食品和临床环境中大肠埃希菌耐药现状及健康风险研究
2024-01-01李梦涵汪庆杨光李思敏刘长振张晓婷齐丽英李书唱申芷瑜
李梦涵 汪庆 杨光 李思敏 刘长振 张晓婷 齐丽英 李书唱 申芷瑜

摘要:抗生素的滥用,导致大量残留的抗生素及抗生素耐药基因在食品环境和临床环境中被检出。大肠埃希菌(Escherichia coli)作为食源性致病菌之一,极易获得和传播耐药基因,对多种抗生素产生耐药性。耐药性大肠埃希菌可以在食品环境和临床环境间传播,使人体内菌群的耐药性增强。大肠埃希菌作为抗生素耐药基因的储存库,其抗生素耐药性已严重威胁到食品安全和人类健康,食品环境与临床环境中耐药菌株的出现成为全球关注的公共卫生问题。本文结合国内外研究进展,综述了食品和临床环境中大肠埃希菌的耐药现状,阐述了食品贸易对耐药性大肠埃希菌全球性传播的推动作用以及临床环境中耐药性大肠埃希菌的驱动因素,探讨了耐药性大肠埃希菌在食品与临床环境间的传播途径以及对人体的健康风险,以期为未来耐药性大肠埃希菌的研究和治理提供参考。
关键词:大肠埃希菌;耐药性;食品环境;临床环境;多重耐药
中图分类号:R978文献标志码:A
Research progress on antibiotic resistance and health risk of Escherichia coli in food and clinical environment
Li Menghan, Wang Qing, Yang Guang, Li Simin, Liu Changzhen,
Zhang Xiaoting, Qi Liying, Li Shuchang, and Shen Zhiyu
(College of Energy and Environmental Engineering, Hebei Key Laboratory of Air Pollution Cause and Impact, Hebei University of Engineering, Handan 056038)
Abstract The abuse of antibiotics leads to the detection of many residual antibiotics and antibiotic resistance genes in food environment and clinical environment. As one of the foodborne pathogenic bacteria, Escherichia coli can easily acquire and spread antibiotic resistance genes and develop resistance to many antibiotics. Antibiotic resistance in Escherichia coli can spread between the food environment and the clinical environment, increasing the resistance of the bacterial community in the human body. As a repository of antibiotic resistance genes, antibiotic-resistant Escherichia coli has seriously threatened food safety and human health, and the emergence of antibiotic resistance strains in food environment and clinical environment have become a global public health concern. Based on the research progress both at home and abroad, this paper introduced the present situation of antibiotic-resistant Escherichia coli in food and clinical environment, elaborated the food trade to promote the spread of antibiotic-resistant Escherichia coli in global clinical environment and the driving factors of antibiotic-resistant strains of Escherichia coli. Escherichia coli resistance in the route of transmission between food and clinical environment and the risk to the health of human body are discussed, in order to provide a reference for future research and management of antibiotic-resistant Escherichia coli.
Key words Escherichia coli; Antibiotic resistance; Food environment; Clinical environment; Multiple
antibiotic resistance
抗生素自問世以来,被广泛应用于临床治疗、动物养殖、农业生产等领域[1-3]。抗生素的过度使用及滥用,导致大量残留的抗生素及抗生素耐药基因在环境中被检出[4-6],加快了耐药性产生和传播的速度[7-8]。耐药菌在世界各地不断出现并蔓延,呈现出耐药水平高,耐药模式复杂的特点[9-11]。
大肠埃希菌是人类和动物肠道中最常见的共生菌,也是重要的病原体之一,可引发多种人畜共患病[12]。同时大肠埃希菌作为抗生素耐药基因的储存库,极易获得和传播耐药基因,从而对多种抗生素产生耐药性[13-15]。耐药性大肠埃希菌可通过水平基因转移随食物链进入人体,对食品安全与人类健康构成严重威胁[16]。因此,加强抗生素的管控,遏制耐药性的发展迫在眉睫。本文结合国内外研究,综述了食品与临床环境中大肠埃希菌的耐药现状,阐述了食品及临床环境中大肠埃希菌耐药性的传播及对人体的健康风险,同时对未来耐药性大肠埃希菌的研究和防治进行了展望。
1 食品环境中大肠埃希菌的耐药现状
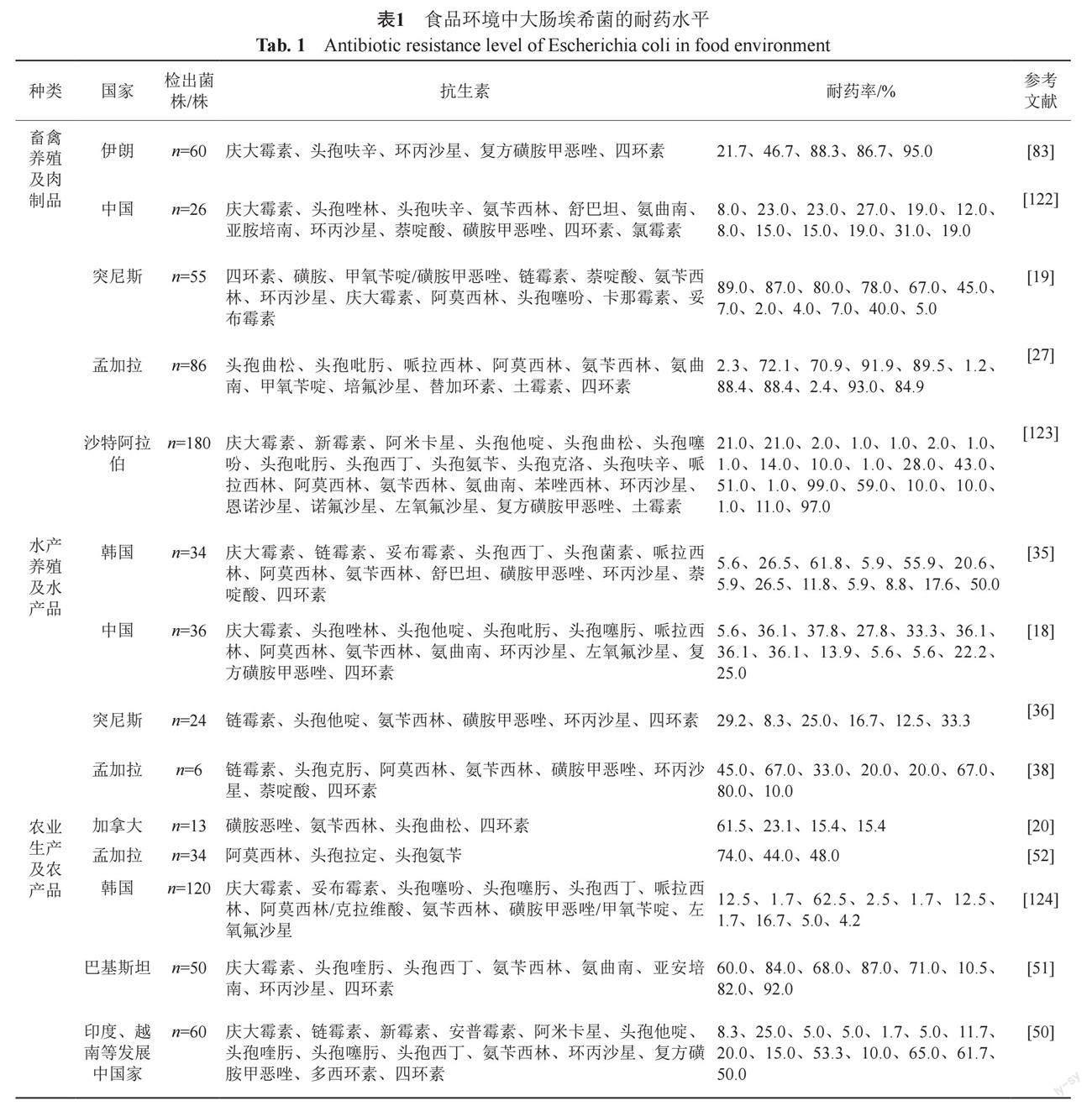
大肠埃希菌极易获得耐药性[17],在抗生素广泛使用的驱动下,耐药性大肠埃希菌普遍存在于肉制品、水产品、农产品等食品环境。如表1所示,各个国家和地区的食品环境中均检测出耐药性大肠埃希菌,检出菌株均具有耐药水平高、范围广、形式复杂等特点[18-20]。
1.1 畜禽养殖及肉制品
畜禽养殖及肉制品中检测出大肠埃希菌耐药的现象十分普遍[21-22]。在美国、巴西、中国等全球主要的鸡肉出口国家,从鸡肉中检测到的大肠埃希菌对四环素、磺胺甲恶唑、链霉素和氨苄西林等抗生素的耐药率均高于40%[23]。中国是全球畜牧业中抗生素的主要使用者[24]。周玲等[25]采用Mate分析对中国2005—2020年猪源大肠埃希菌的耐药性数据进行研究,发现大肠埃希菌对四环素类的耐药率在2005—2010年(94.5%)、2011—2015年(92.3%)、2016—2020年(96.3%)3个时间段均高于90%。中国作为抗生素使用大国,所检出的猪源大肠埃希菌对四环素类抗生素表现出较高的耐药性,这与Abdelgader等[26]的研究结果一致。有研究发现孟加拉[27]和突尼斯[19]肉制品中的大肠埃希菌对土霉素、氨苄西林具有较高耐药率,原因之一是土霉素、氨苄西林在畜禽养殖中被大量用于治疗或预防动物疾病。随着集约化养殖业的兴起与发展,全球各个国家在畜禽养殖中大量使用抗生素,使得动物在聚集性的养殖环境中反复高频率的接触抗生素,最终导致肉制品成为耐药基因传播的媒介,给人类健康带来潜在风险[28-30]。因此,在畜禽养殖中应加强抗生素使用的管控,缓解畜禽养殖及肉制品中大肠埃希菌的耐药现象。
1.2 水产养殖及水产品
四环素、阿莫西林和喹诺酮类等一系列用于提高水产养殖产量的抗生素正在世界范围内被广泛使用[31-33]。水产养殖业不恰当地使用抗生素导致水产品中检测出大量残留的抗生素,加剧了大肠埃希菌耐药性的传播[34]。在中国[18]、韩国[35]采集的水产品中均检测出耐药性大肠埃希菌,分离株对头孢唑林、头孢菌素和哌拉西林等不同种类抗生素表现出不同程度的耐药性。Said等[36]对从食用鱼类和贝类中分离的大肠埃希菌进行耐药性检测,结果表明大肠埃希菌对四环素表现出较高的耐药性,这与李景云等[37]的研究结果一致。Noor等[38]通过对水产品中分离的大肠埃希菌进行研究,发现分离株对头孢克肟(67%)、阿莫西林(33%)、氨苄西林(20%)等抗生素具有耐药性,其中对喹诺酮类抗生素如环丙沙星(67%)、萘啶酸(80%)的耐药率较高。Dib等[39]在阿尔及利亚采集的水产品中发现了多重耐药大肠埃希菌。从世界各地水产养殖及水产品中大肠埃希菌的分离鉴定及耐药性分析来看,耐药性大肠埃希菌在水产品中普遍存在[40-41]。人类食用含有耐药性大肠埃希菌的水产品极易发生耐药菌感染[42],因此应对水产品中耐药性大肠埃希菌的传播机制进行深入研究,保障食品安全,降低人类的感染风险。
1.3 农业生产及农产品
農业生产及农产品中的耐药性大肠埃希菌是威胁人类健康的潜在风险[43-46]。近些年,与新鲜农产品相关的食源性疾病数量一直在增加,其中大肠埃希菌是最常见的病原体[47-48]。Araujo等[49]发现耐药性大肠埃希菌可通过食用蔬菜传播给人类。姚旭等[50]对在印度、越南等发展中国家的蔬菜中分离出的60株大肠埃希菌进行药敏分析,结果表明大肠埃希菌对复方磺胺甲恶唑的耐药率最高为65.0%,对头孢噻肟(20.0%)、氨苄西林(53.3%)、四环素(50.0%)等抗生素表现出不同程度的耐药。Shah等[51]在蔬菜沙拉样品中分离出50株大肠埃希菌,对其进行耐药性研究同样发现大肠埃希菌对β-内酰胺类、四环素类和氯霉素类等抗生素表现出不同程度的耐药。Nawas等[52]从孟加拉国采集的沙拉中分离得到34株大肠埃希菌,发现大肠埃希菌对阿莫西林、头孢拉定和头孢氨苄的耐药率分别为74%、44%和48%。据报道,在加拿大[20]的植物性食品和中国[53]的凉拌菜样品中均分离出耐药性大肠埃希菌,此外,在尼泊尔[54]的蔬菜沙拉样品中检出的大肠埃希菌表现出多重耐药。农产品中大肠埃希菌的耐药及多重耐药问题不容忽视,因此要加强对大肠埃希菌耐药机制的研究,以遏制细菌耐药性的传播扩散。
1.4 食品贸易加速耐药性大肠埃希菌的全球性传播
随着国际贸易规模的扩大,进口食品中的耐药性大肠埃希菌在全球范围内被频繁检出[55-56]。Boss等[41]在瑞士进口海产品中检测到大肠埃希菌对环丙沙星(22%)、四环素(17%)等抗生素耐药。Bergenholtz等[57]在丹麦本地肉制品中未分离到对头孢噻呋耐药的大肠埃希菌,而进口肉类中对其耐药的大肠埃希菌具有较高的流行率。由于食品贸易的全球化,耐药性很容易通过食物链在全球范围内进行传播[58]。产超广谱β-内酰胺酶(extended-spectrum beta-lactamases,ESBLs)大肠埃希菌能水解第三代头孢菌素类药物,对头孢他啶、头孢曲松等多种抗生素产生耐药,并且可经食物链直接或间接传播给人类[59]。产ESBLs大肠埃希菌从全球各地的进口食品中分离出来,成为威胁人类健康的全球性问题。多项研究在瑞典[60]、日本[61]等多个国家的进口食品中检测到产ESBLs大肠埃希菌。Jung等[20]从进口产品中检测出对氨苄西林、头孢曲松耐药的产ESBLs大肠埃希菌,并从中发现一种与多重耐药性相关的可移动遗传元件。Egervarn等[60]在瑞典的进口肉制品中发现携带产ESBLs和对映异构体的大肠埃希菌,此前在丹麦的进口肉类中发现过同样的情况[62]。Muller等[56]研究发现进口食品成为德国本土多重耐药和产ESBLs大肠埃希菌的重要来源。食物中携带的耐药菌及耐药基因会随着食品的进出口贸易传播到全球各个国家和地区,加剧耐药性大肠埃希菌的全球性传播[63]。
2 临床环境中大肠埃希菌的耐药现状
2.1 臨床环境中大肠埃希菌耐药性的驱动因素
致病性大肠埃希菌是食源性疾病的主要病原体之一[64],也是临床感染最常见的病原菌[65],药物不合理的使用是多重耐药菌感染的主要因素[66]。Browne等[67]分析了2000年至2018年间人类抗生素消费报告,发现自2000年以来全球人类对抗生素的消费量增加了46%。临床医学中抗生素的广泛使用使人体微生物暴露在高浓度药物下,这对大肠埃希菌施加了选择性压力,从而增加了耐药性大肠埃希菌 数量增多的风险[58],抗生素的大量消耗是临床环境中大肠埃希菌耐药性增长的主要驱动力。β-内酰胺类抗生素被大量用于临床治疗严重的细菌感染,由大肠埃希菌携带的ESBLs是β-内酰胺类抗生素耐药的重要介体[68-69]。不仅如此,ESBLs基因作为耐药性的决定因素之一,常与其他耐药基因整合在一起,借助可移动遗传元件在不同菌株和菌种间传播,加剧了多重耐药菌株的扩散[70]。大肠埃希菌携带的ESBLs是驱动耐药性增长的重要原因。此外,有研究发现在临床上长期使用低剂量抗生素进行预防性治疗会对大肠埃希菌形成选择性压力[71],诱导耐药菌进行繁殖或者耐药基因发生突变,这是导致大肠埃希菌耐药性增加的潜在驱动因素[72]。临床环境中大肠埃希菌耐药性的增加是不同驱动因素相互作用的结果[73]。
2.2 临床环境中大肠埃希菌耐药性的严峻形势
临床环境中的大肠埃希菌对多种常用抗生素具有不同程度的耐药性(表2)。国内外学者在临床样本中均分离得到耐药性大肠埃希菌,并对这些大肠埃希菌进行了药敏分析。刘洁等[74]和朱文杰等[75]对中国临床样本进行了检测,发现大肠埃希菌对氨苄西林、头孢呋辛、米诺环素和环丙沙星等抗生素的耐药率较高,这与沙特阿拉伯[76]临床样本中的一项大肠埃希菌的研究结果一致。Lee等[77]研究了采集自韩国临床样本的1414株大肠埃希菌,发现大肠埃希菌对头孢唑啉(86.0%)、头孢呋辛(93.6%)、头孢泊肟(99.5%)等多种抗生素具有较高的耐药性。有研究对来自伊朗的临床样本进行分析时发现大肠埃希菌对青霉素的耐药率达到了100%[78]。由此看来,临床环境中的大肠埃希菌耐药形势十分严峻。
大肠埃希菌对临床上常用的一些抗生素的耐药性有所增加,临床上检出的耐药菌株的数量不断攀升。Badura等[79]分离了来自临床患者的12万余株大肠埃希菌,检出耐药菌株的比例随着时间的推移而增加,最突出的是氨苄西林(1998年的25.4%—2013年的40%)、头孢噻肟(0.1%~6.7%)、头孢他啶(0.3%~14.2%)、环丙沙星(4.3%~16.7%)。从2005年开始,ESBLs阳性分离株的数量显著增加(0.1%~6.3%)。孟祥红等[80]对2008年至2010年中国某医院分离的大肠埃希菌进行了药敏分析,研究发现大肠埃希菌对亚胺培南、哌拉西林/他唑巴坦、阿米卡星3种抗生素的耐药率逐年上升。东欧的一项研究中报告了2011—2016年[81]产ESBLs大肠埃希菌(20.1%)的比例高于在2004—2010年[82]报告的比例(15.3%),产ESBLs大肠埃希菌的检出率在东欧呈上升的趋势。这与在伊朗的食品[83]和临床[78]环境中发现的产ESBLs大肠埃希菌的流行趋势一致。
迄今为止,临床环境中多重耐药、泛耐药甚至是全耐药细菌不断被发现,耐药菌通常表现为交叉耐药和多重耐药的特性。Jafri等[84]对巴基斯坦临床样本中的大肠埃希菌进行药敏分析,发现约65%的大肠埃希菌为多重耐药菌株,即对3种或3种以上抗生素耐药,多重耐药的情况十分明显。Mukherjee等[85]分析了来自印度住院患者样品中的大肠埃希菌,发现菌株对呋喃妥因(72.5%)、阿米卡星(70%)、氨苄西林(97.5%)、萘啶酸和头孢氨苄(95%)、阿莫西林(92.5%)、复方磺胺甲恶唑(82.5%)和环丙沙星(80%)耐药率较高,并且几乎所有的菌株都表现出多重耐药性。
临床环境中大肠埃希菌对常用抗生素的耐药性在全球范围内呈上升趋势,尤其是多重耐药菌株的出现增加了临床治疗的难度,严重危害人类健康[86]。因此,应充分了解临床中大肠埃希菌的耐药趋势,合理使用抗生素,积极探索新的治疗方案。
3 耐药性大肠埃希菌在食品与临床环境间的传播
耐药性大肠埃希菌可以在食品环境和临床环境间传播[87]。blaCMY-2和incK是欧洲肉鸡分离出的大肠埃希菌中常见的ESBLs的基因组合[60],这种基因组合已经在瑞典[88]和加拿大[89]的临床环境中传播。Eshrati等[90]研究了产ESBLs大肠埃希菌在食物链(鸡肉样品)和败血症人群之间的传播关系,结果表明鸡肉样品在食物链中的污染是败血症人群存在产ESBLs大肠埃希菌感染的主要原因之一。大肠埃希菌的耐药性面临从食品环境向人类扩散的风险[63]。此外,在人类临床样本和食物来源的大肠埃希菌中发现诸多可移动遗传元件。荷兰一项研究发现,从肉鸡中分离的大肠埃希菌所携带的ESBLs基因和质粒同样存在于临床分离菌株中[91-92]。Sunde等[93]分别对来自 挪威肉类和血液感染的耐药大肠埃希菌进行进一步检测,发现整合子在临床大肠埃希菌中的出现频率显著高于肉源大肠埃希菌。大肠埃希菌可以通过质粒、整合子等可移动遗传元件的水平传播获得耐药基因,耐药性可通过这些可移动遗传元件在各种环境中发生转移[14, 94-96]。
抗生素耐药性是一个生态系统问题,耐药性可以通过水循环[97]、空气[98]和土壤[99]等多种直接和间接的途径在动物、环境和人类等不同的环境中进行传播[100]。大肠埃希菌的耐药性可以通过水循环在临床环境和污水处理厂之间进行传播[97]。有研究在污水处理厂的末端出水中检测到含有携带耐药基因的细菌分离株,这些耐药基因会随着出水被释放到环境中,导致耐药菌的进一步传播[101]。Girijan等[102]在医院污水直接排放点附近的沉积物样本中检测到耐药性大肠埃希菌。医院污水的排放加剧了耐药菌和耐药基因的传播,使得耐药性通过水生生态系统进入食品环境,人类通过食用受污染的食物而获得耐药性。在畜禽养殖、果蔬种植和人类疾病治疗中使用的抗生素大量重叠,这可能导致由大肠埃希菌引起的腹泻、感染等疾病发病率的增加[103]。黏菌素在临床上被广泛用于治疗大肠埃希菌引发的疾病,同时在各种肉类和蔬菜中经常检测出携带黏菌素耐药编码基因的大肠埃希菌[104]。此外,Wang等[105]研究发现由于黏菌素在畜禽养殖和人类医疗中的广泛使用,导致黏菌素耐药基因从环境转移到临床环境中,临床中耐药性分离株的数量不断增加。耐药性大肠埃希菌可通过多种途径在食品与临床环境间进行传播,同时也给人类健康带来潜在的风险[106-107]。因此,应优化食品生产流程,管控临床治疗中抗生素的使用,加强对大肠埃希菌耐药数据的监测,对耐药性传播途径进行深入研究,减少耐药性大肠埃希菌在环境间的传播,遏制大肠埃希菌耐药性不断攀升的局面。
4 人体健康风险
大肠埃希菌是引起腹泻、败血症和尿路感染等疾病的主要病原体[108]。大量的抗生素被用于治疗由大肠埃希菌引起的疾病[109],在减轻了传染病负担的同时导致耐药性大肠埃希菌的出现。在临床上抗生素对疾病的治疗逐渐丧失效力,耐药性大肠埃希菌的传播导致其引起的疾病不能得到有效的治疗,患者会出现严重的并发症从而引发身体机能的损害[24]。抗生素大量用于医疗领域的同时,还被广泛用于食品生产的不同环节,多种类、大剂量的抗生素被添加到饲料中用作食用动物的生长促进剂以提高产量,在果蔬种植中用作农药喷洒以预防虫害等[110-111]。在生产活动中使用的抗生素残留将会扩散到周围水体或者渗入地下水造成水污染,有研究在尼日利亚的鱼塘里检测出耐药性大肠埃希菌,发现大肠埃希菌对呋喃妥因、庆大霉素等抗生素耐药率较高[112],水环境将成为庞大的耐药基因储藏库[97]。同时,在畜禽养殖中动物不能有效地代谢体内的抗生素,含有抗生素残留物的动物粪便通常作为肥料与土壤进行混合用于农业生产,长期施用将会导致土壤中细菌的耐药水平增加[113-114]。大腸埃希菌的耐药性将会通过被污染的水、土壤、食物链等多种途径进行传播,最终传播给人类,耐药基因随之转移到人体肠道内的 细菌,使人体内菌群的耐药性增强[115-116]。
大肠埃希菌是耐药基因在环境和人体间转移的重要媒介[117-118]。抗生素在临床和食品中的大规模使用,对人类的危害表现在多个方面[119],增强了人体内大肠埃希菌的耐药性[115-116],增加了大肠埃希菌感染类疾病的治疗难度[24],提高了临床治疗成本[120]。食品贸易的流通加速了耐药性大肠埃希菌的全球性传播[121],加剧了对人体健康的威胁[47]。人类现代生产和生活方式驱动了细菌耐药性的产生,而人类对抗生素的过度使用及监管的缺失进一步加速了抗生素耐药性在环境中的扩散和传播。在这种严峻的形势下,如何在临床治疗上合理用药,如何在生产活动中有效预防和控制动植物疾病,如何控制耐药菌不断增加、耐药模式愈发复杂的窘境是当今亟待解决的问题。
5 展望
耐药性大肠埃希菌已经严重威胁到了食品安全和人体健康。在经济发展全球化推动下,抗生素耐药性通过食物链以及食品贸易在世界各个国家间传播,其中耐药性在蔬菜、水果等农产品中的传播是一个被低估的耐药来源。食品和临床环境中频繁检测出耐药性大肠埃希菌,特别是多重耐药性大肠埃希菌的出现频率急剧增加,导致临床用药困难,对现有的医疗条件提出了极大的挑战。因此在“后抗生素”时代,对未来耐药性大肠埃希菌的研究和治理进行了一些展望,包括:
(1)应全面加强国际合作,加快构建和完善全球耐药性监测系统,利用大数据对耐药数据进行分析利用,整合资源,共同治理。
(2)要加强对食品生产中抗生素使用的管控,并对大肠埃希菌通过食物链进入人体的传播机制以及其耐药性在环境中的传播途径进行深入探究。
(3)在临床上面对细菌感染,应逐步减少对抗生素的依赖,规范用药的同时积极探索新的治疗方案,加强探索毒力基因与大肠埃希菌耐药性之间的关系,为临床合理用药提供理论依据。
参 考 文 献
Hughes L, Hermans P, Morgan K. Risk factors for the use of prescription antibiotics on UK broiler farms[J]. J Antimicrob Chemother, 2008, 61(4): 947-952.
俞慎, 王敏, 洪有为. 环境介质中的抗生素及其微生物生态效应[J]. 生态学报, 2011, 31(15): 4437-4446.
王冉, 刘铁铮, 王恬. 抗生素在环境中的转归及其生态毒性[J]. 生态学报, 2006, 26(1): 265-270.
张俊华, 陈睿华, 刘吉利, 等. 宁夏养牛场粪污和周边土壤中抗生素及抗生素抗性基因分布特征[J]. 环境科学, 2021, 42(6): 2981-2991.
程森, 路平, 冯启言. 渔业复垦塌陷地抗生素抗性基因与微生物群落[J]. 环境科学, 2021, 42(5): 2541-2549.
付星宇, 汪庆, 毕聪聪, 等. 大气环境中抗生素耐药菌的来源与传播扩散研究进展[J]. 中国抗生素杂志, 2021, 46(9): 821-828.
Goossens H, Ferech M, Vander S R, et al. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study[J]. Lancet, 2005, 365(9459): 579-587.
Aminov R I. The role of antibiotics and antibiotic resistance in nature[J]. Environ Microbiol, 2009, 11(12): 2970-2988.
Sáenz Y, Zarazaga M, Bri?as L, et al. Antibiotic resistance in Escherichia coli isolates obtained from animals, foods and humans in Spain[J]. Int J Antimicrob Agents, 2001, 18(4): 353-358.
Thorsteinsdottir T R, Haraldsson G, Fridriksdottir V, et al. Prevalence and genetic relatedness of antimicrobial-resistant Escherichia coli isolated from animals, foods and humans in Iceland[J]. Zoonoses Public Health, 2010, 57(3): 189-196.
Belotindos L, Villanueva M, Miguel J, Jr., et al. Prevalence and characterization of quinolone-resistance determinants in Escherichia coli isolated from food-producing animals and animal-derived food in the philippines[J]. Antibiotics, 2021, 10(4): 413-428.
Rafique M, Potter R F, Ferreiro A, et al. Genomic characterization of antibiotic resistant Escherichia coli isolated from domestic chickens in Pakistan[J]. Front Microbiol, 2020, 10: 3052-3061.
Wright, Gerard D. The antibiotic resistome: the nexus of chemical and genetic diversity[J]. Nat Rev Microbiol, 2007, 5(3): 175-186.
Carattoli A. Importance of integrons in the diffusion of resistance[J]. Vet Res, 2001, 32(3): 243-259.
Farshad S, Japoni A, Hosseini M. Low distribution of integrons among multidrug resistant E. coli strains isolated from children with community-acquired urinary tract infections in Shiraz, Iran[J]. Pol J Microbiol, 2008, 57(3): 193-198.
Frye J G, Jackson C R. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals[J]. Front Microbiol, 2013, 4: 135-156.
李德喜, 李新生, 杜向黨, 等. 氨基糖苷类药物高水平耐药16S rRNA甲基化酶基因在动物源大肠埃希菌中的检测[J]. 江西农业学报, 2009, 21(8): 4-6.
纪雪, 邢新月, 梁冰, 等. 淡水养殖鱼大肠埃希菌分离鉴定与耐药性研究[J]. 动物医学进展, 2018, 39(12): 80-84.
Soufi L, Abbassi M S, Sáenz Y, et al. Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia[J]. Foodborne Pathog Dis, 2009, 6(9): 1067-1073.
Jung D, Rubin J E. Identification of antimicrobial resistant bacteria from plant-based food products imported into Canada[J]. Int J Food Microbiol, 2020, 319: 1-27.
Mayrhofer S, Paulsen P, Smulders F J, et al. Antimicrobial resistance profile of five major food-borne pathogens isolated from beef, pork and poultry[J]. Int J Food Microbiol, 2004, 97(1): 23-29.
Hagos Y, Gugsa G, Awol N, et al. Isolation, identification, and antimicrobial susceptibility pattern of Campylobacter jejuni and Campylobacter coli from cattle, goat, and chicken meats in Mekelle, Ethiopia[J]. PLoS One, 2021, 16(2): 1-13.
Roth N, Kasbohrer A, Mayrhofer S, et al. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview[J]. Poult Sci, 2019, 98(4): 1791-1804.
Talebi Bezmin Abadi A, Rizvanov A A, Haertlé T, et al. World health organization report: Current crisis of antibiotic resistance[J]. J Bionanosci, 2019, 9(4): 778-788.
周玲, 犹银俊, 龚才伟, 等. 我国健康猪源大肠埃希菌对四环素类药物耐药性的Meta分析[J]. 畜牧与兽医, 2021, 53(4): 61-68.
Abdelgader S A, Shi D, Chen M, et al. Antibiotics resistance genes screening and comparative genomics analysis of commensal Escherichia coli isolated from poultry farms between China and Sudan[J]. Biomed Res Int, 2018, 2018: 1-9.
Parvin M S, Talukder S, Ali M Y, et al. Antimicrobial resistance pattern of Escherichia coli isolated from frozen chicken meat in Bangladesh[J]. Pathogens, 2020, 9(6): 1-17.
Johnson J R, Kuskowski M A, Kirk S, et al. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods[J]. J Infect Dis, 2005, 191(7): 1040-1049.
Caruso G. Antibiotic resistance in Escherichia coli from farm livestock and related analytical methods: A review[J]. J AOAC Int, 2018, 101(4): 916-922.
Thakali A, Macrae J D. A review of chemical and microbial contamination in food: What are the threats to a circular food system?[J]. Environ Res, 2021, 194: 1-16.
Heuer O, Kruse H, Grave K, et al. Human health consequences of use of antimicrobial agents in aquaculture[J]. Clin Infect Dis, 2009, 49(8): 1248-1253.
Cabello F C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment[J]. Environ Microbiol, 2006, 8(7): 1137-1144.
Akinbowale O L, Peng H, Barton M D. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia[J]. J Appl Microbiol, 2010, 100(5): 1103-1113.
Lihan S, Lee S Y, Seng C T, et al. Plasmid-mediated antibiotic resistant Escherichia coli in sarawak rivers and aquaculture farms, northwest of Borneo[J]. Antibiotics, 2021, 10(7): 776-791.
Koo H J, Woo G J. Characterization of antimicrobial resistance of Escherichia coli recovered from foods of animal and fish origin in Korea[J]. J Food Prot, 2012, 75(5): 966-972.
Said L B, Hamdaoui M, Jouini A, et al. First detection of CTX-M-1 in extended-spectrum beta-lactamase-producing Escherichia coli in seafood from Tunisia[J]. J Food Prot, 2017, 80(11): 1877-1881.
李景云, 崔生辉, 马越, 等. 动物及健康人肠道共生的大腸埃希菌耐药性及产生原因分析[J]. 中国抗生素杂志, 2008, 33(9): 552-556.
Noor R, Hasan M F, Rahman M M. Molecular characterization of the virulent microorganisms along with their drug-resistance traits associated with the export quality frozen shrimps in Bangladesh[J]. Springerplus, 2014, 3(1): 1-7.
Dib A L, Agabou A, Chahed A, et al. Isolation, molecular characterization and antimicrobial resistance of Enterobacteriaceae isolated from fish and seafood[J]. Food Control, 2018, 88: 54-60.
Le Q P, Ueda S, Nguyen T N, et al. Characteristics of extended-spectrum beta-lactamase-producing Escherichia coli in retail meats and shrimp at a local market in Vietnam[J]. Foodborne Pathog Dis, 2015, 12(8): 719-725.
Boss R, Overesch G, Baumgartner A. Antimicrobial resistance of Escherichia coli, Enterococci, Pseudomonas aeruginosa, and Staphylococcus aureus from raw fish and seafood imported into Switzerland[J]. J Food Prot, 2016, 79(7): 1240-1246.
Nadimpalli M, Vuthy Y, De Lauzanne A, et al. Meat and fish as sources of extended-spectrum beta-lactamase-producing Escherichia coli, Cambodia[J]. Emerg Infect Dis, 2019, 25(1): 126-131.
Romyasamit C, Sornsenee P, Chimplee S, et al. Prevalence and characterization of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from raw vegetables retailed in Southern Thailand[J]. Peer J, 2021, 9: 1-21.
Richter L, Du Plessis E M, Duvenage S, et al. Occurrence, identification, and antimicrobial resistance profiles of extended-spectrum and AmpC beta-lactamase-producing enterobacteriaceae from fresh vegetables retailed in Gauteng Province, South Africa[J]. Foodborne Pathog Dis, 2019, 16(6): 421-427.
Song J, Oh S S, Kim J, et al. Extended-spectrum beta-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea[J]. Sci Rep, 2020, 10(1): 1-7.
Blaak H, Van Hoek A H, Veenman C, et al. Extended spectrum ss-lactamase- and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment[J]. Int J Food Microbiol, 2014, 168: 8-16.
Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria[J]. Infect Drug Resist, 2015: 49-61.
Newitt S, Macgregor V, Robbins V, et al. Two linked enteroinvasive Escherichia coli outbreaks, Nottingham, UK, June 2014[J]. Emerg Infect Dis, 2016, 22(7): 1178-1184.
Araujo S, Silva I, Tacao M, et al. Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms[J]. Int J Food Microbiol, 2017, 257: 192-200.
姚旭. 市售肉菜產ESBLs和碳青霉烯酶肠杆菌的分子流行病学研究[D]. 广州: 华南农业大学, 2016.
Shah M S, Eppinger M, Ahmed S, et al. Multidrug-resistant diarrheagenic E. coli pathotypes are associated with ready-to-eat salad and vegetables in Pakistan[J]. J Korean Soc Appl Biol Chem, 2015, 58(2): 267-273.
Nawas T, Mazumdar R, Das S, et al. Microbiological quality and antibiogram of E. coli, Salmonella and Vibrio of salad and water from restaurants of chittagong[J]. J Environ Sci Nat Res, 2012, 5(1): 159-166.
周臣清, 張娟, 黄宝莹,等. 广州市蔬菜类凉拌菜中微生物污染状况调查及大肠埃希氏菌耐药现象研究[J]. 食品安全质量检测学报, 2021, 12(6): 2485-2490.
Sapkota S, Adhikari S, Pandey A, et al. Multi-drug resistant extended-spectrum beta-lactamase producing E. coli and Salmonella on raw vegetable salads served at hotels and restaurants in Bharatpur, Nepal[J]. BMC Res Notes, 2019, 12(1): 516-521.
Jansen W, Grabowski N, Gerulat B, et al. Food safety hazards and microbiological zoonoses in European meat imports detected in border inspection in the period 2008–2013[J]. Zoonoses Public Health, 2016, 63(1): 53-61.
Muller A, Jansen W, Grabowski N T, et al. ESBL- and AmpC-producing Escherichia coli from legally and illegally imported meat: Characterization of isolates brought into the EU from third countries[J]. Int J Food Microbiol, 2018, 283: 52-58.
Bergenholtz R D, Jorgensen M S, Hansen L H, et al. Characterization of genetic determinants of extended-spectrum cephalosporinases (ESCs) in Escherichia coli isolates from Danish and imported poultry meat[J]. J Antimicrob Chemother, 2009, 64(1): 207-209.
Holmes A H, Moore L S P, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance[J]. Lancet, 2016, 387(10014): 176-187.
Worku W, Desta M, Menjetta T. High prevalence and antimicrobial susceptibility pattern of Salmonella species and extended-spectrum beta-lactamase producing Escherichia coli from raw cattle meat at butcher houses in Hawassa city, Sidama regional state, Ethiopia[J]. PLoS One, 2022, 17(1): 262-308.
Egervarn M, Borjesson S, Byfors S, et al. Escherichia coli with extended-spectrum beta-lactamases or transferable AmpC beta-lactamases and Salmonella on meat imported into Sweden[J]. Int J Food Microbiol, 2014, 171: 8-14.
Nahar A, Awasthi S P, Hatanaka N, et al. Prevalence and characteristics of extended-spectrum beta-lactamase-producing Escherichia coli in domestic and imported chicken meats in Japan[J]. J Vet Med Sci, 2018, 80(3): 510-517.
Agers Y, Hald T, Hg B B, et al. DANMAP 2010 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark[R], 2011.
Elbediwi M, Li Y, Paudyal N, et al. Global burden of colistin-resistant bacteria: Mobilized colistinresistance genes study (1980—2018)[J]. Microorganisms, 2019, 7(10): 461-478.
Hashempour-Baltork F, Hosseini H, Shojaee-Aliabadi S, et al. Drug resistance and the prevention strategies in food borne bacteria: An update review[J]. Adv Pharm Bull, 2019, 9(3): 335-347.
Johnson J R, Urban C, Weissman S J, et al. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-beta-lactamase-producing E. coli from the United States, 2000 to 2009[J]. Antimicrob Agents Chemother, 2012, 56(5): 2364-2370.
Laxminarayan R, Heymann D L. Challenges of drug resistance in the developing world[J]. BMJ, 2012, 344: 1567-1570.
Browne A J, Chipeta M G, Haines-Woodhouse G, et al. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study[J]. Lancet Planet Health, 2021, 5(12): 893-904.
Kumar D, Singh A K, Ali M R, et al. Antimicrobial susceptibility profile of extended spectrum beta-lactamase (ESBL) producing Escherichia coli from various clinical samples[J]. Infect Dis, 2014, 7: 1-8.
Flament-Simon S C, Garcia V, Duprilot M, et al. High prevalence of ST131 subclades C2-H30Rx and C1-M27 among extended-spectrum beta-lactamase-producing Escherichia coli causing human extraintestinal infections in patients from two hospitals of Spain and France during 2015[J]. Front Cell Infect Microbiol, 2020, 10: 125-133.
Teklu D S, Negeri A A, Legese M H, et al. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia[J]. Antimicrob Resist Infect Control, 2019, 8: 39-50.
Ching C, Zaman M H. Development and selection of low-level multi-drug resistance over an extended range of sub-inhibitory ciprofloxacin concentrations in Escherichia coli[J]. Sci Rep, 2020, 10(1): 8754-8582.
Projahn M, Daehre K, Semmler T, et al. Environmental adaptation and vertical dissemination of ESBL-/pAmpC-producing Escherichia coli in an integrated broiler production chain in the absence of an antibiotic treatment[J]. Microb Biotechnol, 2018, 11(6): 1017-1026.
王磊, 曹巍. 全球抗生素耐藥性现状分析及对策建议[J]. 军事医学, 2017, 41(5): 329-333.
刘洁, 杨晶, 高立芳, 等. 862例医院感染患者病原菌菌种及其耐药性分析[J]. 山东医药, 2019, 59(27): 33-37.
朱文杰, 张翔, 黄友, 等. 2018年江苏省某三甲医院临床细菌分布及耐药性分析[J]. 医学动物防制, 2020, 36(11): 1105-1108.
Azim N S A, Al-Harbi M A, Al-Zaban M I, et al. Prevalence and antibiotic susceptibility among gram negative bacteria isolated from intensive care units at a tertiary care hospital in Riyadh, Saudi Arabia[J]. J Pure Appl Microbiol, 2019, 13(1): 201-208.
Lee S J, Lee D S, Choe H S, et al. Antimicrobial resistance in community-acquired urinary tract infections: Results from the korean antimicrobial resistance monitoring system[J]. J Infect Chemother, 2011, 17(3): 440-446.
Momtaz H, Karimian A, Madani M, et al. Uropathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties[J]. Ann Clin Microbiol Antimicrob, 2013, 12(8): 1-12.
Badura A, Feierl G, Pregartner G, et al. Antibiotic resistance patterns of more than 120 000 clinical Escherichia coli isolates in Southeast Austria, 1998—2013[J]. Clin Microbiol Infect, 2015, 21(6): 569-575.
孟祥紅, 孙红宁, 孙敏霞, 等. 2008—2010年我院下呼吸道感染革兰氏阴性杆菌的耐药分析[J]. 标记免疫分析与临床, 2011, 18(6): 353-356.
Dowzicky M J, Chmelarova E. Antimicrobial susceptibility of Gram-negative and gram-positive bacteria collected from Eastern Europe: Results from the tigecycline evaluation and surveillance trial (T.E.S.T.), 2011-2016[J]. J Glob Antimicrob Resist, 2019, 17: 44-52.
Balode A, Punda-Polic V, Dowzicky M J. Antimicrobial susceptibility of Gram-negative and Gram-positive bacteria collected from countries in Eastern Europe: Results from the tigecycline evaluation and surveillance Trial (T.E.S.T.) 2004-2010[J]. Int J Antimicrob Agents, 2013, 41(6): 527-535.
Jahantigh M, Samadi K, Dizaji R E, et al. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran[J]. BMC Vet Res, 2020, 16(1): 267-272.
Jafri S A, Qasim M, Masoud M S, et al. Antibiotic resistance of E. coli isolates from urine samples of Urinary Tract Infection (UTI) patients in Pakistan[J]. Bioinformation, 2014, 10(7): 419-422.
Mukherjee M, Basu S, Mukherjee S K, et al. Multidrug-resistance and extended spectrum beta-lactamase production in uropathogenic E. coli which were isolated from hospitalized patients in Kolkata, India[J]. J Clin Diagn Res, 2013, 7(3): 449-453.
Murray C J L, Ikuta K S, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis[J]. The Lancet, 2022, 399(10325): 629-655.
Aminov R I, Mackie R I. Evolution and ecology of antibiotic resistance genes[J]. FEMS Microbiol Lett, 2010, (2): 147-161.
Borjesson S, Jernberg C, Brolund A, et al. Characterization of plasmid-mediated AmpC-producing E. coli from Swedish broilers and association with human clinical isolates[J]. Clin Microbiol Infect, 2013, 19(7): 309-311.
Baudry P J, Mataseje L, Zhanel G G, et al. Characterization of plasmids encoding CMY-2 AmpC beta-lactamases from Escherichia coli in Canadian intensive care units[J]. Diagn Microbiol Infect Dis, 2009, 65(4): 379-383.
Eshrati B, Baradaran H R, Motevalian S A, et al. Investigating the relationship between extended spectrum beta-lactamase producing Escherichia coli in the environment and food chains with the presence of this infection in people suspected of septicemia: Using the fuzzy set qualitative comparative analysis[J]. J Environ Health Sci Eng, 2020, 18(2): 1509-1520.
Hall L V, Dierikx C M, Stuart J C, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains[J]. Clin Microbiol Infect, 2011, 17(6): 873-880.
Overdevest, Ilse. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands[J]. Emerg Infect Dis, 2011, 17(7): 1216-1222.
Sunde M, Simonsen G S, Slettemeas J S, et al. Integron, plasmid and host strain characteristics of Escherichia coli from humans and food included in the Norwegian antimicrobial resistance monitoring programs[J]. PLoS One, 2015, 10(6): 1-14.
Solberg O D, Ajiboye R M, Riley L W. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli[J]. J Clin Microbiol, 2006, 44(4): 1347-1351.
Blahna M T, Zalewski C A, Reuer J, et al. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada[J]. J Antimicrob Chemother, 2006, 57(4): 666-672.
Mazel D. Integrons: agents of bacterial evolution[J]. Nat Rev Microbiol, 2006, 4(8): 608-620.
Perez-Etayo L, Gonzalez D, Vitas A I. The aquatic ecosystem, a good environment for the horizontal transfer of antimicrobial resistance and virulence-associated factors among extended spectrum beta-lactamases producing E. coli[J]. Microorganisms, 2020, 8(4): 568-580.
Sanz S, Olarte C, Martinez-Olarte R, et al. Airborne dissemination of Escherichia coli in a dairy cattle farm and its environment[J]. Int J Food Microbiol, 2015, 197: 40-44.
Chen C, Pankow C A, Oh M, et al. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments[J]. Environ Int, 2019, 128: 233-243.
Dorado-Garcia A, Smid J H, Van Pelt W, et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: A pooled analysis[J]. J Antimicrob Chemother, 2018, 73(2): 339-347.
Szczepanowski R, Linke B, Krahn I, et al. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics[J]. Microbiology, 2009, 155(7): 2306-2319.
Girijan S K, Paul R, V J R, et al. Investigating the impact of hospital antibiotic usage on aquatic environment and aquaculture systems: A molecular study of quinolone resistance in Escherichia coli[J]. Sci Total Environ, 2020, 748: 1-62.
Szmolka A, Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health[J]. Front Microbiol, 2013, 4: 258-270.
Clemente L, Leao C, Moura L, et al. Prevalence and characterization of ESBL/AmpC producing Escherichia coli from fresh meat in Portugal[J]. Antibiotics, 2021, 10(11): 1333-1348.
Wang Z, Koirala B, Hernandez Y, et al. A naturally inspired antibiotic to target multidrug-resistant pathogens[J]. Nature, 2022, 601(7894): 606-611
Ben Y, Fu C, Hu M, et al. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review[J]. Environ Res, 2019, 169: 483-493.
Pormohammad A, Nasiri M J, Azimi T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis[J]. Infect Drug Resist, 2019, 12: 1181-1197.
Allocati N, Masulli M, Alexeyev M F, et al. Escherichia coli in Europe: An overview[J]. Int J Environ Res Public Health, 2013, 10(12): 6235-6254.
Boeckel T V, Gandra S, Ashok A, et al. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data[J]. Lancet Infect Dis, 2014, 14(8): 742-750.
Bengtsson-Palme J. Antibiotic resistance in the food supply chain: Where can sequencing and metagenomics aid risk assessment?[J]. Curr Opin Food Sci, 2017, 14: 66-71.
Van T T H, Yidana Z, Smooker P M, et al. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses[J]. J Glob Antimicrob Resist, 2020, 20: 170-177.
Njoku O, Agwa O, Ibiene A. Antibiotic susceptibility profile of bacteria isolates from some fishponds in Niger delta region of Nigeria[J]. Br Microbiol Res J, 2015, 7(4): 167-173.
Ghosh S, Lapara T M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria[J]. ISME J, 2007, 1(3): 191-203.
Rahube T O, Yost C K. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application[J]. J Appl Microbiol, 2012, 112(6): 1123-1133.
Day M J, Hopkins K L, Wareham D W, et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study[J]. Lancet Infect Dis, 2019, 19(12): 1325-1335.
White D G, Zhao S, Simjee S, et al. Antimicrobial resistance of foodborne pathogens[J]. Microbes Infect, 2002, 4(4): 405-412.
Angulo F J, Baker N L, Olsen S J, et al. Antimicrobial use in agriculture: Controlling the transfer of antimicrobial resistance to humans[J]. Semin Pediatr Infect Dis, 2004, 15(2): 78-85.
Guanghui W, Day M J, Mafura M T, et al. Comparative analysis of ESBL-positive Escherichia coli isolates from animals and humans from the UK, the Netherlands and Germany[J]. PLoS One, 2013, 8(9): 1-10.
Pakbin B, Allahyari S, Amani Z, et al. Prevalence, phylogroups and antimicrobial susceptibility of Escherichia coli isolates from food products[J]. Antibiotics, 2021, 10(11): 1291-1301.
Livermore D M. Current epidemiology and growing resistance of gram-negative pathogens[J]. Korean J Intern Med, 2012, 27(2): 128-142.
Davies J, Davies D. Origins and evolution of antibiotic resistance[J]. Microbiol Mol Biol Rev, 2010, 74(3): 417-433.
Dong P, Xiao T, Nychas G E, et al. Occurrence and characterization of Shiga toxin-producing Escherichia coli (STEC) isolated from chinese beef processing plants[J]. Meat Sci, 2020, 168: 1-6.
Abo-Amer A E, Shobrak M Y, Altalhi A D. Isolation and antimicrobial resistance of Escherichia coli isolated from farm chickens in Taif, Saudi Arabia[J]. J Glob Antimicrob Resist, 2018, 15: 65-68.
Kim S M, Oh T, Kim H J. Antimicrobial resistance, molecular, and phenotypic diversity of Escherichia coli isolates from fresh vegetable products in Korea[J]. J Korean Soc Appl Biol Chem, 2015, 58(5): 745-750.
Najwa D, Salah A M, Yolanda S, et al. Low antibiotic resistance rates and high genetic heterogeneity of Escherichia coliisolates from urinary tract infections of diabetic patients in Tunisia[J]. J Chemother, 2016, 28(2): 89-94.
Mollick S, Dasgupta T, Hasnain M, et al. Isolation and characterization of pathogens responsible for urinary tract infection in bangladesh and determination of their antibiotic susceptibility pattern[J]. J Appl Pharm Sci, 2016: 72-76.
