Development and characterization of new allohexaploid resistant to web blotch in peanut
2021-12-14WANGSiyuLlLinaFULiuyangLlUHuaQlNLiCUlCaihongMlAOLijuanZHANGZhongxinGAOWeiDONGWenzhaoHUANGBingyanZHENGZhengTANGFengshouZHANGXinyouDUPei
WANG Si-yu ,Ll Li-na,FU Liu-yang ,LlU Hua,QlN Li,CUl Cai-hong,MlAO Li-juan,ZHANG Zhong-xin,GAO Wei,DONG Wen-zhao,HUANG Bing-yan,ZHENG Zheng,TANG Feng-shou,ZHANG Xin-you ,DU Pei
1 School of Life Sciences,Zhengzhou University,Zhengzhou 450001,P.R.China
2 Industrial Crops Research Institute,Henan Academy of Agricultural Sciences/Key Laboratory of Oil Crops in Huang-Huai-Hai Plains,Ministry of Agriculture and Rural Affairs/Henan Provincial Key Laboratory for Oil Crops Improvement,Zhengzhou 450002,P.R.China
Abstract Peanut diseases seriously threaten peanut production,creating disease-resistant materials via interspecific hybridization is an effective way to deal with this problem. In this study,the embryo of an interspecific F1 hybrid was obtained by crossing the Silihong (Slh) cultivar with Arachis duranensis (ZW55),a diploid wild species. Seedlings were generated by embryo rescue and tissue culture. A true interspecific hybrid was then confirmed by cytological methods and molecular markers.After treating seedlings with colchicine during in vitro multiplication,the established interspecific F1 hybrid produced seeds which were named as Am1210. With oligonucleotide fluorescence in situ hybridization (Oligo FISH),molecular marker evaluations,morphological and web blotch resistance characterization,we found that:1) Am1210 was an allohexaploid between Slh and ZW55; 2) the traits of spreading lateral branches,single-seeded or double-seeded pods and red seed coats were observed to be dominant compared to the erect type,multiple-seeded pods and brown seed coats; 3) the web blotch resistance of Am1210 was significantly improved than that of Slh,indicating the contribution of the web blotch resistance from the wild parent A.duranensis. In addition,69 dominant and co-dominant molecular markers were developed which could be both used to verify the hybrid in this study and to identify translocation or introgression lines with A.duranensis chromosome fragments in future studies as well.
Keywords:peanut,interspecific hybridization,allohexaploid,Oligo FISH,molecular marker,web blotch
1.lntroduction
Cultivated peanut (Arachis hypogaeaL.2n=4x=40) is an important oil crop worldwide. However,peanut diseases seriously threaten peanut production,which can cause peanut yield losses of more than 20%. Peanut web blotch(Phoma arachidicolaMarasas),which usually spreads rapidly and causes severe yield losses,is one of the most serious foliar diseases in peanut (Phipps 1985; Zhanget al.2019). Therefore,creating disease-resistant materials including web-blotch-resistant materials are important for peanut production.
WildArachisspecies have developed superior traits during evolution because of long-term exposure to natural abiotic and biotic threats,such as resistance to pathogens of web blotch and rust. They may differ from peanut cultivars in their mechanisms of disease resistance,and are important genetic resources for improving cultivated peanut (Stalker 2017).
Interspecific hybridization is an effective measure to utilize elite genes of wild species. However,interspecific hybridization of peanut has been extremely difficult and time-consuming owing to the low percentage of successful pollinations that result in hybrids,the sterility of progenies and genomic incompatibilities (Gregory and Gregory 1979;Stalker 1981; Nagyet al.2010; Zhanget al.2013; Fuet al.2016). In order to effectively utilize the beneficial genes of wild species,special hybridization pathways have been established,such as autotetraploid method,in which crosses can be made between cultivated peanut and artificial autotetraploids obtained with the chromosomes doubling of wild species (Mallikarjunaet al.2011); heterotetraploid method,in which crosses can be made between cultivated peanut and a heteroploid derived through chromosome doubling the F1hybrids of two diploid wild species (Churchet al.2005); and triploid-hexaploid method,in which crosses are first made between cultivated peanuts and diploid species to produce triploids,followed by chromosome doubling of triploids to yield hexaploid; backcrossing is further made between hexaploid and cultivated peanuts to obtain interspecific progenies (Heet al.2013). Using interspecific hybridization technology,the chromosome introgression lines ofA.duranensis,A.stenosperma,A.cardenasii,etc.,have been successfully developed,and new peanut germplasms resistant to leaf spot,the root-knot nematode,rust,stripe virus,and bud necrosis virus have been created (Dwivediet al.2002; Mallikarjuna and Sastri 2002; Stalkeret al.2002; Mallikarjuna and Hoisington 2009;Pandeyet al.2012; Pasupuletiet al.2013).
Arachis duranensisis an A-genome diploid wild species of the sectionArachiswith small chromosomes and bright centromeric 4´,6-diamidino-2-phenylindole (DAPI) bands(Seijoet al.2004; Robledoet al.2009). Its chromosomes could be easily distinguished from those of cultivated peanut by oligonucleotide fluorescencein situhybridization (Oligo FISH),which facilitates the accurate identification of hybrid progeny between these wild species and cultivated peanut(Duet al.2018,2019).
Previous studies have shown thatA.duranensisis resistant to aflatoxin,rust and late leaf spot disease (Sharmaet al.2003; Xueet al.2004). In this study,with the aim to improve the resistance of cultivated peanut,the following was performed:1) developing interspecific F1hybrid and allohexaploid of crossing between cultivated peanut andA.duranensis; 2) characterizing the hybrids by molecular markers and cytology; and 3) evaluating its phenotype and reaction to web blotch.
2.Materials and methods
2.1.Plant materials
The peanut cultivar,Silihong (Slh) and the diploidArachisspecies,A.duranensis(ZW55) were used to develop interspecific F1hybrid and an allohexaploid.
2.2.lnterspecific hybridization,embryo rescue and chromosome doubling
Interspecific hybridization,embryo rescue and chromosome doubling were performed according to the methods of Liet al.(2017). Briefly,artificial hybridization was conducted at the flowering stage. Pods from the pollinated flowers were then harvested when they reached maturity. After treating the pods with 75% alcohol,the embryos were dissected and put on solid media of Murashige and Skoog (MS)+0.075 mg L-1indole-acetic-acid (IAA)+0.01 mg L-1kinetin (Kn)+5% sucrose at 25°C. Seedlings of F1hybrids were also multiplied on media of MS+0.8 mg L-1naphthalene acetic acid (NAA)+5% sucrose,and MS+0.1 mg L-1NAA+0.4 mg L-16-benzylaminopurine (6-BA)+5%sucrose. For the allohexaploid,chromosome doubling was performed by treating the seedlings in agar media with MS+0.1 mg L-1NAA+0.4 mg L-16-BA+5% sucrose+0.05%colchicine for 11 days.
2.3.Chromosome preparation and Oligo FlSH
Chromosome preparation was performed according to the method of Duet al.(2013). First,the root tips were excised and pretreated with 2 mmol L-18-hydroxyquinoline for 3 h,and fixed in absolute ethanol (three parts):glacial acetic acid(one part) for 12 h at 4°C. A length of 0.3-0.5 mm of the root tip meristem was then excised and squashed in 45%glacial acetic acid,and frozen at -80°C for 12 h. The spread chromosomes were dehydrated in 100% ethanol and dried in air after the cover slips were removed.
BecauseA.duranensisis assumed to be the A-genome donor to the cultivated parent,it is impossible to distinguish the A-genome of cultivated peanut from that of theA.duranensisgenome through genomicin situhybridization(GISH). Sequential FISH with oligos (DP-1,DP-2,DP-3,DP-4,DP-5,DP-6,and DP-7) and rDNA (45S rDNA and 5S rDNA) probes were therefore conducted to accurately identify the hybrid w1210. The procedures were performed according to the methods of Duet al.(2018).
The DP-1,DP-3,DP-4,and DP-6 oligonucleotides were modified at the 5´-ends with 6-carboxytetramethylrhodamine(TAMRA),and DP-2,DP-5 and DP-7 were modified at the 5´-ends with FAM (6-carboxyfluorescein) by the Invitrogen Company (Shanghai,China). The hybridization solution,consisting of 7.5 μL formamide (Sigma,Germany),1.5 μL 20× saline-sodium citrate (SSC) buffer,1.0 μL of each probe,and 2.0 μL of 50% dextran sulfate (Sigma) was denatured at 105°C for 13 min. The spread chromosomes were denatured in 70% formamide at 75°C for 70 s,and then hybridized overnight at 37°C.
After hybridization,the slides were washed three times in 2×SSC at 25°C,and mounted with VECTASHIELD®Mounting Medium,following which DAPI staining was performed.Slides were examined using a Leica DM6000 fluorescence microscope (Leica,Germany). Separate images from each filter set were captured using a cooled charge-coupled device (CCD) camera (Leica). Slides were denatured after FISH using repetitive oligonucleotide probes,and then washed to remove all signals,and subsequently dried.
A second FISH procedure using 45S rDNA and 5S rDNA probes was conducted on the same slides to develop karyotypes. Clones of 5S rDNA and 45S rDNA of wheat(Triticum aestivumL.),provided by Dr.Bikram S Gill from Kansas State University,USA,were employed. The 5S rDNA clone and total genomic DNA ofA.ipaensiswere labeled with biotin-16-dUTP (Roche,Germany) by nick translation and detected with fluorescein anti-biotin (Roche);whereas 45S rDNA and total genomic DNA ofA.ipaensiswere labeled with digoxigenin-11-dUTP (Roche) and detected with anti-digoxigenin-rhodamine (Roche).
2.4.Molecular marker analysis
To confirm the interspecific hybrids,the specific marker E66 (F:TCCTTCCCACAATAACAATGAA,R:GAGGAGAAAACATGGCCTAAAA) ofA.duranensiswas used,as previously described byFu (2017). To further develop more specific markers for the allohexaploid,1 115 pairs of simple sequence repeat (SSR) and transposable element (TE) markers (Heet al.2003; Moretzsohnet al.2005; Zhaoet al.2017) were amplified and screened by polyacrylamide gel electrophoresis using Am1210 and its parental genomic DNA.
2.5.Evaluation of resistance to web blotch
Web blotch was inoculated by Wanget al.(2015).Web blotch resistance was evaluated in the Yuanyang Experimental Field of Henan Academy of Agricultural Sciences using a mixture ofPhoma arachidicolaisolates collected in Yuanyang,China. The plants of Slh,w1210 and Am1210 were inoculated in two growing seasons (2017-2018) at their flowering and pegging stages. Twenty days after inoculation,adult plant reactions were investigated.Adult plant reactions were scored at heading on a scale of 0-9,where readings of 0 were interpreted as immunity;1-3,high resistance; 4-6,moderate resistance; and 7-9,high susceptibility.
2.6.Pollen stainability (pollen viability) and phenotypic trait investigation
Flowers of F1hybrids were collected between 8:00 and 8:30 a.m.from each plant to assess pollen viability. Pollen was squeezed out of the anthers onto a glass slide and stained with 2% acetocarmine for about 2 min. Using a light microscope,darkly stained,well-formed,and large pollen grains were classified as viable,whereas unstained,and misshapen pollen grains were classified as non-viable.
The girl accepted her offer with joy, and she was at once set to work in a corner of the kitchen,15 where all the farm servants came and made fun of her, and the ass’s skin in which she was wrapped. But by-and-by they got so used to the sight of it that it ceased to amuse them, and she worked so hard and so well, that her mistress grew quite fond of her. And she was so clever at keeping sheep and herding24 turkeys that you would have thought she had done nothing else during her whole life !
The proportion of viable over total pollen grains counted was expressed as a percentage and used as an indicator of pollen viability. Two flowers were examined for each plant and the average was calculated.Phenotypic traits of four plants of the allohexaploid and its cultivated parent,including the petiole length,height of the main stem,the top 3rd leaf length on the main stem,the top 3rd leaf width on the main stem,length of first primary branch,number of branches,seed coat color,and plant type,were investigated during the mature period.
3.Results
3.1.Establishment and characterization of the interspecific F1 hybrid
Morphologically well-developed pods were obtained in the interspecific cross (w1210) between Slh and ZW55.No significant differences in pod length were observed between the w1210 and its cultivated peanut parents;however,F1embryos from w1210 were abnormal. The embryo length of cultivated peanut was (22.06±5.01) mm,but that of w1210 F1was (3.15±0.03) mm,which was significantly shorter than that of its female parent. Embryo rescue operations were conducted over the interspecific cross and F1hybrids established. Pollen stainability of the cultivated parent of Slh was 98.00%,whereas that of w1210 was only 6.15%. No pods setting was observed in plants (Table 1; Fig.1-A).
To confirm whether the F1progeny were true interspecific hybrids at both the cytological and molecular levels,thechromosome numbers of w1210 were checked using cytological slides (Fig.1-B). Furthermore,we analyzed the amplifications of w1210 and its parents using the SSR marker,E66. The marker produced specific bands associated with both the cultivated and wild parents,revealing the true hybrid nature of w1210 (Fig.2).

Table 1 Crossability data of the interspecific hybrid F1

Fig.1 Cytological analysis and pollen viability of the interspecific hybrid,w1210. A,pollen viability of w1210. Yellow arrows show fertile pollen and blue arrows show sterile pollen. B,cytological analysis of w1210.
3.2.Establishment and cytological characterization of the allohexaploid
To restore fertility and maintain continuation of the progenies,in vitromultiplication and colchicine treatment of the F1hybrid were performed. The F1plants treated with colchicine were established and observed to bear pods and were subsequently referred to as Am1210. The karyotypes of Am1210 and their parents (Slh and ZW55) were established by Oligo FISH,45S rDNA and 5S rDNA plasmid FISH,and DAPI staining (Fig.3-D-I).
Based on these karyotypes,we found that the chromosome numbers of Slh and ZW55 were 40 and 20,respectively,and the genomes of Slh (Ahyand Bhy) and ZW55 (Adu) showed unique FISH patterns. Firstly,both Ahyand Aducontained the small chromosome 9 and showed strong centromeric DAPI bands,while the Bhygenome did not.
Secondly,45S rDNA sites in Ahyand Aduwere distributed mainly on chromosomes 2 and 10,whereas in Bhy,they were distributed on chromosomes 3,7 and 10. Thirdly,Ahyand Adushowed significant differences in their oligonucleotide bands,including green signals in the telomeric regions of chromosome Ahy7 and centromeric regions of Bhy5.

Fig.2 PCR amplification pattern of interspecific hybrids F1 and their parents using the SSR marker E66. M,DL 2000 marker.Red arrow shows specific loci of Shilihong (Slh); blue arrow shows specific loci of ZW55.
By comparing the chromosomes of two A sub-genomes,we observed significant differences in the Ahyand Adugenomes among chromosomes 1,3,5,and 10. On chromosome 1,Ahy1 showed red oligonucleotide signals in the long arm of Ahy1,whereas no signals were observed with Adu1. On chromosome 3,Adu3 showed green oligonucleotide signals at the end of the long arm,but Ahy3 did not. Furthermore,Adu5 and Adu10 showed strong green oligonucleotide signals in the centromeric regions,whereas the signals of Ahy5 and Ahy10 were weak. The karyotype of Am1210 revealed its 60 chromosomes,all of which had features of Slh and ZW55,indicating that Am1210 was an amphiploid of w1210.
3.3.Specific molecular marker development for the allohexaploid
In order to develop specific markers to track chromosome fragments from its progenitors in the hybrids,1 115 SSR markers were used to analyze Slh,ZW55 and Am1210.As a result,69 markers (6.2%) were obtained,including 41 co-dominant markers and 28 dominant markers (Table 2).
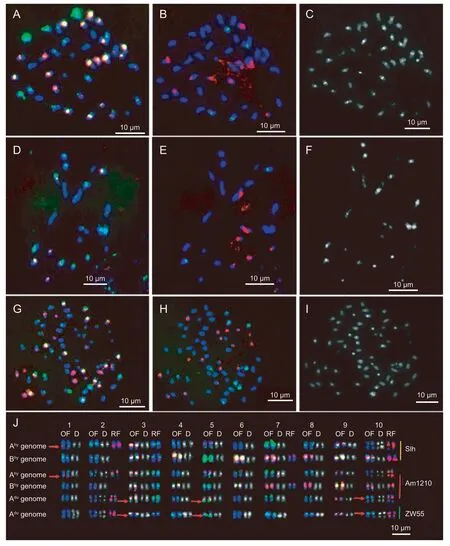
Fig.3 Sequential fluorescence in situ hybridization (FISH) and 4´,6-diamidino-2-phenylindole (DAPI) staining of Silihong (Slh),Arachis duranensis (ZW55) and Am1210,and their karyotypes. A-C,sequential FISH using Oligo#1 (A,green),Oligo#2 (A,red),5S rDNA (B,green),45S rDNA (B,red),and DAPI staining (C) in Slh. D-F,sequential FISH using Oligo#1 (D,green),Oligo#2(D,red),5S rDNA (E,green),45S rDNA (E,red),and DAPI staining (F) in ZW55. G-I,sequential FISH using Oligo#1 (G,green),Oligo#2 (G,red),5S rDNA (H,green),45S rDNA (H,red),and DAPI staining (I) in Am1210. J,OF,signal of Oligo#1 (green) and Oligo#2 (red),respectively; D,DAPI staining (gray white); RF,signals of 5S rDNA (green) and 45S rDNA (red),respectively.

Table 2 Dominant or co-dominant molecular markers of Arachis duranensis in Am1210
(Continued on next page)

Table 2 (Continued from preceding page)
3.4.Phenotypic trait analysis of the allohexaploid
Based on the analysis of phenotypic traits,we found that Am1210 and Slh showed significant differences in petiole length,main stem height and thickness of the main stem,but no significant differences in the length of the first primary lateral branches,height of the main stem and total number of branches observed,indicating that Am1210 had more characteristics resembling the wild parent (Table 3).Regarding the plant types,Slh was erect,and both ZW55 and Am1210 showed spreading characteristics,indicating the dominant feature of spreading (Fig.4-A).
In sharp contrast with w1210,the pollen fertility of Am1210 was significantly increased and well-developed pods were harvested from this hybrid (Table 1). The Slh cultivar had multiple-seeded pods,ZW55 and Am1210 had single-and double-seeded pods,which also reflected a feature of the male parent (Fig.4-B). In addition,although ZW55 had pink seeds,Slh and Am1210 had red seeds,which reflected a feature of the female parent (Fig.4-C).In addition,the leaves of Am1210 were much greener and thicker,as compared with those of Slh and ZW55 (Fig.4-D),reflecting the impact of ploidy level.
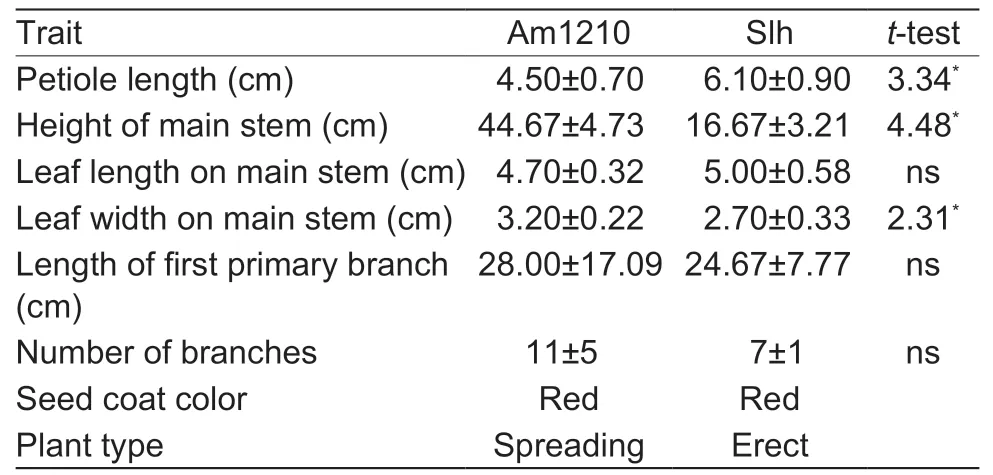
Table 3 Comparison of specific agronomic traits in the Am1210 allohexaploid and Shilihong (Slh)
3.5.Web blotch resistance
Resistance to web blotch of F1hybrid and its allohexaploid was investigated. In 2017 and 2018,w1210 was both scored grade 2,and Am1210 was both scored grade 1. But the scores of Slh were grades 7 and 8 in the two years,indicating its high susceptibility (Table 4). It is quite obvious that the resistance of both the F1hybrid and the allohexaploid were significantly improved compared with their cultivated peanut parent in two years due to the genomic enclosure of wild species (Fig.5).
4.Discussion
4.1.Allohexaploid provides new germplasm for peanut improvement
Interspecific hybridization is an effective way to improve the resistance of peanut,as it can transfer beneficial resistance genes from wild species to cultivated peanut. In the present study,the F1hybrid and its allohexaploid were established through interspecific hybridization followed by embryo rescue and artificial chromosome doubling. Based on observation of morphological traits of growth habit,number of seeds per pod and seed coat color,we speculate that characters of spreading growth habit,single-seeded or double-seeded pods resembling the male parent were dominant over those of erect growth habit and multipleseeded pods,and the red seed coat color from the female parent is dominant over a brown seed coat color. The results also showed that Am1210 retained more wild characteristics in growth,plant height and lateral branch length,which may restrict the utilization of important genes ofA.duranensisto some extent. The seed coat color of Am1210 was red,consistent with the seed coat color of Slh. This suggests that the red seed genetic material of introgressiveA.duranensisplants can be propagated.

Fig.4 Morphologies of plants (A),pods (B) and seeds (C) of Shilihong (Slh),ZW55 and Am1210,and leaves (D) of Slh,w1210,ZW55,and Am1210.

Fig.5 Web blotch reaction of interspecific hybrids Shilihong(Slh). F1,w1210; Am1210,allohexaploid.
In previous studies,Slh was reportedly highly susceptible to web blotch (Fuet al.2013; Liet al.2017),which is consistent with the results of our web blotch analyses.However,the high resistance of the F1hybrids w1210 and allohexaploid Am1210 which were derived from the cross between Slh and ZW55,indicated that the resistance of peanut cultivars to web blotch might be improved by introducing the genomes ofA.duranensisinto cultivated peanut. To our knowledge,this is the first finding thatA.duranensiswas resistant to web blotch,which might make allohexaploid Am1210 become a new resistance germplasm for peanut breeding in the future.
4.2.Oligonucleotide probes and molecular markers facilitate the identification of interspecific hybridization of peanut
At present,interspecific hybridization between cultivated peanut and wildArachisspecies by conventional methods is believed to be restricted only to members of the sectionArachis(Stalker 2017). Previous cytological studies have suggested thatA.duranensisis the donor of the A-genome of peanut (Robledoet al.2009; Bertioliet al.2016). Close relationships with cultivated peanut make the identification of interspecific hybrid progenies by GISH difficult. In this study,theA.duranensisgenome in the allohexaploid Am1210 was effectively identified by Oligo FISH,demonstrating the great potentials of Oligo FISH in the identification of chromosomes in interspecific hybrid progenies.
Nevertheless,most chromosomes ofA.duranensishad similar bands to those of the A-genome of cultivated peanut,making it difficult to identify alien chromosomes in the future. Therefore,more oligonucleotide probes need to be developed to enrich the chromosome features ofA.duranensis. In addition,69 dominant and co-dominant markers ofA.duranensiswere obtained in this study,which could be used both to verify the hybrid in this study and to identify translocation or introgression lines ofA.duranensischromosome fragments in future studies as well.
5.Conclusion
One interspecific F1hybrid and its allohexaploid from an interspecific cross between a peanut cultivar and a diploid wildArachisspecies were developed in this study. They were effectively identified by molecular markers and Oligo FISH. Evaluation of resistance to web blotch revealed that F1hybrid and allohexaploid were significantly more resistant than their cultivated parent,indicating that the resistance might be from the wild parentA.duranensis. High resistance to web blotch of allohexaploid Am1210 and its progenies will provide new resistance germplasm resources for peanut breeding. And the Oligo FISH employed and new molecular markers developed in this study will be very useful for identification and utilization of allohexaploid Am1210 and its progenies.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31801397),the Henan Province Young Talents Lifting Project,China (2018HYTP003),the Independent Innovation Foundation of Henan Academy of Agricultural Sciences,China (2019ZC13),the earmarked fund for China Agriculture Research System (CARS-13),and the Henan Provincial Agriculture Research System,China (S2012-05).
杂志排行
Journal of Integrative Agriculture的其它文章
- The dynamic impact of income and income distribution on food consumption among adults in rural China
- Driving factors of direct greenhouse gas emissions from China’s pig industry from 1976 to 2016
- ATP regulates the phosphorylation and degradation of myofibrillar proteins in ground ovine muscle
- Use of two-stage dough mixing process in improving water distribution of dough and qualities of bread made from wheatpotato flour
- lmpact of climate change on maize yield in China from 1979 to 2016
- Estimating daily actual evapotranspiration of a rice-wheat rotation system in typical farmland in the Huai River Basin using a two-step model and two one-step models
