MOLECULAR CLONING, CHARACTERIZATION, AND EXPRESSION ANALYSIS OF TWO ISOFORMS OF ANTI-LIPOPOLYSACCHARIDE FACTOR FROM THE ORIENTAL RIVER PRAWN, MACROBRACHIUM NIPPONENSE
2017-09-12WANGYingHuiXIUYunJiGUWeiMENGQingGuoandWANGWen
WANG Ying-Hui, XIU Yun-Ji, GU Wei, MENG Qing-Guoand WANG Wen
(1. Jiangsu Key Laboratory for Biodiversity & Biotechnology and Jiangsu Key Laboratory for Aquatic Crustacean Diseases, College of Life Sciences, Nanjing Normal University, Nanjing 210023, China; 2. Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China; 3. Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266200, China)
MOLECULAR CLONING, CHARACTERIZATION, AND EXPRESSION ANALYSIS OF TWO ISOFORMS OF ANTI-LIPOPOLYSACCHARIDE FACTOR FROM THE ORIENTAL RIVER PRAWN, MACROBRACHIUM NIPPONENSE
WANG Ying-Hui1, XIU Yun-Ji2,3, GU Wei1, MENG Qing-Guo1and WANG Wen1
(1. Jiangsu Key Laboratory for Biodiversity & Biotechnology and Jiangsu Key Laboratory for Aquatic Crustacean Diseases, College of Life Sciences, Nanjing Normal University, Nanjing 210023, China; 2. Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China; 3. Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266200, China)
Anti-lipopolysaccharide factors (ALFs), a type of the potent antimicrobial peptide, can bind and neutralize lipopolysaccharide (LPS) and exhibit broad spectrum antimicrobial activities. In order to study the function of ALFs in congenital immunization of Macrobrachium nipponense, two isoforms of the ALF homologues (MnALF1 and MnALF2) were cloned and characterized from the oriental river prawn M. nipponense. The full-length cDNA sequences of MnALF1 and MnALF2 were 1008 and 836 bp, encoding 121 and 124 amino acids, respectively. All of these sequences contained one signal peptide and an LPS-binding domain with two conserved cysteine residues at both ends of the domain. The deduced peptide of MnALF1 and MnALF2 was highly similar to previously identified ALFs in crustaceans. qRT-PCR showed that MnALFs were expressed in all detected tissues. MnALF1 transcript was predominantly detected in heart and intestine and MnALF2 transcript was predominantly detected in hemocytes and hepatopancreas. After challenge with Aeromonas hydrophila, two MnALF transcripts in heart, intestine, hemocytes and hepatopancreas showed a clear time-dependent response expression pattern (the expression levels of MnALF1 present an trend of downregulated first and then increasing, which reached to the highest level at 24h, 12h, 36h, 24h in heart, intestine, hemocytes and hepatopancreas respectively. However, for MnALF2, the expression level present an downregulated trend and then increased in heart, intestine; In hemocytes and hepatopancreas, the expression level present an increasing trend and then down-regulated. MnALF2 transcripts reached to the top at 24h post-challenge). These results suggest that two MnALF isoforms have different tissue specificity and might provide multiple protective functions in immune defense against invading bacteria.
Macrobrachium nipponense; Anti-lipopolysaccharide factor; Expression analysis; Aeromonas hydrophila
The oriental river prawn Macrobrachium nipponense is a freshwater or brackish prawn and it is one of the important economic aquaculture and commercial species in China, Japan and Vietnam[1]. M. nipponense is an indigenous species and the frequency of disease outbreaks has been very low in thepast. However, with rapid development of large-scale culture, various diseases caused by genetic retrogression and viruses, bacteria has become increasingly higher[2]. It is necessary to understand the immune defense mechanisms of M. nipponense to provide new insights into disease prevention in prawn aquaculture.
Invertebrates, which lack adaptive immune systems, depend completely on the innate immune system that is composed of humoral and cellular responses[3]. Antimicrobial peptides (AMPs), components of the humoral immune responses, play essential functions in the innate immune system[4,5]. They distributed broadly in the whole living kingdom with killing activities against bacteria, fungi or viral pathogens[6].
ALF is one of the group of AMPs, exhibiting binding and neutralizing activities to lipopolysaccharides[7]. The first ALF was found in Limulus polyphemus (LpALF)[8]and showed strong antimicrobial effect on Gram-negative R-type bacteria[9]. From crystal structure analysis, the LPS binding domain of LpALF contains two highly conserved-cysteine residues[10,11]. The LpALF derived peptide modulates cytokine gene expression and promotes resolution of bacterial acute infection in mice[12]. More recently, a number of crustacean ALFs with similar functions have been reported in penaeid shrimps[13,14], freshwater prawns[15—17], crayfish[18], lobster[19], and crabs[20,21]. Antibacterial activity against both Gram-negative and Gram-positive bacteria which was confirmed by the in vitro assays in decapods[22,23]. Many investigations have revealed that ALFs presented multiple biological activities against fungi and virus[24,25].
Although AMPs have been widely investigated in invertebrates, ALF has not been isolated from M. nipponense. In the current study, two ALFs from M. nipponense were identified and characterized. Here, the full length cDNAs of MnALF1 and MnALF2 were cloned. The deduced amino acid sequences were compared with other known crustacean ALFs. The expressions of MnALF1 and MnALF2 in different tissues were investigated, and their expression profiles in heart, intestine, hemocytes and hepatopancreas were studied after challenge with Aeromonas hydrophila.
1 Materials and methods
1.1 Animal and RNA extraction
The oriental river prawns, M. nipponense (2—3 g per prawn) were purchased from an aquaculture market in Nanjing, Jiangsu Province, China. They were cultured in tanks at 20℃ with freshwater and an aeration system. Prawn health status was assessed daily during acclimation by monitoring both general activity and food intake. Total RNA from different tissues was extracted by using TRIzol Reagent following the manufacturer’s instructions. RNA quality was assessed by electrophoresis on 1.2% agarose gel and the RNA concentration was measured by the absorbance at 260 nm on a spectrophotometer.
1.2 cDNA library construction and EST analysis
A cDNA library was constructed from the hemocytes of M. nipponense (Shanghai Hanyu Bio-Tech). Random sequencing of the library using illumina Hiseq 2000 genome analyzer yielded 43289 ESTs. BLASTx analysis revealed that two ESTs were homologous to ALF in Macrobrachium rosenbergii (ADI80708 and ACG60660). These two ESTs were selected for further cloning of MnALFs.
1.3 Cloning the full-length cDNA of MnALFs
SMARTerTMRACE cDNA Amplification Kit (Takara, Japan) was used for Rapid amplification of cDNA ends (RACE). Two pairs of gene specific primers were designed based on the corresponding EST sequences. For the 5′-RACE, The PCR reactions were performed with MnALF1-R1 or MnALF2-R1 and Universal Primer A Mix (UPM). For the 3′-RACE, the PCR reactions were performed with MnALF1-F1 or MnALF2-F1 and UPM. The polymerase chain reaction was performed as follows: 1 cycle at 94℃ for 2min; 30 cycles at 94℃ for 30s, 68℃ for 30s, 72℃ for 3min, and 1 cycle at 72℃ for 10min. The PCR fragments were cloned into pMD19-T vector and sequenced by Springen (Nanjing) Biotechnology Company.
1.4 Sequence analysis
The homology searches for nucleotide and amino acid sequence similarities were conducted with BLAST programs (http://blast.ncbi.nlm.nih.gov/ Blast.cgi). The deduced amino acid sequence was analyzed with the Expert Protein Analysis System (http://www.expasy.org/). SignalP 4.1 program was utilized to predict the presence and location of signal peptide (http://www.cbs.dtu.dk/services/SignalP/). Multiple sequences alignment was performed using the ClustalW2 (http://www.ebi.ac.uk/Tools/msa/ clustalw2/). A cladogram was constructed based on the amino sequences alignment by the neighbor-joining (NJ) algorithm embedded in MEGA 5 program.
1.5 Bacterial challenge and sample preparation
During the experiment, the prawns were fed once daily with commercial feed. The prawns were randomly divided into two groups and 50 prawns injected individually with 50 μL live A. hydrophila sus-pension (104cells/mL) which used as a challenge group. For the control group, 50 prawns were injected with 50 μL saline (0.85% NaCl) (pH=7.0). Every three individuals were randomly sampled to eliminate individual differences at 0, 1h, 12h, 24h and 36h post challenge. Heart, intestine, hemocytes and hepatopancreas were collected, respectively, and all of samples extracted at different times were stored at–80℃ for subsequent total RNA extraction.
1.6 Expression analysis of MnALFs transcript
The mRNA expressions of MnALFs transcript in different tissues, including hemocytes, heart, hepatopancreas, gill, intestine, nerve and muscle of untreated prawns, and the temporal expressions of MnALFs in heart, intestine, hemocytes and hepatopancreas of prawns challenged with A. hydrophila were determined by quantitative real-time RT-PCR.
The first-strand cDNA was synthesized by using PrimeScriptTM1st Strand cDNA Synthesis Kit (TaKaRa) with 1 µg of total RNA. Two pairs of gene-specific primer (MnALF1-RT-F, MnALF1-RT-R and MnALF2-RT-F, MnALF2-RT-R) (Tab. 1) were used to amplify MnALF1 and MnALF2 genes, respectively. The primers Mnβ-actinF and Mnβ-actinR (Tab. 1) were used to amplify β-actin for internal standardization[26,27].
qRT-PCR was carried out in a total volume of 20 µL (10 µL of 2×SYBR Premix Ex Taq, 1 µL cDNA mix, 0.5 µL of each primer (10 µmol/L), and 8 µL of sterile distilled H2O) . The PCR program was 95℃for 30s, followed by 40 cycles of 95℃ for 5s and 60℃ for 30s. All samples were run thrice. All datawere given in terms of relative mRNA expression as mean±S.E. Statistical significance was determined by one-way ANOVA and post-hoc Duncan multiple range tests. Significance was set at P<0.05.

Tab. 1 Primers used in the present study
2 Results
2.1 cDNA cloning and sequence analysis of MnALF1 and MnALF2
In this research, two different forms of ALF cDNA were identified from the oriental river prawn, M. nipponense. These ALF genes were designated as MnALF1 and MnALF2. The complete cDNA sequence of MnALFs were obtained by overlapping the corresponding EST with the amplified fragments. The complete cDNA sequence of MnALF1 is 1008 bp, including a 146 bp 5′ UTR, a 496 bp 3′ UTR, and a 366 bp ORF encoding a protein of 121 amino acids. The poly (A) tail was found in MnALF1, while no poly (A) signal (AATAAA) was detected. MnALF1 protein contains a signal peptide of 20 residues and an LPS-binding domain of 22 residues with 2 conserved cysteines. The theoretical pI of MnALF1 is 6.29 and molecular weight (Mw) is 13869.14 Da. The theoretical pI of LPS-binding domain is 9.14. The sequence data was deposited in GenBank with accession number KF696705.
The complete sequence of MnALF2 cDNA is 836 bp in length consisting of a 120 bp 5′ UTR, a 341 bp 3′ UTR, and a 375 bp ORF encoding a protein of 124 amino acids. A canonical polyadenylation signalsequence AATAAA was present at 15 nucleotides upstream of the poly (A) tail. A signal peptide of 25 amino acids and an LPS-binding domain were also detected in MnALF2. MnALF2 has a pI of 9.12 and an Mw of 13868.11 Da. The LPS-binding domain of MnALF2 has a pI of 9.14. The sequence data was deposited in GenBank with accession number KF696706.
2.2 Similarity and phylogenetic analyses
BLAST search indicated that two forms exhi bited similarity with other crustacean ALFs. MnALF1 showed the highest similarity with ALF2 and ALF3 from M. rosenbergii (81% identity). MnALF2 has 90% identity with ALF4 from M. rosenbergii and 86% identity with ALF from Macrobrachium olfersii, respectively.
Multiple sequence alignment of MnALF1 and MnALF2 with ALFs in crustaceans revealed that all these ALFs were conserved; they manifested a signal peptide and an LPS-binding domain, especially the two conserved cysteine residues at both ends of the domain.
A phylogenetic tree was constructed with ALF homologs from crustaceans to identify their evolutionary relationships (Fig. 1). Based on the tree topo-logy, ALF homologs could be classified into three main clades: A, B, and C. MnALF1 together with MrAlf3, FcAlf5 and PtAlf4 forming a small group belonged to Clade C. MnALF2 has a closer evolutionary relationship with MrAlf4 grouped into Clade A.
2.3 Tissue distributions of MnALF1 and MnALF2
qRT-PCR was employed to quantify mRNA expressions of MnALF1 and MnALF2 in the tissues of healthy prawns, including hemocytes, heart, hepatopancreas, gill, intestine, nerve and muscle (Fig. 2). The mRNA transcripts of MnALF1 and MnALF2 were detected in all the selected tissues. Fig. 2A showed that MnALF1 was mainly expressed in heart and intestine. The transcript of MnALF2 was detected mainly in the hemocytes and hepatopancreas (Fig. 2B).
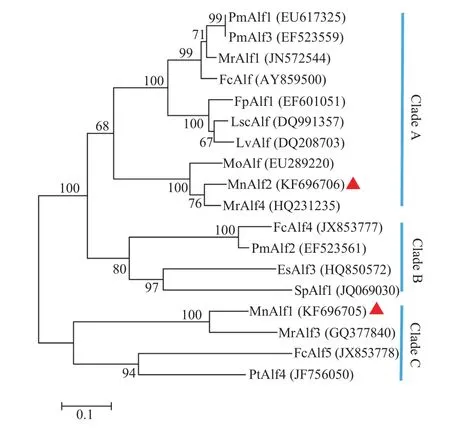
Fig. 1 Neighbor-joining phylogenetic tree of crustaceans ALF genes. The reliability of the branching was tested by bootstrap resampling (1000 pseudoreplicates)
2.4 Expression profiles of MnALF1 and MnALF2 after A. hydrophila challenge
The temporal mRNA expressions of MnALF1 and MnALF2 transcript in heart, intestine, hemocytes and hepatopancreas post A. hydrophila challenge are shown in Fig. 3.

Fig. 2 Tissue distributions of MnALF1 (A) and MnALF2 (B) transcripts. Vertical bars represent the mean±S.E.

Fig. 3 After challenge with A. hydrophila, the time-course expression levels of MnALF1 and MnALF2 in heart (A), intestine (B), hemocytes (C) and hepatopancreas (D). Vertical bars represent the mean±S.E. (n=3). β-actin was used as internal control
In heart, MnALF1 was down-regulated signifi-cantly at 1h post-challenge (26%). As time progressed, the subsequent expression was up-regulated and returned to the normal level at 24h post-challenge. For MnALF2, its expression level decreased during the first 12h after A. hydrophila challenge, and the lowest level occurred at 12h (49%). MnALF2 increased from 12h to 24h and reached the highest level at 24h, which was a 2.91-fold increase compared to that in the internal group.
In intestine, both MnALF1 and MnALF2 decreased initially at 1h post challenge. Afterward, MnALF1 transcript expression increased to the peak at 12h post injection (3.14-fold). The peak of MnALF2 occurred at 24h, which was a 8.15-fold increase compared to that in the control group.
In hemocytes, the temporal increase was detected for MnALF2 at 1h post challenge. Then, MnALF1 and MnALF2 transcript expressions decreased significantly at 12h. As time progressed, the expression of MnALF2 reached the peak at 24h.
In hepatopancreas, two MnALFs transcript expression fluctuated slightly during the first 12h. At 24h, both expression levels of MnALF1 and MnALF2 increased significantly and reached to their highest levels, which were 3.62-fold and 4.29-fold higher than that in the control group, respectively.
3 Discussion
ALFs play important roles in the innate immune system of crustaceans. Previous studies reported that the potent antimicrobial activity against both bacterial and viral made ALFs becoming potential therapeutic agents[24,28]. In this study, two isoforms of ALF were identified and characterized from the oriental river prawn, M. nipponense. The deduced protein sequences of MnALF1 and MnALF2 revealed a conserved structure composed of a signal peptide, an LPS-binding domain and two conserved cysteine residues. The two cysteine residues formed a disulfide loop, within which the positively charged amino acid residues were mainly clustered. All data demonstrate that the LPS-binding domain and the positively charged amino acids are essential for ALFs biological activities[10,29,30].
Together with other ALFs, the sequences of the MnALFs were used to construct a neighbor-joining phylogenetic tree to reveal the relationships of crustacean ALFs. On the tree topology, the MnALF1 and MnALF2 cluster together with MrALF3 and MrALF4, respectively. Based on the tree topology, crustacean ALFs could be classified into three main clades: A, B, and C. MnALF1 was classified within Clade C and MnALF2 was grouped into Clade A (Fig. 1). Meanwhile, three isoforms of M. rosenbergii ALFs were classified within two Clades. In previous research, ALFs from P. monodon were also divided into two different groups according to their genomic loci[15]. It suggests that the common ancestor of decapods proba bly had two ALF homologues.
The hypothetical LPS-binding domain is stabi lized by a disulfide bond. For MnALF1, this binding site consists of six positively charged amino acid residues (three Lys, two Arg and one His). For MnALF2, this binding site consists of five positively charged amino acid residues (four Lys and one Arg). An amphipathic loop, which possesses an alternating series of hydrophilic and hydrophobic residues, is generated by the disulfide bond. It was predicted that the hydrophobic residues forming one hydrophobic side can enter into the lipid bilayer, leaving the positively charged residues on the other side pointing out to bind to the hydrophilic and phosphate groups of lipid A on the bacterial membrane[20,30].
MnALF1 was mainly expressed in the heart and intestine and MnALF2 was mainly expressed in the hemocytes and hepatopancreas. Previous studies showed that most ALF transcripts were mainly situated in hemocytes. However, inconsistent results were obtained in some crustaceans, where ALF transcripts were prominently expressed in hepatopancreas, intestines, gills, or other tissues[24]. Previous researches have demonstrated that hemocytes are the major sites of AMP production, and the intestine or gonad is the secondary expression site for AMPs[31]. In this study, the results showed that the heart and hepatopancreas are also important production sites for MnALFs.
In this study, the expression patterns of MnALF1 and MnALF2 under bacterial challenge were investigated. The expression transcripts of MnALFs mostly decreased during the initial hours of A. hydrophila challenge, which is similar to the pattern observed in PtALF5[32]and PtALF7[33]. ALF isoforms exhibit clear time-dependent expression patterns[34], but the activation times are different. For MnALF1, its expression level was down-regulated after A. hydrophila stimulation and then increased and reached to the highest level at 24h, 12h, 36h, 24h in heart, intestine, hemocytes and hepatopancreas respectively. Moreover, in heart and intestine MnALF2 was down-regulated significantly at 1h post-challenge and reached the highest level at 24h. However, in hemocytes and hepatopancreas, the expression of MnALF2 start to increase after A. hydrophila stimulation and then downregulated significantly and reached their highest levelat 24h. In short, the expression level of MnALF1 and MnALF2 transcripts reached the top at 24h post-challenge, which is the same as observed in PtALF5 and PtALF7. This result is consistent with the conclusion that ALFs possess potent antimicrobial activity against Gram-negative bacteria.
In conclusion, we cloned and characterized two isoforms of the ALF homologues (MnALF1, MnALF2) from M. nipponense. Both of them contained one signal peptide and an LPS-binding domain with two conserved cysteine residues at both ends of the domain. MnALF1 and MnALF2 had different tissue distribution and after challenge with Aeromonas hydrophila, the two MnALF transcripts in heart, intestine, hemocytes and hepatopancreas showed a clear time-dependent response expression pattern. The present results might provide multiple protective functions in immune defense against invading bacteria.
[1]Xiu Y J, Hou L B, Liu X Q, et al. Isolation and characterization of two novel C-type lectins from the oriental river prawn, Macrobrachium nipponense [J]. Fish and Shellfish Immunology, 2015, 46(2): 603—611
[3]Vazquez L, Alpuche J, Maldonado G, et al. Review: Immunity mechanisms in crustaceans [J]. Innate Immunity, 2009, 15(3): 179—188
[4]Lin M C, Lin S B, Lee S C, et al. Antimicrobial peptide of an anti-lipopolysaccharide factor modulates of the inflammatory response in RAW264.7 cells [J]. Peptides, 2010, 31(7): 1262—1272
[5]Lin M C, Pan C Y, Hui C F, et al. Shrimp anti-lipopolysaccharide factor (SALF), an antimicrobial peptide, inhibits proinflammatory cytokine expressions through the MAPK and NF-kB pathways in LPS-induced HeLa cells [J]. Peptides, 2013, 40: 42—48
[6]Arockiaraj J, Kumaresan V, Bhatt P, et al. A novel singledomain peptide, anti-LPS factor from prawn: Synthesis of peptide, antimicrobial properties and complete molecular characterization [J]. Peptides, 2014, 53: 79—88
[7]Li S H, Guo S Y, Li F H, et al. Characterization and function analysis of an anti-lipopolysaccharide factor (ALF) from the Chinese shrimp Fenneropenaeus chinensis [J]. Developmental and Comparative Immunology, 2014, 46(2): 349—355
[8]Tanaka S, Nakamura T, Morita T, et al. Limulus anti-LPS factor: an anticoagulant which inhibits the endotoxin mediated activation of Limulus coagulation system [J]. Biochemical and Biophysical Research Communications, 1982, 105(2): 717—723
[9]Warren H, Glennon M, Wainwright N, et al. Binding and neutralization of endotoxin by Limulus antilipopolysaccharide factor [J]. Infection and Immunity, 1992, 60(6): 2506—2513
[10]Ried C, Wahl C, Miethke T, et al. High affinity endotoxinbinding and neutralizing peptides based on the crystal structure of recombinant Limulus anti-lipopolysaccharide factor [J]. Journal of Biological Chemistry, 1996, 271(8): 28120—28127
高速的退场,已能看出相关逻辑所在。那么为何选择西王?为何将打造了多年的“准冠军”球队转让给一家民营企业——西王集团?
[11]Hoess A, Watson S, Siber G, et al. Crystal structure of an endotoxin-neutralizing protein from the horseshoe crab, Limulus anti-LPS factor, at 1.5 A resolution [J]. EMBO Journal, 1993, 12(9): 3351—3356
[12]Vallespi M, Alvarez-Obregon J, Rodriguez-Alonso I, et al. A Limulus anti-LPS factor-derived peptide modulates cytokine gene expression and promotes resolution of bacterial acute infection in mice [J]. International Immunopharmacology, 2003, 3(2): 247—256
[13]Ponprateep S, Tharntada S, Somboonwiwat K, et al. Gene silencing reveals a crucial role for anti-lipopolysaccharide factors from Penaeus monodon in the protection against microbial infections [J]. Fish and Shellfish Immunology, 2012, 32(1): 26—34
[14]Tassanakajon A, Amparyup P, Somboonwiwat K, et al. Cationic antimicrobial peptides in penaeid shrimp [J]. Marine Biotechnology, 2011, 13(4): 639—657
[15]Ren Q, Du Z Q, Li M, et al. Cloning and expression analysis of an anti-lipopolysaccharide factor from giant freshwater prawn, Macrobrachium rosenbergii [J]. Molecular Biology Reports, 2012a, 39(7): 7673—7680
[16]Ren Q, Zhang Z, Li X C, et al. Three different anti-lipopolysaccharide factors identified from giant freshwater prawn, Macrobrachium rosenbergii [J]. Fish and Shellfish Immunology, 2012b, 33(4): 766—774
[17]Lu K Y, Sung H J, Liu C L, et al. Differentially enhanced gene expression in hemocytes from Macrobrachium rosenbergii challenged in vivo with lipopolysaccharide [J]. Journal of Invertebrate Pathology, 2009, 100(1): 9—15
[18]Sun C, Xu W T, Zhang H W, et al. An anti-lipopolysaccharide factor from red swamp crayfish, Procambarus clarkii, exhibited antimicrobial activities in vitro and in vivo [J]. Fish and Shellfish Immunology, 2011, 30(1): 295—303
[19]Beale K M, Towle D W, Jayasundara N, et al. Anti-lipopolysaccharide factors in the American lobster Homarus americanus: Molecular characterization and transcriptional response to Vibrio fluvialis challenge [J]. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 2008, 3(4): 263—269
[20]Zhu L, Lan J F, Huang Y Q, et al. SpALF4: A newly identified anti-lipopolysaccharide factor from the mud crab Scylla paramamosain with broad spectrum antimicrobial activity [J]. Fish and Shellfish Immunology, 2014, 36(1): 172—180
[21]Imjongjirak C, Amparyup P, Tassanakajon A. Molecular cloning, genomic organization and antibacterial activity of a second isoform of antilipopolysaccharide factor (ALF) from the mud crab, Scylla paramamosain [J]. Fish and Shellfish Immunology, 2011, 30(1): 58—66
[22]Li C, Zhao J, Song L, et al. Molecular cloning, genomic organization and functional analysis of an anti-lipopolysaccharide factor from Chinese mitten crab Eriocheir sinensis [J]. Developmental and Comparative Immunology, 2008, 32(7): 784—794
[23]Yedery R, Reddy K. Identification, cloning, characterizationand recombinant expression of an anti-lipopolysaccharide factor from the hemocytes of Indian mud crab, Scylla serrata [J]. Fish and Shellfish Immunology, 2009, 27(2): 275—284
[24]Zhang Y, Wang L L, Wang L, et al. The second anti-lipopolysaccharide factor (EsALF-2) with antimicrobial activity from Eriocheir sinensis [J]. Developmental and Comparative-Immunology, 2010, 34(9): 945—952
[25]Tharntada S, Ponprateep S, Somboonwiwat K, et al. Role of anti-lipopolysaccharide factor from the black tiger shrimp, Penaeus monodon, in protection from white spot syndrome virus infection [J]. Journal of General Virology, 2009, 90(6): 1491—1498
[26]Zhao W H, Chen L Q, Qin J G, et al. MnHSP90 cDNA characterization and its expression during the ovary development in oriental river prawn, Macrobrachium nipponense [J]. Molecular Biology Reports, 2011, 38(2): 1399—406
[27]Xiu Y J, Wu T, Du J, et al. Molecular characterization and expression analysis of extracellular copper/zinc superoxide dismutase (ecCuZnSOD) from oriental river prawn, Macrobrachium nipponense [J]. Aquaculture, 2013, 380(4): 23—28
[28]de la Vega E, O’Leary N, Shockey J, et al. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): A broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection [J]. Molecular Immunology, 2008, 45(7): 1916—1925
[29]Somboonwiwat K, Bachere E, Rimphanitchayakit V, et al. Localization of anti-lipopolysaccharide factor (ALFPm3) in tissues of the black tiger shrimp, Penaeus monodon, and characterization of its binding properties [J]. Developmental and Comparative Immunology, 2008, 32(10): 1170—1176
[30]Yang Y S, Boze H, Chemardin P, et al. NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: model of the possible lipid A-binding site [J]. Biopolymers, 2009, 91(3): 207—220
[31]Supungul P, Klinbunga S, Pichyangkura R, et al. Antimicrobial peptides discovered in the black tiger shrimp Penaeus monodon using the EST approach [J]. Diseases of Aquatic Organisms, 2004, 61(1—2): 123—135
[32]Liu Y, Cui Z X, Li X H, et al. Molecular cloning, expression pattern and antimicrobial activity of a new isoform of antilipopolysaccharide factor from the swimming crab Portunus trituberculatus [J]. Fish and Shellfish Immunology, 2012, 33(1): 85—91
[33]Liu Y, Cui Z X, Li X H, et al. Molecular cloning, genomic structure and antimicrobial activity of PtALF7, a unique isoform of anti-lipopolysaccharide factor from the swimming crab Portunus trituberculatus [J]. Fish and Shellfish Immunology, 2013, 34(2): 652—659
[34]Liu Y, Cui Z X, Luan W S, et al. Three isoforms of anti-lipopolysaccharide factor identified from eyestalk cDNA library of swimming crab Portunus trituberculatus [J]. Fish and Shellfish Immunology, 2011, 30(2): 583—591
日本沼虾两种抗脂多糖因子的特性研究
王英慧1修云吉2,3顾 伟1孟庆国1王 文1
(1. 南京师范大学生命科学学院, 江苏省生物多样性与生物技术重点实验室, 江苏省水生甲壳动物病害重点实验室, 南京210023; 2. 中国水产科学研究院黄海水产研究所, 农业部海洋渔业可持续发展重点实验室, 青岛 266071; 3. 海洋科学与技术国家实验室,海洋渔业科学与食物产出过程功能实验室, 青岛 266200)
为了研究抗脂多糖因子ALFs在日本沼虾先天性免疫中的功能作用, 研究从日本沼虾中克隆了2种抗脂多糖因子MnALF1、MnALF2。MnALF1 cDNA 全长1008 bp, 编码121个氨基酸; MnALF2 cDNA 全长836 bp,编码124个氨基酸。这2种氨基酸均包含有一个信号肽序列和一个LPS结合位点, 并且在结合位点的两端(N-端和C-端)都有2个保守的半胱氨酸残基。这2种MnALFs与之前发现的甲壳动物的ALFs是非常相似的。qRTPCR结果显示MnALFs在所有被检测的组织中均有表达。其中MnALF1主要在心脏和小肠内表达, 而MnALF2则主要在血细胞和肝胰脏中表达。在用嗜水气单胞菌刺激之后发现2种MnALFs在心脏、小肠、血细胞、肝胰脏中都呈现出明显的时间依赖表达模式(MnALF1在刺激之后呈现出先减少后增加的趋势, 之后分别在不同组织的不同时间点达到最大值; 然而, 对于MnALF2, 在心脏和小肠中先减少后增加, 在血细胞和肝胰脏中呈现出先增加后减少, 最后都在24h达到最大值)。结果提示这2种MnALF具有不同的组织特异性, 并且在细菌侵染的免疫防御中起着重要的保护作用。
日本沼虾; 抗脂多糖因子; 表达分析; 嗜水气单胞菌
Q344+.1
A
1000-3207(2017)05-0977-07
10.7541/2017.122
date: 2016-06-08; Accepted date: 2016-11-12
Supported by the National Natural Science Foundation of China (31570176, 31602198); the Natural Science Foundation of Jiangsu Province (BK20151545); Project for Aquaculture in Jiangsu Province (D2015-13, Y2016-28)
Brief introduction of author: WANG Ying-Hui (1990—), Female, Heze, Shandong; Master; Research field: Disease of aquatic organism. E-mail: 1223775449@qq.com
WANG Wen (1957—), Female, Shihezi, Xinjiang; Doctor; Research field: Disease of aquatic organism. E-mail: wenwang@njnu.edu.cn
