在维持端粒稳定过程中的作用
人染色体端粒DNA一般由“TTAGGG”六聚体重复序列组成,可形成包括G-四链体、D(t)环等在内的非B型DNA结构。尽管这些DNA结构为端粒稳定所必需,但如果它们出现在DNA复制过程中则会造成DNA复制叉停滞,使端粒变短[45~47]。维持端粒稳定是保证细胞增殖能力的关键。肿瘤细胞的端粒长度的维持机制有两种,一是重新激活端粒酶,一是利用同源重组“替补”机制维持(统称为端粒延长替代机制,alterative lengthening of telomere, ALT)[48]。ALT细胞(缺乏端粒酶而需要ALT维持端粒长度的细胞)中可见由端粒DNA的额外染色体形成的C-环(C-circles),可用于ALT活性的标记[43]。
DNA损伤修复系统对端粒酶复制稳定端粒发挥着重要的调控作用[49]。Cox等[49]发现SMARCAL1参与端粒的ALT维持机制。在ALT细胞内,SMARCAL1与端粒DNA结合,协助停滞的DNA复制叉重启,应对端粒DNA复制应激[49]。而缺失SMARCAL1的ALT细胞则会出现端粒DNA复制困难,表现为DNA复制叉持续停滞,并最终形成DSBs,表现出高强度染色体融合(图3)[49]。与此同时,SMARCAL1缺失会影响端粒的长度异质性,推测与ALT细胞中SMARCAL1水平过低催生了更多的C-环有关[49]。
此外,Poole等[50]发现SMARCAL1在端粒处发挥作用时无需RPA协助,因为端粒部位重复DNA序列更倾向于形成G-四链体等非B型DNA二级结构,而G-四链体本身就可有效招募SMARCAL1。但是,当SMARCAL1与DNA复制叉内的前导链模板结合时则需要RPA激活[33,50]。
1.3 SMARCAL1与DNA双链断裂修复
真核细胞内与DNA DSBs修复有关的机制有两类,一类是依赖DNA同源性的同源重组修复(homologous recombination, HR),一类则是不严格依赖DNA同源性(无需模板)的非同源末端连接(NHEJ)[51]。HR常用于细胞周期的S、G2等时相,而NHEJ则可用于整个细胞周期,特别当细胞处于G0/G1和S期早期,由于缺乏“同源染色体”,NHEJ对DSBs修复起着关键的作用[52~54]。
Keka等[55]发现SMARCAL1参与G1期DSBs的NHEJ修复过程。SMARCAL1的退火解旋酶活性可以协助DNA末端结合蛋白Ku70/80“获取”双链DNA末端,有利于进一步依序招募参与NHEJ修复的蛋白因子,以完成NHEJ或MMEJ (小同源末端连接,microhomology-mediated end-joining,MMEJ)在G1时相对DNA DSBs修复。在此过程中,SMARCAL1的ATP依赖的退火能力可能避免了RPA在DSB末端与ssDNA结合,因此促进Ku70/Ku80/DNA- PKcs复合体对DNA双链末端识别、结合和保护,为后续XRCC4和DNA连接酶IV在断裂末端的积聚提供便利,从而提供了NHEJ修复的精确度[55~60](图4)。
SMARCAL1在Ku70/Ku80/DNA-PKcs聚积的DNA末端协助Mre11、RAD50、Nbs1组装成MRN复合物(Mre11-RAD50-Nbs1)[65,66]。具体过程如下:DSB信号被ATM“捕获”,ATM将H2AX磷酸化成γ-H2AX。γ-H2AX与Nbs1作用进一步促进RAD50和Mre11在DSB断端处形成MRN复合体(RAD50- Mre11-Nbs1, MRN)[67~69]。其中,RAD50二聚体上的ATP酶(Walker A和Walker B)负责与DSB两个末端结合,避免末端错位或漂移。Nbs1则协助SMARCAL1退火可被NHEJ修复的DNA断端,使之保持双链状态,并由RAD50的绞链区将两个断端“固定”[70]。

图3 SMARCAL1在维持端粒稳定中的作用
ALT细胞内的染色体端粒易出现复制逆境,SMARCAL1帮助重塑端粒DNA上停滞的复制叉,确保端粒序列的完全复制[46,47]。在SMARCAL1缺陷的情况下,ALT端粒上持续停滞的复制叉会催生DNA双链断裂,出现端粒DNA复制障碍及形成C-环,此时,染色体易融合,造成基因组不稳定[49,50]。

图4 SMARCAL1在NHEJ修复DNA双链断裂中的可能作用
A:DSB形成后,RPA识别并结合DNA断端,之后,招募SMARCAL1。SMARCAL1的退火解旋酶活性保证DSB断口DNA呈双链状态;B:Ku70/Ku80组成的异源二聚体与DNA末端结合;C:DNA-Ku复合体招募DNA-PKcs,形成Ku70/Ku80/DNA-PKcs复合体;D:SMARCAL1促进Ku70/Ku80/DNA-PKcs复合体稳定结合在DSB端口,并激活DNA-PKcs。DNA-PKcs对包括自身在内的蛋白进行磷酸化修饰;E:DSB断端的SMARCAL1促进Ku70/Ku80/DNA-PKcs复合体招募DNA连接酶IV和XRCC4复合体,进一步形成Ku70/Ku80/DNA-PKcs/XRCC4/DNA连接酶IV修复复合体;F:含有连接酶的复合体完成两个DNA断端的连接[55~64]。
此外,SMARCAL1也和RAD50一起参与阻止复制叉反转,通过调节Mre11的核酸酶活性,防止Mre11过度降解新生DNA区段[71]。
1.4 SMARCAL1与基因转录
SMARCAL1作为SWI/SNF家族(负责催化核小体重塑)中的成员同时拥有解旋酶(helicase)和ATPase活性,因此有可能参与某些基因转录过程中的核小体重塑。当前研究较多的是SMARCAL1对基因转录的调节。编码一种亮氨酸拉链蛋白,参与人类基因组中5%~15%的基因的转录,在细胞增殖、分化、生长和凋亡中发挥重要作用[72~74]。SMARCAL1作为辅因子参与基因的转录[72~74]。Heravi等[75]发现SMARCAL1通过ATP依赖性方式改变DNA结构,调节的转录[75]。Tapan等[76]发现SMARCAL1是转录的负调控因子。通过与激活蛋白BRG1和RNA聚合酶Ⅱ(RNAPⅡ)“争夺”基因P1启动子上游的一段富含GC碱基的159 bp DNA区域(-B159),当激活蛋白BRG1和RNAPⅡ占据时,基因转录;当该区域被SMARCAL1占据时,基因关闭[76]。SMARCAL1与结合后使得相应部位的染色质结构更难与BRG1和RNAPⅡ结合,故可抑制的转录[76]。
2 SMARCAL1在三核苷酸重复序列扩增中的潜在作用
人基因组中特定基因部位处的三核苷酸重复序列(CAG)n·(CTG)n的“动态”扩增与多种遗传性脊髓–小脑共济失调、亨廷顿疾病、阿尔兹海默综合征等神经–肌肉系统退行性病变的发生密切相关[77~79]。已有的研究结果表明,(CAG)n重复序列的扩增与DNA剪切修复过程中产生的单链DNA错误、DNA复制过程中出现DNA链断裂、DNA复制过程中新生链和模板链间发生“滑动”、链转换(strand switching)、蛋白质与重复DNA序列结合等许多因素有关。上述过程可能起因于(CAG)n重复DNA形成的含错配碱基对的DNA发卡结构,DNA发卡结构可能会直接干扰DNA复制、修复和重组。不仅如此,在基因转录过程中(CAG)n三核苷酸重复序列一旦形成R环结构(DNA杂交体与非模板链形成)也可影响DNA复制叉的移动,导致非模板链DNA断裂,甚至可被用于DNA重新复制的引物[77~79]。本实验室已有的研究结果表明,基因的正本基因()功能丧失的大肠杆菌细胞内,疾病相关的(CAG)n·(CTG)n序列呈特异性扩增(待发表)。鉴于SMARCAL1在催化DNA复制叉重塑及避免DNA复制叉崩解的过程中与RecG功能一致,推测出现在人类患者细胞内的(CAG)n·(CTG)n序列扩增有可能与DNA复制叉重塑和DNA复制重起始失败有关(图5A),或出现在(CAG)n重复序列内的R环结构影响了DNA复制,并在RecG/SMARCAL1功能异常的情况下诱生DNA链重排或造成该处DNA局部额外复制[77~79](图5B)。最近,类似图5A的工作机制已被Kononenko等[80]的工作证实。他们利用小鼠幼红细胞白血病细胞系验证了可导致人脆性X染色体综合征(马丁–贝尔综合征)的三核苷酸重复序列(CGG)n的扩增不稳定与SMARCAL1所具有的功能直接有关[80]。但是,目前尚无有工作表明SMARCAL1自身是否具有类似于RecG的RNA解旋酶活性或与其他RNA解旋酶一起参与RNA转录,故图5B所描述的机制依然有待进一步检验。
为了阐释长期困扰国际医学界的亨廷顿疾病、阿尔兹海默综合征和多种遗传性脊髓–小脑共济失调综合征等发病过程中出现的(CAG)n扩增的原因,以及明确其他与三核苷酸重复序列失稳定引发的人类神经–肌肉退行性病变的病理机制,当前亟需深入分析SMARCAL1在包括(CAG)n重复序列在内的三核苷酸重复序列稳定维护过程中可能发挥的作用[77~79]。
3 SMARCAL1的基因突变型与SIOD的症状关联
世界范围内SIOD的发病率约为1/300万~1/100万[8,9,81,82]。导致SIOD发生的基因突变多为双等位基因功能缺失、错义突变、插入/缺失(insertion and deletion, Ins/Del)、大片段缺失以及SMARCAL1 mRNA拼接错误(表1)[9,83]。上述基因改变常出现在SMARCAL1的RecA样结构域I中(图1),由于突变影响了SMARCAL1的ATP酶活性,故常见疾病的严重程度与突变体SMARCAL1所表现出的ATP酶活性成反比[82]。SMARCAL1等位基因缺失、无义或移码突变常见于重症患者。重症SIOD患者的症状在孕期外显,表现为胎儿生长迟缓、甲状腺功能减退症、骨髓衰竭、短暂性脑缺血发作、中风和肾衰竭等,一般死于5岁前。SMARCAL1等位基因错义突变患者症状较轻,多数发病较晚,常见8~13岁以后发病,数年后进展至肾衰[84,85](表1)。

图5 SMARCAL1及其可能的突变体基因在三核苷酸重复序列稳定维护中的潜在作用
A:SMARCAL1参与三核苷酸重复序列部位处DNA复制叉的重塑。根据参考文献[80]修改绘制;B:SMARCAL1自身或引导其他RNA解旋酶清除在三核苷酸重复序列转录过程中形成的R-环结构[77~79]。

表1 Smarcal1基因突变型与SIOD的症状关联

续表
“?”核苷酸或氨基酸突变的位置待确认。
SIOD的共有临床症状是T细胞免疫缺陷。在淋巴细胞发育过程中,编码免疫球蛋白和T细胞受体抗原结合域的功能基因需要经过与NHEJ类似的V(D)J重组才能形成[98,99]。与NHEJ类似,V(D)J重排也需产生DNA双链断裂,并由NHEJ机制完成断链末端的连接[98,99]。而突变体SMARCAL1常影响NHEJ在V(D)J重排重组的连接效率,这可能是SIOD患者常见T细胞免疫缺陷的原因之一[96,100]。此外,突变体SMARCAL1有可能通过影响-基因表达进一步影响SIOD的征候。研究发现成年小鼠的肾脏和脑组织中,不表达而高表达[13,101]。暗示SIOD患者表现出的肾功能不全和脑发育受损可能与SMARCAL1突变体不能正确调节表达有关[19]。Tapan等[76]发现SIOD患者的染色质也会出现异常,这种情况暗示SMARCAL1可能依然具有染色质重塑活性。可能由于trxG和PcG的复合体诱导组蛋白的翻译后修饰,影响染色体的结构。而SMARCAL1通过结合染色质直接影响基因表达。
4 结语与展望
目前,DNA退火酶SMARCAL1的功能异常与SIOD之间的关联已经得到了明确,但SIOD患者所能表现的症状征候差异与不同基因突变型的对应关系的细节依然处于不断更新状态。由于临床上SIOD症状可累及人体多个系统,轻重患者之间的临床表现并不尽然一致,也很难呈现一种症状的渐进发展特征,暗示基因突变可能依附携带突变的组织和器官,一种可能的情形是基因突变对不同组织细胞内的染色质结构、基因转录、DNA损伤修复依细胞类型不同而具有差异。因此,目前亟需对有关基因表达的组织特异性机制加深了解。在鉴定出的分子机制方面,SMARCAL1的退火酶活性在复制叉重塑过程中的作用已被体外实验证实,但尚缺乏体内实验证据的支持。不仅如此,不同突变型对其活性的影响呈现多样性(表1),如何关联SIOD症状也需要进一步积累体内实验数据。近年来本实验室对SMARCAL1的原核生物功能类似物DNA解旋酶RecG的研究发现,RecG缺陷突变后的大肠杆菌细胞容许与亨廷顿疾病、阿尔兹海默综合症以及多种遗传型小脑–脊髓共济失调在内的40多种神经-肌肉系统退行性疾病直接有关的三核苷酸重复序列CAG出现特异性扩增,类似的情形是否也有可能出现在携带某些突变类型的患者体内需要给予重视。当前关于SMARCAL1分子机制的研究主要集中于其对DNA复制叉重塑、DNA双链断裂损伤修复及端粒完整性维持等方面。作为SWI/SNF相关、基质相关和激动蛋白依赖的染色质调节因子家族成员的SMARCAL1在染色质重塑、组蛋白修饰编码(histone code)方面是否依然具有作用尚缺乏更多了解。综上所述,深入开展基因及其突变型的分子遗传学和SMARCAL1蛋白的体内分子生物学研究将不仅有助于系统理解SIOD发病机制,而且对破解长期困扰人类健康的40余种进行性神经-肌肉系统退行性疾病的致病原因提供借鉴和参考。
[1] Pan XF ed. The Molecular Biology of Gene and Diseases. Beijing: Chemical Industry Press, 2014, 1–450.潘学峰. 基因疾病的分子生物学. 北京: 化学工业出版社, 2014, 1–450.
[2] Pan XF, Xiao P, Li HQ, Zhao DX, Duan F. The gratuitous repair on undamaged DNA misfold,, 2011: 401–430.
[3] Poole LA, Cortez D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability., 2017, 52(6): 696–714.
[4] Bansal R, Arya V, Sethy R, Rakesh R, Muthuswami R. RecA-like domain 2 of DNA-dependent ATPase A domain, a SWI2/SNF2 protein, mediates conformational integrity and ATP hydrolysis., 2018, 38(3): BSR20180568.
[5] Bansbach CE, Bétous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks., 2009, 23(20): 2405–2414.
[6] Bétous R, Mason AC, Rambo RP, Bansbach CE, Badu- Nkansah A, Sirbu BM, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication., 2012, 26(2): 151–162.
[7] Lugli N, Sotiriou SK, Halazonetis TD. The role of SMARCAL1 in replication fork stability and telomere maintenance., 2017, 56: 129–134.
[8] Severino M, Giacomini T, Verrina E, Prato G, Rossi A.Reversible cerebral vasoconstriction complicating cerebral atherosclerotic vascular disease in Schimke immuno- osseous dysplasia., 2018, 60(9): 885– 888.
[9] Driscoll R, Cimprich KA. HARPing on about the DNA damage response during replication., 2009, 23(20): 2359–2365.
[10] Kakar S, Fang X, Lubkowska L, Zhou YN, Shaw GX, Wang YX, Kashlev M, Ji X. Allosteric activation of bacterial Swi2/Snf2 (Switch/Sucrose non-fermentable) protein RapA by RNA polymerase: biochemical and structural studies., 2015, 290(39): 23656– 23669.
[11] Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms., 2011, 21(3): 396–420.
[12] Morimoto M. Characterization of the disease pathogenesis of Schimke immuno-osseous dysplasia., 2016: 27–46.
[13] Baradaran-Heravi A, Raams A, Lubieniecka J, Cho KS, DeHaai KA, Basiratnia M, Mari PO, Xue Y, Rauth M, Olney AH, Shago M, Choi K, Weksberg RA, Nowaczyk MJ, Wang W, Jaspers NG, Boerkoel CF. SMARCAL1 deficiency predisposes to non-Hodgkin lymphoma and hypersensitivity to genotoxic agents., 2012, 158A(9): 2204–2213.
[14] Dekel B, Metsuyanim S, Goldstein N, Pode-Shakked N, Kovalski Y, Cohen Y, Davidovits M, Anikster Y.Schimke immuno-osseous dysplasia: expression of SMARCAL1 in blood and kidney provides novel insight into disease phenotype., 2008, 63(4): 398–403.
[15] Haokip, Thangminlen D, Kumari.Human SMARCAL1-a member of the SWI2/SNF2 family-is required for cell division., 2011, 25(12): 3515–3524.
[16] Elizondo LI, Cho KS, Zhang W, Yan J, Huang C, Huang Y, Choi K, Sloan EA, Deguchi K, Lou S, Baradaran- Heravi A, Takashima H, Lücke T, Quiocho FA, Boerkoel CF. Schimke immuno-osseous dysplasia: SMARCAL1 loss-of-function and phenotypic correlation., 2009, 46(1): 49–59.
[17] Patne K, Rakesh R, Arya V, Chanana UB, Sethy R, Swer PB, Muthuswami R. BRG1 and SMARCAL1 transcriptionally co-regulate DROSHA, DGCR8 and DICER in response to doxorubicin-induced DNA damage., 2017, 1860(9): 936– 951.
[18] Baradaran-Heravi A, Lange J, Asakura Y, Cochat P, Massella L, Boerkoel CF. Bone marrow transplantation in Schimke immuno‐osseous dysplasia., 2013, 161A(10): 2609–2613.
[19] Zhang L, Fan S, Liu H, Huang C. Targeting SMARCAL1 as a novel strategy for cancer therapy., 2012, 427(2): 232–235.
[20] Carroll C, Hunley TE, Guo Y, Cortez D. A novel splice site mutation in SMARCAL1 results in aberrant exon definition in a child with schimke immunoosseous dysplasia., 2015, 167A(10): 2260–2264.
[21] Prato G, Grandis ED, Mancardi MM, Croci C, Pisciotta D, Uccella S, Costanzo C, Severino S, Tortora D, Pavanello M, Venesellividovits E. Schimke Immuno-osseous dysplasia: a peculiar EEG pattern., 2018, 49(S 01): S1–S12.
[22] Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase., 2008, 322(5902): 748–750.
[23] Ghosal G, Yuan J, Chen J. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1., 2011, 12(6): 574–580.
[24] Ciccia A, Bredemeyer AL, Sowa ME,Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart., 2009, 23(20): 2415–2425.
[25] Hauk G, Bowman GD. Structural insights into regulation and action of SWI2/SNF2 ATPases., 2011, 21(6): 719–727.
[26] Aydin ÖZ, Vermeulen W, Lans H. ISWI chromatin remodeling complexes in the DNA damage response., 2014, 13(19): 3016–3025.
[27] Kohashi K, Yamamoto H, Yamada Y, Kinoshita I, Taguchi T, Iwamoto Y, Oda Y. SWI/SNF Chromatin-remodelingComplex Status in SMARCB1/INI1-preserved Epithelioid Sarcoma., 2018, 42(3): 312–318.
[28] Han JJ, Song ZT, Sun JL, Yang ZT, Xian MJ, Wang S, Sun L, Liu JX. Chromatin remodeling factor CHR18 interacts with replication protein RPA1A to regulate the DNA replication stress response in., 2018, 220(2): 476–487.
[29] Bétous R, Couch FB, Mason AC,Eichman BF, Manosas M, Cortez D. Substrate-selective repair and restart of replication forks by DNA translocases., 2013, 3(6): 1958–1969.
[30] 芦广庆, 段金志, 张昱. 哺乳动物DNA连接酶在DNA双链断裂修复通路中的作用. 遗传, 2016, 38(2): 178–179.
[31] Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response., 2015, 25(1): 9–23.
[32] Holsclaw JK, Sekelsky J. Annealing of complementary DNA sequences during Double-Strand Break repair inis mediated by the ortholog of SMARCAL1., 2017, 206(1): 467–480.
[33] Bhat KP, Bétous R, Cortez D. High-affinity DNA-bindingdomains of replication protein A (RPA) direct SMARCAL1- dependent replication fork remodeling., 2015, 290(7): 4110–4117.
[34] Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair., 2010, 15: 883–900.
[35] Xie S, Lu Y, Jakoncic J, Sun H, Xia J, Qian C. Structure of RPA32 bound to the N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins., 2014, 281(15): 3382– 3396.
[36] Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Ré C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S, Ochs F, Lukas C, Lukas J, Gorgoulis VG, Scapozza L, Halazonetis TD. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication Forks., 2016, 64(6): 1127–1134.
[37] Feldkamp MD, Mason AC, Eichman BF, Chazin WJ. Structural analysis of replication protein A recruitment of the DNA damage response protein SMARCAL1., 2014, 53(18): 3052–3061.
[38] Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation., 2009, 8(9): 1004–1008.
[39] Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, Carroll CM, Jung SY, Qin J, Cimprich KA, Cortez D. ATR phosphorylates SMARCAL1 to prevent replication fork collapse., 2013, 27(14): 1610–1623.
[40] Bétous R, Glick GG, Zhao R, Cortez D. Identification and characterization of SMARCAL1 protein complexes., 2013, 8(5): e63149.
[41] Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability., 2009, 37(11): 3475–3492.
[42] Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response., 2015, 16(4): 207–220.
[43] Margalef P, Kotsantis P, Borel V, Bellelli R, Panier S, Boulton S J. Stabilization of reversed replication forks by telomerase drives telomere catastrophe., 2018, 172(3): 439–453.
[44] Bianco PR, Lyubchenko YL. SSB and the RecG DNA helicase: an intimate association to rescue a stalled replication fork., 2017, 26(4): 638–649.
[45] Wu XM, Tang WR, Luo Y. ALT-Alternative lengthening of telomere., 2009, 31(12): 1185– 1191.吴晓明, 唐文如, 罗瑛. ALT-端粒延长替代机制. 遗传, 2009,31(12): 1185–1191.
[46] Ying SY, Xiong JX, Mai HX, Lin JJ, Jiang LN, Cheng L, Ye QN. Advances on the regulation of telomerase., 2016, 38(4): 289–299营孙阳, 熊加秀, 麦洪旭, 林佳佳, 姜丽娜, 程龙, 叶棋浓. 端粒酶调控研究进展. 遗传, 2016,38(4): 289–299.
[47] Sfeir A, Kosiyatrakul ST, Hockemeyer D,MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication., 2009, 138(1): 90–103.
[48] Sobinoff AP, Pickett HA. Alternative lengthening of telomeres: DNA repair pathways converge., 2017, 33(12): 921–932.
[49] Cox KE, Maréchal A, Flynn RL. SMARCAL1 resolves replication stress at ALT telomeres., 2016, 14(5): 1032–1040.
[50] Poole LA, Cortez D. SMARCAL1 and telomeres: replicating the troublesome ends., 2016, 7(3): 270–274.
[51] Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks., 2018, 293(27): 10512–10523.
[52] Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J,Escribano-Diaz C, Durocher D. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions., 2014, 344(6180): 189–193.
[53] Sims J, Copenhaver GP, Schloegelhofer P. Meiotic DNA repair in the nucleolus employs a non-homologous end joining mechanism., doi: https://doi.org/ 10.1101/553529.
[54] Lu HM, Shamanna RA, de Freitas JK, Okur M, Khadka P, Kulikowicz T,Holland PP, Tian J, Croteau DL, Davis AJ, Bohr VA. Cell cycle-dependent phosphorylation regulates RECQL4 pathway choice and ubiquitination in DNA double-strand break repair., 2017, 8(1): 2039.
[55] Keka IS, Mohiuddin, Maede Y, Rahman MM, Sakuma T, Honma M, Yamamoto T, Takeda S, Sasanuma H. Smarcal1 promotes double-strand-break repair by nonhomologous end-joining., 2015, 43(13): 6359–6372.
[56] Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2., 2010, 29(19): 3358–3369.
[57] Ling AK, So CC, Le MX, Chen AY, Hung L, Martin A. Double-stranded DNA break polarity skews repair pathway choice during intrachromosomal and interchromosomal recombination., 2018, 115(11): 2800–2805.
[58] Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis., 2015, 117(2–3): 194–205.
[59] Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair., 2017, 18(8): 495–506.
[60] Hammel M, Yu Y, Radhakrishnan SK, Chokshi C, Tsai MS, Matsumoto Y, Kuzdovich M, Remesh SG, Fang S, Tomkinson AE, Lees-Miller SP, Tainer JA. An intrinsically disordered APLF links Ku, DNA-PKcs, and XRCC4-DNA ligase IV in an extended flexible non- homologous end joining complex., 2016, 291(53): 26987–27006.
[61] Hinde E, Kong X, Yokomori K,Gratton E. Chromatin dynamics during DNA repair revealed by pair correlation analysis of molecular flow in the nucleus., 2014, 107(1): 55–65.
[62] Sibanda BL, Chirgadze DY, Ascher DB,Blundell TL. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair., 2017, 355(6324): 520–524.
[63] Ma Y, Pannicke U, Lu H,Niewolik D, Schwarz K, Lieber MR.The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis.2005, 280(40): 33839–33846.
[64] Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells., 2014, 42(3): e19.
[65] Yin X, Liu M, Tian Y, Wang J, Xu Y. Cryo-EM structure of human DNA-PK holoenzyme., 2017, 27(11): 1341–1350.
[66] Hiom K. Coping with DNA double strand breaks., 2010, 9(12): 1256–1263.
[67] Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities., 2014, 53(1): 7–18.
[68] Casari E, Rinaldi C, Marsella A, Gnugnoli M, Colombo CV, Bonetti D, Longhese MP. Processing of DNA double-strand breaks by the MRX complex in a chromatin context.2019, 6: 43.
[69] Korsholm LM, Gál Z, Lin L, Quevedo O, Ahmad DA, Dulina E, Luo YL, Bartek J, Larsen DH. Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair., 2019, 47(15): 8019–8035.
[70] Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme., 2008, 321(5897): 1807–1812.
[71] Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Técher H, Baldi G, Shen R, Ciccia A, Pellegrini L, Krejci L, Costanzo V. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments., 2017, 67(5): 867–881.
[72] Levens DL. Reconstructing myc., 2003, 17(9): 1071–1077.
[73] Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression ofoncogenic activity by ribosomal protein haploinsufficiency., 2008, 456(7224): 971–975.
[74] Sutherland C, Cui YX, Mao HB, Hurley LH. A mechanosensor mechanism controls the G-quadruplex/ i-motif molecular switch in the MYC promoter NHE III1., 2016, 138(42): 14138–14151.
[75] Baradaran-Heravi A, Cho KS, Tolhuis B, Sanyal M, Morozova O, Morimoto M, Elizondo LI, Bridgewater D, Lubieniecka J, Beirnes K, Myung C, Leung D, Fam HK, Choi K, Huang Y, Dionis KY, Zonana J, Keller K, Stenzel P, Mayfield C, Lücke T, Bokenkamp A, Marra MA, van Lohuizen M, Lewis DB, Shaw C, Boerkoel CF. Penetrance of biallelic SMARCAL1 mutations is associated with environmental and genetic disturbances of gene expression., 2012, 21(11): 2572– 2587.
[76] Sharma T, Bansal R, Haokip DT, Goel I, Muthuswami R. SMARCAL1 negatively regulates c-Myc transcription by altering the conformation of the promoter region., 2015, 5: 17910.
[77] Pan XF. Mechanism of trinucleotide repeats instabilities: the necessities of repeat non-B secondary structure formation and the roles of cellular trans-acting factors., 2006, 33(1): 1–11.
[78] Pan XF, Jiang N, Chen XF, Zhou XH, Ding L, Duan F. R-loop structure: the formation and the effects on genomic stability.2014, 36(12): 1185–1194.潘学峰, 姜楠,陈细芳,周晓宏,丁良,段斐. R环的形成及对基因组稳定性的影响. 遗传,2014,36(12): 1185–1194.
[79] Wang XH, Pan XF, Li HQ. Advances in the studies of the expansion of (CAG) n·(CTG) n trinucleotide repeats and mecha-nisms underlying its related diseases., 2016, 39(15): 274–281.王希恒, 潘学峰, 李红权.(CAG)n·(CTG)n三核苷酸重复序列扩增及相关疾病机制研究进展. 国际遗传学杂志, 2016,39(15): 274–281.
[80] Kononenko AV, Ebersole T, Vasquez KM, Mirkin SM.Mechanisms of genetic instability caused by (CGG)n repeats in an experimental mammalian system., 2018, 25(8): 669–676.
[81] 王晓慧, 方方, 丁昌红, 黄昱, 王旭, 李楠. Schimke免疫–骨发育不良一例. 中华儿科杂志, 2015, 53(8): 631–632.
[82] Lücke T, Franke D, Clewing JM, Boerkoel CF, Ehrich JH, Das AM, Zivicnjak M. Schimke versus non-Schimke chronic kidney disease: an anthropometric approach., 2006, 118(2): e400–407.
[83] Gupta M, Mazumder M, Dhatchinamoorthy K, Nongkhlaw M, Haokip DT, Gourinath S, Komath SS, Muthuswami R. Ligand-induced conformation changes drive ATP hydrolysis and function in SMARCAL1., 2015, 282(19): 3841–3859.
[84] Saraiva JM, Dinis A, Resende C, Faria E, Gomes C, Correia AJ, Gil J, da Fonseca N. Schimke immuno- osseous dysplasia: case report and review of 25 patients., 1999, 36(10): 786–789.
[85] Boerkoel CF, O'Neill S, André JL, Benke PJ, Bogdanovíć R, Bulla M, Burguet A, Cockfield S, Cordeiro I, Ehrich JH, Fründ S, Geary DF, Ieshima A, Illies F, Joseph MW, Kaitila I, Lama G, Leheup B, Ludman MD, McLeod DR, Medeira A, Milford DV, Ormälä T, Rener- Primec Z, Santava A, Santos HG, Schmidt B, Smith GC, Spranger J, Zupancic N, Weksberg R. Manifestations and treatment of Schimke immuno-osseous dysplasia: 14 new cases and a review of the literature., 2000, 159(1–2): 1–7.
[86] Wang W, Song H, Wei M, Qiu Z, Wang C, Zhang Y, Li M, Yuan YH, Tang XY. SMARCAL1 gene analysis of 2 Chinese Schimke immuno-osseous dysplasia children.2015, 53(1): 45–50.王薇, 宋红梅, 魏珉, 邱正庆, 王晨, 张玉, 李明, 袁裕衡, 唐晓艳. Schimke免疫–骨发育不良儿童基因分析. 中华儿科杂志, 2015, 53(1): 45–50.
[87] Liu ZQ,Song FY, Liu Y, Qiu MF, Qian Y, Chen XB. Schimke immuno-osseous dysplasia (SIOD): A case report and review of literatures.. 2017, 33(2): 111–115.刘子勤, 宋福英, 刘颖, 邱明芳, 钱晔, 陈晓波. Schimke免疫–骨发育不良一例并文献复习. 中华内分泌代谢杂志, 2017, 33(2): 111–115.
[88] Liu SM, Zhang MC, Ni MX, Zhu PR, Xia XY.A novel compound heterozygous mutation of the SMARCAL1 gene leading to mild Schimke immune-osseous dysplasia: a case report., 2017, 17(1): 217.
[89] Barraza-García J, Rivera-Pedroza CI, Belinchón A, Fernández-Camblor C, Valenciano-Fuente B, Lapunzina P, Heath KE. A novel SMARCAL1 missense mutation that affects splicing in a severely affected Schimke immunoosseous dysplasia patient., 2016, 59(8): 363–366.
[90] Santangelo L, Gigante M, Netti GS, Diella S, Puteo F, Carbone V, Gesualdo L.A novel SMARCAL1 mutation associated with a mild phenotype of Schimke immuno- osseous dysplasia (SIOD)., 2014, 15(1): 41.
[91] Carroll C, Badu-Nkansah A, Hunley T, Baradaran- Heravi A, Cortez D, Frangoul H. Schimke Immunoosseous Dysplasia associated with undifferentiated carcinoma and a novel SMARCAL1 mutation in a child., 2013, 60(9): E88–90.
[92] Yue Z, Xiong S, Sun L, Huang W, Mo Y, Huang L, Jiang X, Chen S, Hu B, Wang Y. Novel compound mutations of SMARCAL1 associated with severe Schimke immuno- osseous dysplasia in a Chinese patient, 2010, 25(5):1697–1702.
[93] Bökenkamp A, deJong M, van Wijk JA, Block D, van Hagen JM, Ludwig M.R561C missense mutation in the SMARCAL1 gene associated with mild Schimke immuno- osseous dysplasia., 2005, 20(12): 1724– 1728.
[94] Zivicnjak M, Franke D, Zenker M, Hoyer J, Lücke T, Pape L, Ehrich JH.SMARCAL1 mutations: a cause of prepubertal idiopathic steroid-resistant nephrotic syndrome., 2009, 65(5): 564–568.
[95] Kilic SS, Donmez O, Sloan EA, Elizondo LI, Huang C, André JL, Bogdanovic R, Cockfield S, Cordeiro I, Deschenes G, Fründ S, Kaitila I, Lama G, Lamfers P, Lücke T, Milford DV, Najera L, Rodrigo F, Saraiva JM, Schmidt B, Smith GC, Stajic N, Stein A, Taha D, Wand D, Armstrong D, Boerkoel CF.Association of migraine- like headaches with Schimke immuno-osseous dysplasia., 2005, 135(2): 206–210.
[96] Simon AJ, Lev A, Jeison M, Borochowitz ZU, Korn D, Lerenthal Y, Somech R. Novel SMARCAL1 bi-allelic mutations associated with a chromosomal breakage phenotype in a severe SIOD patient., 2014, 34(1): 76–83.
[97] Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, André JL, Bogdanovic R, Burguet A, Cockfield S, Cordeiro I, Fründ S, Illies F, Joseph M, Kaitila I, Lama G, Loirat C, McLeod DR, Milford DV, Petty EM, Rodrigo F, Saraiva JM, Schmidt B, Smith GC, Spranger J, Stein A, Thiele H, Tizard J, Weksberg R, Lupski JR, Stockton DW. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia., 2002, 30(2): 215–220.
[98] Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end., 2004, 200: 132–141.
[99] Buck D, Moshous D, de Chasseval R, Ma Y, le Deist F, Cavazzana-Calvo M, Fischer A, Casanova JL, Lieber MR, de Villartay JP. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV., 2006, 36(1): 224–235.
[100] Lev A, Amariglio N, Levy Y,Spirer Z, Anikster Y, Rechavi G, Dekel B, Somech R. Molecular assessment of thymic capacities in patients with Schimke immuno- osseous dysplasia., 2009, 133(3): 375– 381.
[101] Dutta P, Tanti GK, Sharma S, Goswami SK, Komath SS, Mayo MW, Hockensmith JW, Muthuswami R. Global epigenetic changes induced by SWI2/SNF2 inhibitors characterize neomycin-resistant mammalian cells., 2012, 7(11): e49822.
SMARCAL1, roles and mechanisms in genome stability maintenance
Yalei Wen1, Kenao Lü2, Xiaokang Xu1, Xin Zhang1, Liang Ding1, Xuefeng Pan1,2,3
SMARCAL1 is an ATP-driven DNA annealing helicase that is similar in structure to the chromatin regulators in the subfamily A group of the SWI/SNF-related matrix-associated actin-dependent chromatin regulators. SMARCAL1 catalyzes the formation of dsDNA by annealing the single-stranded binding protein RPA coated ssDNA with its complementary strand bothand. In humans, different mutations ofgene are found to be closely related to different symptoms shown in individuals with Schimke immuno-osseous dysplasia (SIOD). This paper reviews the recent research progress of SMARCAL1 functions in remodeling DNA replication forks at damaged DNA sites, working in classical non-homologous end joining (NHEJ) repair of DNA double-stranded breaks, and in maintaining chromosomal telomere integrity. The relationships between the mutations ofgene in different SIOD symptoms, and the possible involvements of SMARCAL1 in neuromuscular degenerative diseases associated with trinucleotide repeats expansions are also updated and discussed to better understand the roles and mechanisms of the annealing helicase in genome stability maintenance.
SMARCAL1; RPA; DNA replication fork; SIOD; expansions of trinucleotide repeats
2019-07-07;
2019-09-04
北京自然科学基金项目(编号:5132014)和河北省医学科学重点项目(编号:20160051)资助[Supported by the Beijing Natural Science Foundation (No. 5132014) and the Hebei Provincial Key Research Programs for Medical Science (No. 20160051)]
文雅蕾,硕士研究生,专业方向:分子药理学。E-mail: 491574395@qq.com
丁良,博士,教授,研究方向:分析化学。E-mail: 345823685@qq.com
潘学峰,博士,教授,研究方向:生物化学、分子遗传学和药理等。E-mail: xuefengpancam@aliyun.com
10.16288/j.yczz.19-158
2019/9/27 9:18:00
URI: http://kns.cnki.net/kcms/detail/11.1913.R.20190926.1625.002.html
(责任编委: 卢大儒)
