Interaction between insulin-like growth factor binding protein-related protein 1 and transforming growth factor beta 1 in primary hepatic stellate cells
2017-08-16XiuQingLiQianQianZhangHaiYanZhangXiaoHongGuoHuiQinFanandLiXinLiu
Xiu-Qing Li, Qian-Qian Zhang, Hai-Yan Zhang, Xiao-Hong Guo, Hui-Qin Fan and Li-Xin Liu
Taiyuan, China
Interaction between insulin-like growth factor binding protein-related protein 1 and transforming growth factor beta 1 in primary hepatic stellate cells
Xiu-Qing Li, Qian-Qian Zhang, Hai-Yan Zhang, Xiao-Hong Guo, Hui-Qin Fan and Li-Xin Liu
Taiyuan, China
BACKGROUND:We previously showed that insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) is a novel mediator in liver fi brosis. Transforming growth factor beta 1 (TGFβ1) is known as the strongest effector of liver fibrosis. Therefore, we aimed to investigate the detailed interaction between IGFBPrP1 and TGFβ1 in primary hepatic stellate cells (HSCs).
METHODS:We overexpressed TGFβ1 or IGFBPrP1 and inhibited TGFβ1 expression in primary HSCs for 6, 12, 24, 48, 72, and 96 hours to investigate their interaction and observe the accompanying expressions of α-smooth muscle actin (α-SMA), collagen I, fi bronectin, and phosphorylated-mothers against decapentaplegic homolog 2/3 (p-Smad2/3).
RESULTS:We found that the adenovirus vector encoding the TGFβ1 gene (AdTGFβ1) induced IGFBPrP1 expression while that of α-SMA, collagen I, fi bronectin, and TGFβ1 increased gradually. Concomitantly, AdIGFBPrP1 upregulated TGFβ1, α-SMA, collagen I, fi bronectin, and p-Smad2/3 in a time-dependent manner while IGFBPrP1 expression was decreased at 96 hours. Inhibition of TGFβ1 expression reduced the IGFBPrP1-stimulated expression of α-SMA, collagen I, fi bronectin, and p-Smad2/3.
CONCLUSIONS:These fi ndings for the fi rst time suggest the existence of a possible mutually regulation between IGFBPrP1 and TGFβ1, which likely accelerates liver fi brosis progression. Furthermore, IGFBPrP1 likely participates in liver fi brosis in a TGFβ1-depedent manner, and may act as an upstream regulatory factor of TGFβ1 in the Smad pathway.
(Hepatobiliary Pancreat Dis Int 2017;16:395-404)
insulin-like growth factor binding proteinrelated protein 1; transforming growth factor β1; primary hepatic stellate cells; α-smooth muscle actin; extracellular matrix; Smad pathway
Introduction
The insulin-like growth factor binding proteinrelated protein 1 (IGFBPrP1), also known as insulin-like growth factor binding protein 7 (IGFBP7),[1]is a secreted protein.[2]IGFBPrP1 has distinct characteristics[3]and shows tumor suppressor functions by participating in cell proliferation, senescence, and apoptosis in numerous cancers.[4-6]IGFBPrP1 increases the epithelial-to-mesenchymal transition (EMT) phenotype of malignant mesenchymal and epithelial cells.[7]There is evidence that IGFBPrP1 stimulates α-smooth muscle actin (α-SMA) expression in human brain endothelial cell (HBEC).[8]In addition, IGFBPrP1 plays a rolein hepatic morphogenesis and shows the highest expression in the mid phase of the hepatic stellate cell (HSC) transdifferentiation process.[9,10]We previously found that IGFBPrP1 activated and transdifferentiated HSCsin vitro.[11]Moreover, IGFBPrP1 protein levels markedly increased in the fi brotic and cirrhotic human liver specimensin vivo.[12]
Liver fi brosis is a progressive pathological process characterized by remodeling of the extracellular matrix (ECM) and excessive deposition of collagen.[13]Multiple cytokines participate in HSC activation and liver fi brosis, and the most established mediator is transforming growth factor beta 1 (TGFβ1).[14]TGFβ1 acts as a major prof i brotic cytokine that potently promotes fi broblast recruitment, proliferation, differentiation into myof i broblasts and the production of ECM.[15]We previously also found that the expressions of both IGFBPrP1 and TGFβ1 were enhanced in mouse liver samples with thioacetamide (TAA)-induced fi brosis.[16]Moreover, IGFBPrP1 overexpression induced the excessive expressions of both ECM and TGFβ1 in the HSC-T6 cell line or mice.[12,17]However, the interaction between IGFBPrP1 and TGFβ1 and the contribution of IGFBPrP1 to ECM expression in HSCs is unknown.
IGFBPrP1 induces liver fi brosis through Smaddependent and independent pathways.[17]We observed that phosphorylated-mothers against decapentaplegic homolog 2/3 (p-Smad2/3) expression was upregulated by an adenoviral vector encoding the IGFBPrP1 gene (AdIGFBPrP1) in cultured HSC-T6 cells.[18]Currently, it is not clear whether IGFBPrP1 directly induces p-Smad2/3 expression, thereby inhibiting TGFβ1 expression in primary HSCs. The present study was to investigate the relationship between IGFBPrP1 and TGFβ1, and the effect of their interaction on liver fi brogenesis.
Methods
Primary cell isolation, culture, and identif i cation
The animals were obtained from Shanxi Medical University Laboratory Animal Center (Taiyuan, China). All the animal protocol procedures were approved by the Shanxi Medical University Animal Care and Use Committee (SCXK2009-0001). The healthy male Sprague-Dawley rats were anesthetized with chloral hydrate, their livers were sequentially perfusedin situusing a pronase E and type IV collagenase solution via the portal vein, and then the primary HSCs were isolated and purif i ed by Nycodenze density gradient centrifugation. The cell viability was determined by Trypan blue staining. The HSCs were seeded on plastic dishes in high glucose Dulbecco’s modif i ed Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and 1000 U/mL penicillin/streptomycin and incubated in a humidif i ed atmosphere containing 5% CO2at 37 ℃ for 48 hours. Then, the medium was replaced with 10% FBS-DMEM, which was changed every 48-72 hours. The antibodies to both desmin (TransGen Biotech, Beijing, China) and α-SMA (Abcam, Cambridge, UK) were used to identify HSCs.
Cell transfection
The HSCs were transfected with AdIGFBPrP1 or AdTGFβ1 (GenePharma Company, Shanghai, China), with enhanced green fl uorescent protein (EGFP) at a multiplicity of infection (MOI) of 10, 20, or 40 (number of virus/number of cells). Four short-hairpin RNAs (shRNAs) targeting the rat TGFβ1 mRNA (NM 021578) were designed and synthesized by Sangon Biotech Company (Shanghai, China). The most effective shTGFβ1 was used to construct the LvshTGFβ1 with red fl uorescent protein (RFP). The HSCs were transfected with the LvshTGFβ1 at different MOI (1, 10, or 100). The transfection eff i cacy was assessed by detecting the number of EGFP- or RFP-positive cells. The optimized MOI was used in subsequent experiments.
Quantitative real-time polymerase chain reaction (qPCR) assays
Total RNA was extracted from the HSCs using a Qiagen Rneasy Mini kit (Qiagen, Dusseldorf, Germany) following the manufacturer’s instructions. Then, 1 μg of total RNA was reverse transcribed by using the PrimeScript RT Master Mix (TransGen Biotech, Beijing, China). The quantitative real-time polymerase chain reaction (qPCR) was performed with a Step One PCR System using SYBR Green PCR kit (Invitrogen, Carlsbad, CA, USA). The primer sequences were as follows; TGFβ1: 5’-ATT CCT GGC GTT ACC TTG G-3’ (forward) and 5’-AGC CCT GTA TTC CGT CTC CT-3’ (reverse); IGFBPrP1: 5’-TCA CCC AGG TCA GCA AAG-3’ (forward) and 5’-TCA CCA GGC AAG AGT TCT GT-3’ (reverse); and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH): 5’-ACA GCA ACA GGG TGG TGG AC-3’ (forward) and 5’-TTT GAG GGT GCA GCG AAC TT-3’ (reverse). The PCR cycle used the following program: 95 ℃ for 10 minutes, followed by 40 cycles at 95 ℃ for 10 seconds, and 60 ℃ for 30 seconds.
Immunocytochemical staining
The HSCs were cultured on glass coverslips in a 6-well plate and incubated with the various treatments. The cells were washed with phosphate-buffered saline (PBS), fi xed in 4% paraformaldehyde for 30 minutes at 4 ℃,permeabilized with 0.5% Triton X-100 at 22 ℃ for 20 minutes. Then, the cells were incubated in 3% hydrogen peroxide (H2O2) for 15 minutes, blocked with 5% bovine serum albumin (BSA) for 20 minutes, and subsequently incubated with a rabbit polyclonal anti-IGFBPrP1 or TGFβ1 antibody [(1:400) Abcam, Cambridge, UK] overnight at 4 ℃. In a parallel experiment, cells were incubated with PBS alone without the primary antibody as a negative control. After incubation, the cells were washed with PBS, incubated with the secondary antibody at 37 ℃ for 30 minutes,followed by incubation with 3, 3-diaminobenzidine (DAB) solution for 5-8 minutes, and the nuclei were stained with hematoxylin. The positive staining was visualized under a microscope.
Western blotting analysis
The total protein from the HSCs was obtained using a kit according to the protocol provided by the manufacturer. Equal amounts of proteins were separated on sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene dif l uoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were incubated with primary antibodies against IGFBPrP1, TGFβ1, α-SMA, collagen I, fi bronectin, p-Smad2/3 (Abcam, Cambridge, UK), Smad2/3 (Santa Cruz Biotechnology, CA, USA), or β-actin (TransGen Biotech, Beijing, China), followed by the horseradish peroxidase (HRP)-conjugated secondary antibodies. The immunoblots were detected with the super enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech, NJ, USA).
Enzyme-linked immunosorbent assay (ELISA)
The TGFβ1 levels of the culture supernatant were detected with a TGFβ1 (rat) ELISA kit (YiHan Biology, Shanghai, China) following the manufacturer’s protocol. Brief l y, all the supernatant samples were centrifuged at 1000 rpm for 10 minutes to discard the particulates, and then the absorbance was measured at 450 nm with a microplate reader.
Statistical analysis
The statistical analysis was performed using the statistical package for the social sciences (SPSS) version 16.0 software. All the results were presented as the mean± standard deviation (SD). An analysis of variance (ANOVA) and SNK-q test were used to compare the different treatment groups, and aP≤0.05 was considered statistically signif i cant.
Results
Identif i cation of HSCs
The harvest rate of the cultured HSCs was 2.5 to 3×107per rat with a cell viability of more than 90%. The freshly isolated HSCs from the Sprague-Dawley rats were nonadherent and round (Fig. 1A). The fl uorescence induced by the retinol esters was observed by using an inverted fl uorescence microscope (Fig. 1B) and after 72 hours, the cells were adherent and stretched into a heterogeneous morphology (Fig. 1C). Most of the HSCs showed myof ibroblast-like morphology after 7 days (Fig. 1D). Desminwas used to identify the HSCs while α-SMA was detected in the activated HSCs, which were analyzed by using immunocytochemical staining. As shown in Fig. 1E and F, the positive cells were over 95%, which indicated that the purity of the isolated HSCs exceeded 95%.
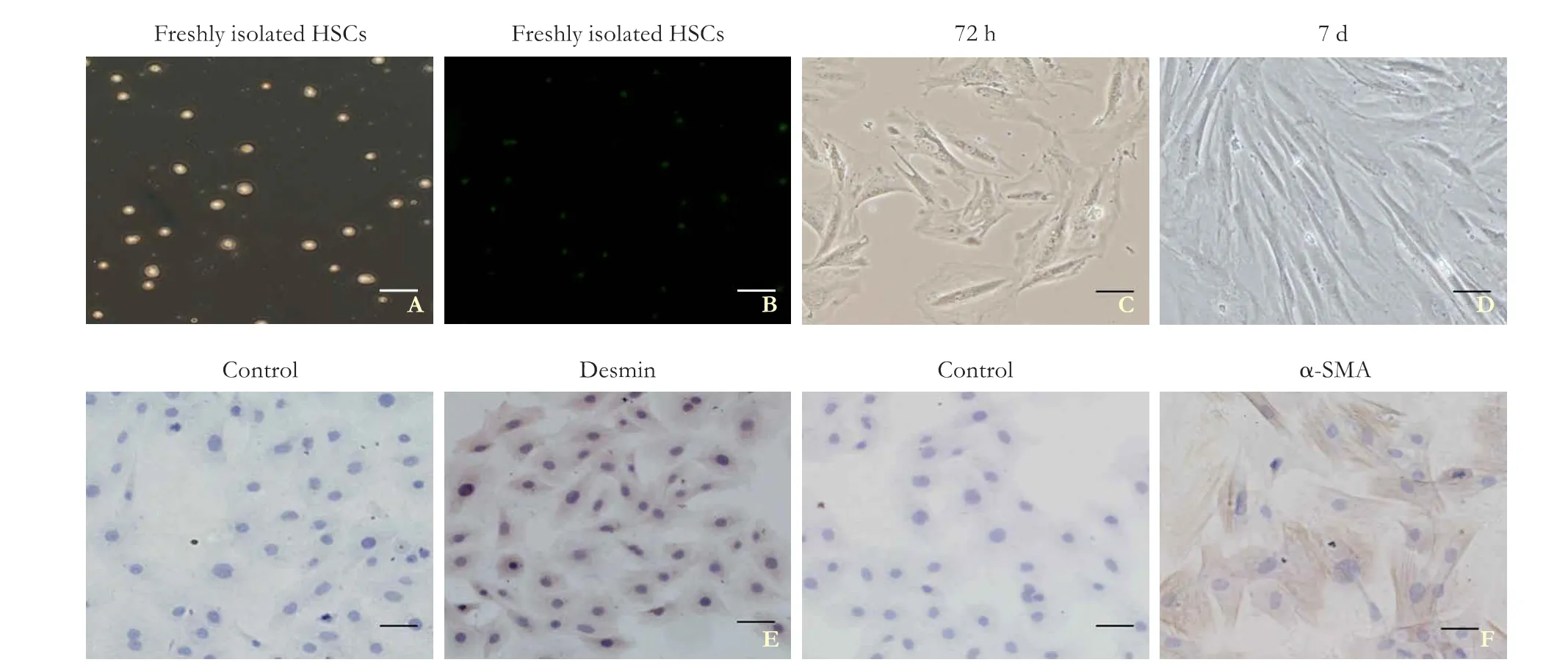
Fig. 1. Representative micrographs of primary rat hepatic stellate cells (HSCs) at different stages. Freshly isolated HSCs were visualized using light (A) and fl uorescence microscopy (B). Primary HSCs were cultured for 72 hours (C) and 7 days (D). Expression of desmin (E) and α-smooth muscle actin (SMA) (F) in HSCs was analyzed using immunocytochemical staining; scale bar, 50 μm.
Optimization of MOI with AdTGFβ1, AdIGFBPrP1, or LvshTGFβ1 in HSCs
After a 48-hour transfection with AdIGFBPrP1 or AdTGFβ1, the HSCs showed a gradual increase in MOI of 10-40, observed using a confocal laser scanning microscope (Fig. 2A-D). A similar increase in fl uorescence intensity was also observed when the concentration of LvshTGFβ1 was increased from MOI 1 to 100 (Fig. 2E and F), and, therefore, MOI values of 40 and 100 were used for the subsequent experiments.
Effect of TGFβ1 overexpression on IGFBPrP1, α-SMA, collagen I, and fi bronectin expressions in primary HSCs
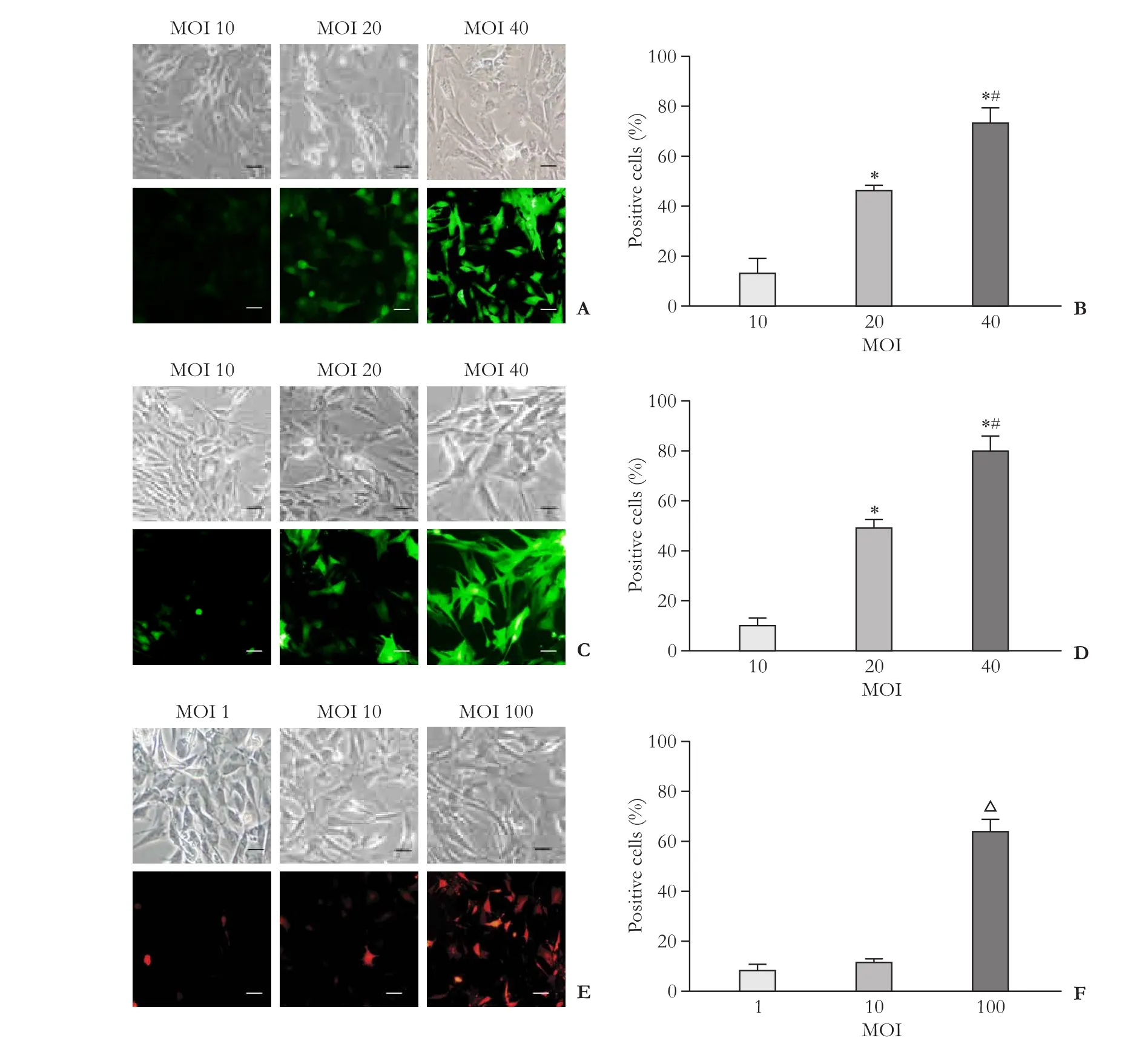
Fig. 2. Optimization of multiplicity of infection (MOI) using adenoviral vectors coding transforming growth factor beta 1 (TGFβ1) or insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) genes (AdTGFβ1 and AdIGFBPrP1, respectively) or lentiviral vectors containing small interfering RNA (siRNA) targeting TGFβ1 (LvshTGFβ1) in primary rat hepatic stellate cells (HSCs). Enhanced green fl uorescent protein (EGFP) or red fl uorescent protein (RFP) expression was detected using confocal microscopy in HSCs after 48-hour transfection. A and B: AdTGFβ1 transfection and eff i cacy; C and D: AdIGFBPrP1 transfection and eff i cacy. *:P<0.05, vs MOI 10 and #:P<0.05, vs MOI 20. E and F: LvshTGFβ1 transfection and eff i cacy. △:P<0.05, vs MOI 1 and 10. Data are presented as mean± SD (n=3 per group); scale bar, 50 μm.
After the HSCs were treated with AdTGFβ1, the TGFβ1 protein level gradually increased from 6 to 96 hours, which showed that the AdTGFβ1 transfection was effective. As shown in Fig. 3A, the IGFBPrP1 mRNA levels of the treated group were increased at 6, 12, and 24 hours compared with that of the control group, which peakedat 12 hours. Immunocytochemical staining and western blotting analysis showed that the increase in the IGFBPrP1 protein was observed at 12, 24, and 48 hours, but peaked at 24 hours. There were no changes in the normal, adenoviral vectors carrying no cDNA (CAd), 6, 72, and 96 hours groups (Fig. 3B and C). The α-SMA and ECM (collagen I and fi bronectin) commenced expression at 6 and 12 hours, respectively, and then both gradually increased (Fig. 3D-F).
Effect of IGFBPrP1 overexpression on expressions of TGFβ1, α-SMA, collagen I, fi bronectin, p-Smad2/3, and Smad2/3 in primary HSCs
Following treatment with AdIGFBPrP1, the HSCs showed an increase in IGFBPrP1 protein at 6 hours, which peaked at 72 hours, indicating the eff i ciency of the AdIGFBPrP1 transfection. As shown in Fig. 4A, the TGFβ1 mRNA gradually increased from 12 to 96 hours, and then peaked at 96 hours. No changes in the TGFβ1 mRNA were observed in both the CAd and 6 hours groups in compared to the normal group. The immunocytochemical staining and western blotting analysis showed increases in TGFβ1 protein at 24, 48, 72, and 96 hours, at which the value peaked. There was no signif i cant increase in the normal, CAd, 6 hours, and 12 hours groups (Fig. 4B and C). The ELISA revealed that the TGFβ1 levels in the supernatant gradually increasedfrom 48 to 96 hours while no changes where observed in the normal and CAd, as well as the 6, 12, and 24 hours groups (Fig. 4D). The western blotting analysis showed that the α-SMA expression was gradually upregulated from 6 to 96 hours (Fig. 4E). In addition, the expressions of collagen I, fi bronectin, and p-Smad2/3, and the ratioof p-Smad2/3 to Smad2/3 were markedly upregulated from 24 to 96 hours. There was no signif i cant increase in the normal, CAd, 6-hour, and 12-hour groups while the expression of the Smad2/3 protein was not signif i cantly different in all the groups (Fig. 4F-I).
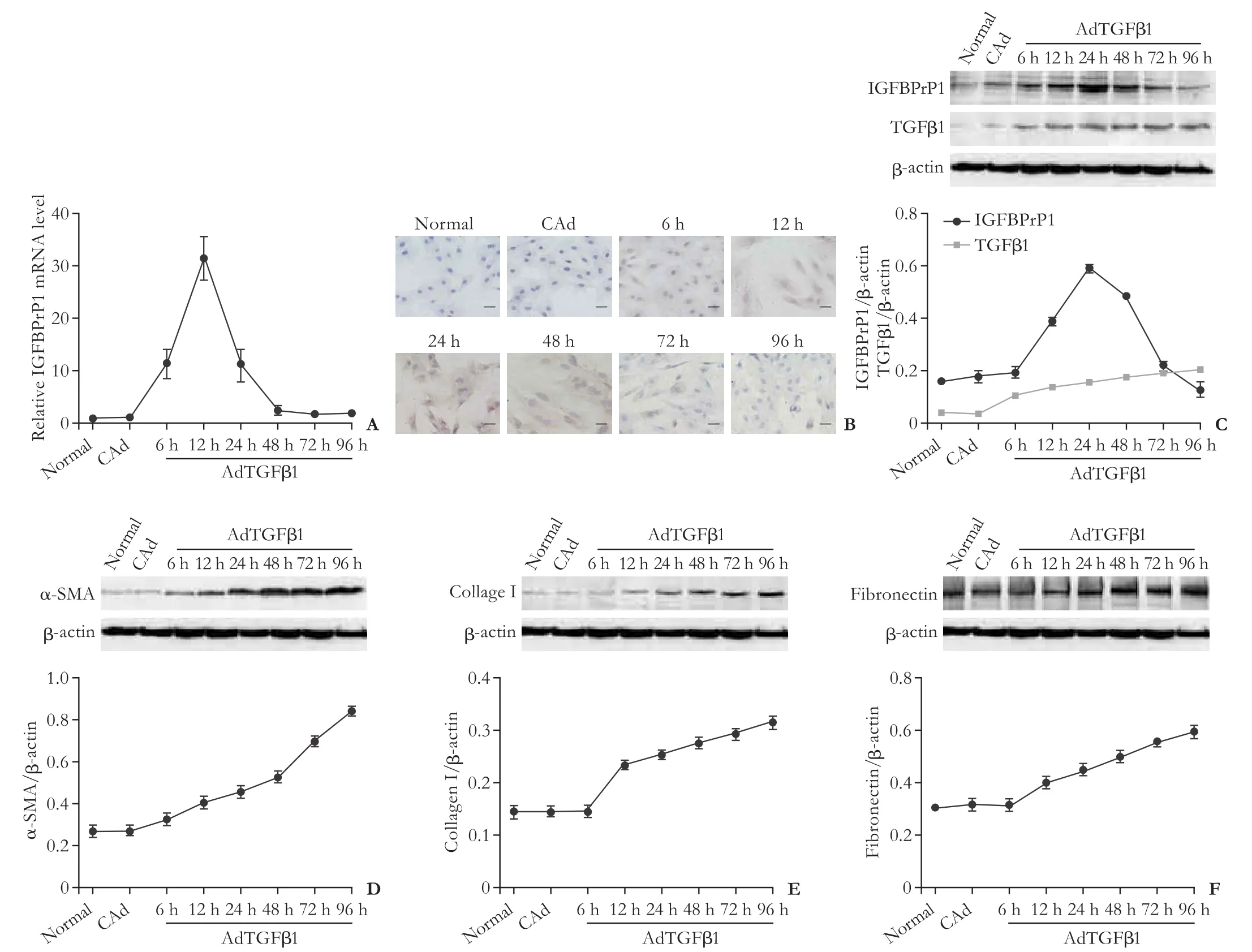
Fig. 3. Effect of adenoviral vectors coding transforming growth factor beta 1 (TGFβ1) gene (AdTGFβ1) on TGFβ1, insulin-like growth factor binding protein-related protein 1 (IGFBPrP1), α-smooth muscle actin (SMA), collagen I, and fi bronectin expressions in primary rat hepatic stellate cells (HSCs). IGFBPrP1 mRNA and protein, and TGFβ1 protein in HSCs were analyzed using qPCR (A), immunocytochemical staining (B) (scale bar, 50 μm), and Western blotting analyses (C) after AdTGFβ1 treatment for 6, 12, 24, 48, 72, and 96 hours. Protein expression of α-smooth muscle actin (SMA) (D), collagen I (E), and fi bronectin (F) were examined by Western blotting after AdTGFβ1 treatment for 6, 12, 24, 48, 72, and 96 hours. β-actin and adenoviral vectors carrying no cDNA (CAd) acted as internal and negative controls, respectively. Data are presented as mean±SD (n=3 per group).
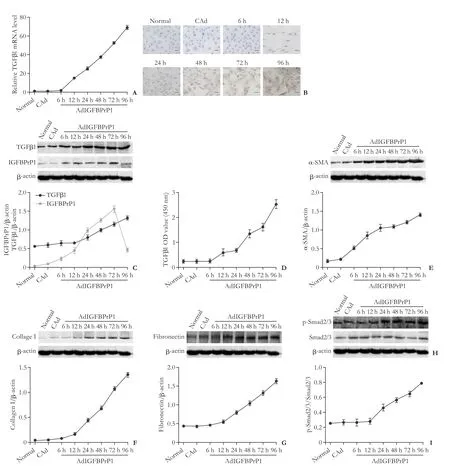
Fig. 4. Effect of adenoviral vectors encoding insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) gene (AdIGFBPrP1) on expressions of IGFBPrP1, transforming growth factor beta 1 (TGFβ1), α-smooth muscle actin (SMA), collagen I, fi bronectin, phosphorylated-mothers against decapentaplegic homolog 2/3 (p-Smad2/3), and Smad2/3 in primary rat hepatic stellate cells (HSCs). A: qPCR for TGFβ1 mRNA; B: Immunocytochemical staining for TGFβ1, scale bar, 50 μm; C: Western blotting analysis for TGFβ1 and IGFBPrP1. β-actin served as an internal control; D: Enzyme-linked immunosorbent assay (ELISA) for TGFβ1. Western blotting for α-SMA (E), collagen I (F), fi bronectin (G) and p-Smad2/3 and Smad2/3 (H), β-actin served as an internal control. I: p-Smad2/3 and Smad2/3 ratio. Adenoviral vectors carrying no cDNA (CAd) acted as a negative control. Data are presented as mean±SD (n=3 per group).
Inhibition of TGFβ1 expression reduces expressions of α-SMA, collagen I, fi bronectin, and p-Smad2/3 in IGFBPrP1-treated HSCs
After the HSCs were treated with the LvshTGFβ1, the TGFβ1 protein level gradually decreased from 6 to 96 hours, which showed that the LvshTGFβ1 transfection was effective (Fig. 5A). As shown in Fig. 5B, the expression of α-SMA increased at 6 and 12 hours, and then gradually decreased from 24 to 96 hours. The expressions of collagen I, fi bronectin, p-Smad2/3, and the ratio of p-Smad2/3 to Smad2/3 were gradually downregulated from 24 to 96 hours while there were no changes in the CAd plus lentiviral vectors carrying negative shRNA (LvshNC), 6 hours, and 12 hours groups compared with the normal group. The expression of Smad2/3 protein did not differ signif icantly between all the groups (Fig. 5C-F).
Discussion
To our knowledge, this is the fi rst study to investigate the detailed interaction between IGFBPrP1 and TGFβ1 in primary HSCs. HSC activation is an essential event in the process of liver fi brosis.[19]During hepatic injury, quiescent HSCs undergo profound phenotypic changes including enhanced cell proliferation, loss of lipid droplets, expression of α-SMA, and excessive deposition of ECM, which all constitute HSC activation.[20]Desmin protein is expressed in quiescent HSCs and is maintained during their activation while α-SMA is a marker of HSC activation. Our immunocytochemical staining clearly showed that both desmin and α-SMA were positive in HSCs, demonstrating that the primary HSCs were successfully isolated and cultured. Because the replicationdef i cient recombinant adenovirus and lentivirus show a highly eff i cient transfection into target cells,[21]we used their vectors to establish gene overexpression and silencing, respectively in HSCsin vitro. The adenovirus and lentivirus show different transfection eff i cacy in different cells. In our experiments, the optimization of the MOI with a high transfection eff i cacy and low impairment of cell viability was conf i rmed and used for the subsequent experiments.
IGFBPrP1 expression was induced by TGFβ1 in HBECs,[22]human fi broblasts,[23]and a colorectal cancer cell line (SW620)[24]in a time-dependent manner. Our experiments showed that IGFBPrP1 expression initially increased and then gradually decreased in AdTGFβ1-treated HSCs. This was not consistent with previous reports and, therefore, further studies are required to clarify these differences. However, in contrast to IGFBPrP1, the expression of TGFβ1 protein gradually increased in a time-dependent manner in HSCs treated with AdTGFβ1. These results indicate that TGFβ1 upregulated IGFBPrP1 expression not only in other cell lines but also in the primary HSCs. Particularly, the level of IGFBPrP1 protein was not consistent with the TGFβ1 expression. Activated HSCs are the main source of IGFBPrP1 in the liver.[10]We also observed that α-SMA was expressed before IGFBPrP1. Based on these fi ndings, we propose that TGFβ1 activates HSCs, which induces IGFBPrP1 in the early phase. Furthermore, IGFBPrP1 is likely degraded in the later stage of HSCs activation.
ECM components change from normal basement matrix components such as type IV collagen to a fi brotic matrix, which includes type I collagen[25]and fi bronectin in liver fi brosis. It is well known that TGFβ1 strongly stimulates HSC to express α-SMA and ECM proteins. Our results also showed that the expressions of α-SMA, collagen I, and fi bronectin increased gradually in a timedependent manner in the primary HSCs treated with AdTGFβ1, which was consistent with TGFβ1 expression but differed from that of IGFBPrP1. Based on the above data, we concluded that the effects of TGFβ1 on both HSC activation and ECM production were persistent and predominant. The changes in both α-SMA and ECM were independent of IGFBPrP1 concentration in the later phase in primary HSCs.
In addition, we used the AdIGFBPrP1 to observe TGFβ1 expression in primary HSCs. We previously reported that the expression of TGFβ1 was gradually increased even after the IGFBPrP1 protein level decreased in the liver of rats treated by AdIGFBPrP1in vivo.[26]In this study, we found that the mRNA and protein level of TGFβ1 gradually increased in a time-dependent manner, suggesting that IGFBPrP1 stimulated TGFβ1 production in the HSCsin vitro. However, IGFBPrP1 protein expression increased before 72 hours and then obviously decreased at 96 hours. This was consistent with previous report.[26]We also observed that the expressions of α-SMA, collagen I, and fi bronectin gradually increased in a timedependent manner, which suggested that they were upregulated following the increase in TGFβ1 expression in HSCs treated with AdIGFBPrP1. Moreover, we found that the HSCs were activated before TGFβ1 expression while the protein expression trends of both collagen I and fi bronectin were similar to that of TGFβ1. HSCs are an important source of cytokines, and following their activation they express a number of prof i brogenic mediators including TGFβ1 and different cytokine receptors including the TGFβ1 receptor.[27]Moreover, HSCs are widely regarded as key fi brogenic cells that co-ordinate hepatic ECM formation.[28]TGFβ1 is a powerful activator of HSCs[29]and potent inducer of type I collagen.[30]Komiya et al[23]reported that endogenous TGFβ1 in fibroblasts might be involved in the induction of α-SMA and fi bronectin by IGFBPrP1, which suggests that the expressions of α-SMA and fi bronectin were induced by IGFBPrP1 in a TGFβ1-depedent manner. Considering the above results, we suggest that IGFBPrP1 may activate HSCs in the early phase, leading to the secretion of TGFβ1 and successive activation of the HSCs in the later stage. This progression was independent of IGFBPrP1. Additionally, TGFβ1 induced the expressions of both collagen I and fi bronectin in the HSCs. This positive feedback mechanism between TGFβ1 and HSCs possibly accounts for the excessive and persistent expressions of TGFβ1, α-SMA, collagen I, and fi bronectin when the IGFBPrP1 protein decreased. Considering the above data, we suggest that IGFBPrP1 likely acts as a critical factor in the activation of HSCs, which induces TGFβ1 expression, thereby causing ECM deposition in the HSCs.
Furthermore, we used LvshTGFβ1 to inhibit TGFβ1expression in primary HSCs with the overexpression of IGFBPrP1. The TGFβ1 receptor inhibitor SB431542 inhibited the expressions of α-SMA and fi bronectin induced by IGFBPrP1 in fi broblasts.[23]Our results showed that the inhibition of TGFβ1 expression obviously attenuated the IGFBPrP1-stimulated induction of α-SMA, collagen I, and fi bronectin at the late stage, and had no obvious effect on the early phase of α-SMA expression. These results demonstrated that IGFBPrP1 suppressed both α-SMA and ECM expressions in the HSCs in the absence of TGFβ1, which was consistent with the report on fi broblasts. These data suggest that TGFβ1 possibly performs an important function in IGFBPrP1-induced HSCs activation and ECM production at the later stage.
Finally, we also observed that the overexpression of IGFBPrP1 gradually upregulated p-Smad2/3 expression in a time-dependent manner in the primary HSCs. The expression trend of the p-Smad2/3 was similar to that of TGFβ1, which was consistent with another report on HSCs.[31]The expression of p-Smad2/3 was signif i cantly decreased by LvshTGFβ1 in the AdIGFBPrP1-treated group of HSCs. Our early study showed the Smad2/3 pathway was activated by IGFBPrP1 in HSCs bothin vivoandin vitro.[18]Recent studies by other groups showed that IGFBP7 might act as a downstream effector of TGFβ1/Smad pathway.[8]Our data showed that the inhibition of TGFβ1 expression possibly affected the IGFBPrP1-induced Smad pathway. Taken together, these results indicate that IGFBPrP1 likely serves as an upstream factor of TGFβ1 in the activation of the Smad pathway in HSCs. This was not in keeping with the previous report and, therefore, further studies are needed to investigate this difference.
The present study has a limitation. Our investigation was performed onlyin vitro. Therefore, we were unable to identify the specif i c interaction between IGFBPrP1 and TGFβ1in vivo. In conclusion, this study showed that there was a potentially mutual regulation between IGFBPrP1 and TGFβ1, which activates HSC and ECM deposition. IGFBPrP1 may act as a potential therapeutic target for liver fi brosis treatment. Further investigations are needed to elucidate thein vivomechanisms of the IGFBPrP1-induced liver fi brosis.
Acknowledgement: We thank Zhang-Feng Dou for assisting with the primary cell isolation.
Contributors: LLX designed the experiments. LXQ analyzed the data and wrote the paper. ZQQ, ZHY, GXH and FHQ performed the experiments. All authors contributed to the interpretation of the study and to further drafts. LLX is the guarantor.
Funding: This work was supported by a grant from the Shanxi Province Foundation for Returness (2012-4).
Ethical approval: All institutional and national guidelines for the care and use of laboratory animals were followed. The study strictly conformed to the ethical rules of standard experimental animal studies, and was approved by the Ethics Committee of Shanxi Medical University (SCXK2009-0001).
Competing interest: No benef i ts in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factorbinding protein (IGFBP) superfamily. Endocr Rev 1999;20:761-787.
2 Chen D, Yoo BK, Santhekadur PK, Gredler R, Bhutia SK, Das SK, et al. Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin Cancer Res 2011;17:6693-6701.
3 Rosenfeld RG, Hwa V, Wilson L, Lopez-Bermejo A, Buckway C, Burren C, et al. The insulin-like growth factor binding protein superfamily: new perspectives. Pediatrics 1999;104:1018-1021.
4 Vizioli MG, Possik PA, Tarantino E, Meissl K, Borrello MG, Miranda C, et al. Evidence of oncogene-induced senescence in thyroid carcinogenesis. Endocr Relat Cancer 2011;18:743-757.
5 Sullivan L, Murphy TM, Barrett C, Loftus B, Thornhill J, Lawler M, et al. IGFBP7 promoter methylation and gene expression analysis in prostate cancer. J Urol 2012;188:1354-1360.
6 Benatar T, Yang W, Amemiya Y, Evdokimova V, Kahn H, Holloway C, et al. IGFBP7 reduces breast tumor growth by induction of senescence and apoptosis pathways. Breast Cancer Res Treat 2012;133:563-573.
7 Rupp C, Scherzer M, Rudisch A, Unger C, Haslinger C, Schweifer N, et al. IGFBP7, a novel tumor stroma marker, with growth-promoting effects in colon cancer through a paracrine tumor-stroma interaction. Oncogene 2015;34:815-825.
8 Pen A, Durocher Y, Slinn J, Rukhlova M, Charlebois C, Stanimirovic DB, et al. Insulin-like growth factor binding protein 7 exhibits tumor suppressive and vessel stabilization properties in U87MG and T98G glioblastoma cell lines. Cancer Biol Ther 2011;12:634-646.
9 Chen L, Goryachev A, Sun J, Kim P, Zhang H, Phillips MJ, et al. Altered expression of genes involved in hepatic morphogenesis and fi brogenesis are identif i ed by cDNA microarray analysis in biliary atresia. Hepatology 2003;38:567-576.
10 Boers W, Aarrass S, Linthorst C, Pinzani M, Elferink RO, Bosma P. Transcriptional prof i ling reveals novel markers of liver fi brogenesis: gremlin and insulin-like growth factor-binding proteins. J Biol Chem 2006;281:16289-16295.
11 Liu LX, Huang S, Zhang QQ, Liu Y, Zhang DM, Guo XH, et al. Insulin-like growth factor binding protein-7 induces activation and transdifferentiation of hepatic stellate cells in vitro. World J Gastroenterol 2009;15:3246-3253.
12 Guo XH, Liu LX, Zhang HY, Zhang QQ, Li Y, Tian XX, et al. Insulin-like growth factor binding protein-related protein 1 contributes to hepatic fi brogenesis. J Dig Dis 2014;15:202-210.
13 Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I, et al. TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J 2011;30:783-795.
14 Bataller R, Brenner DA. Liver fi brosis. J Clin Invest 2005;115:209-218.
15 Sheppard D. Transforming growth factor beta: a central modulator of pulmonary and airway inf l ammation and fi brosis. Proc Am Thorac Soc 2006;3:413-417.
16 Liu LX, Zhang HY, Zhang QQ, Guo XH. Effects of insulin-like growth factor binding protein-related protein 1 in mice with hepatic fi brosis induced by thioacetamide. Chin Med J (Engl) 2010;123:2521-2526.
17 Guo Y, Zhang Y, Zhang Q, Guo X, Zhang H, Zheng G, et al. Insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) contributes to liver inf l ammation and fi brosis via activation of the ERK1/2 pathway. Hepatol Int 2015;9:130-141.
18 Zhang Y, Zhang QQ, Guo XH, Zhang HY, Liu LX. IGFBPrP1 induces liver fi brosis by inducing hepatic stellate cell activation and hepatocyte apoptosis via Smad2/3 signaling. World J Gastroenterol 2014;20:6523-6533.
19 Tao LL, Zhai YZ, Ding D, Yin WH, Liu XP, Yu GY. The role of C/EBP-α expression in human liver and liver fi brosis and its relationship with autophagy. Int J Clin Exp Pathol 2015;8:13102-13175.
20 Lin J, Zheng S, Chen A. Curcumin attenuates the effects of insulin on stimulating hepatic stellate cell activation by interrupting insulin signaling and attenuating oxidative stress. Lab Invest 2009;89:1397-1409.
21 Nakatani T, Kuriyama S, Tominaga K, Tsujimoto T, Mitoro A, Yamazaki M, et al. Assessment of eff i ciency and safety of adenovirus mediated gene transfer into normal and damaged murine livers. Gut 2000;47:563-570.
22 Pen A, Moreno MJ, Durocher Y, Deb-Rinker P, Stanimirovic DB. Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene 2008;27:6834-6844.
23 Komiya E, Furuya M, Watanabe N, Miyagi Y, Higashi S, Miyazaki K. Elevated expression of angiomodulin (AGM/IGFBP-rP1) in tumor stroma and its roles in fi broblast activation. Cancer Sci 2012;103:691-699.
24 Rao C, Lin SL, Ruan WJ, Wen H, Wu DJ, Deng H. High expression of IGFBP7 in fi broblasts induced by colorectal cancer cells is co-regulated by TGF-β and Wnt signaling in a Smad2/3-Dvl2/3-dependent manner. PLoS One 2014;9:e85340.
25 Lakner AM, Moore CC, Gulledge AA, Schrum LW. Daily genetic prof i ling indicates JAK/STAT signaling promotes early hepatic stellate cell transdifferentiation. World J Gastroenterol 2010;16:5047-5056.
26 Guo X, Zhang H, Zhang Q, Li X, Liu L. Screening for and validation of a hepatic fi brosis-related pathway induced by insulin-like growth factor-binding protein-related protein 1. Eur J Gastroenterol Hepatol 2016;28:762-772.
27 Duval F, Moreno-Cuevas JE, González-Garza MT, Rodríguez-Montalvo C, Cruz-Vega DE. Protective mechanisms of medicinal plants targeting hepatic stellate cell activation and extracellular matrix deposition in liver fi brosis. Chin Med 2014;9:27.
28 Lewindon PJ, Pereira TN, Hoskins AC, Bridle KR, Williamson RM, Shepherd RW, et al. The role of hepatic stellate cells and transforming growth factor-beta(1) in cystic fi brosis liver disease. Am J Pathol 2002;160:1705-1715.
29 Yao QY, Xu BL, Wang JY, Liu HC, Zhang SC, Tu CT. Inhibition by curcumin of multiple sites of the transforming growth factor-beta1 signalling pathway ameliorates the progression of liver fi brosis induced by carbon tetrachloride in rats. BMC Complement Altern Med 2012;12:156.
30 Chen A. Acetaldehyde stimulates the activation of latent transforming growth factor-beta1 and induces expression of the type II receptor of the cytokine in rat cultured hepatic stellate cells. Biochem J 2002;368:683-693.
31 Liang TJ, Yuan JH, Tan YR, Ren WH, Han GQ, Zhang J, et al. Effect of ursodeoxycholic acid on TGF beta1/Smad signaling pathway in rat hepatic stellate cells. Chin Med J (Engl) 2009;122:1209-1213.
June 20, 2016
Accepted after revision December 30, 2016
Author Aff i liations: Department of Gastroenterology and Hepatology (Li XQ, Zhang QQ, Zhang HY, Guo XH, Fan HQ and Liu LX), Experimental Center of Science and Research (Zhang QQ, Zhang HY, Guo XH, Fan HQ and Liu LX), The First Clinical Hospital of Shanxi Medical University; and Key Laboratory of Cell Physiology, Provincial Department of the Ministry of Education, Shanxi Medical University (Zhang QQ, Zhang HY, Guo XH, Fan HQ and Liu LX), Taiyuan 030001, China
Li-Xin Liu, MD, PhD, Department of Gastroenterology and Hepatology and Experimental Center of Science and Research, The First Clinical Hospital of Shanxi Medical University; and Key Laboratory of Cell Physiology, Provincial Department of the Ministry of Education, Shanxi Medical University, 85 Jiefang South Road, Taiyuan 030001, China (Tel: +86-351-4639075; Fax: +86-351-8263169; Email:lixinliu6@hotmail.com)
© 2017, Hepatobiliary Pancreat Dis Int. All rights reserved.
10.1016/S1499-3872(17)60013-4
Published online April 24, 2017.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Predictive value of C-reactive protein/albumin ratio in acute pancreatitis
- IgG4-related sclerosing cholangitis and chronic sclerosing sialadenitis mimicking cholangiocarcinoma and neck malignancy
- The International Study Group of Pancreatic Surgery def i nition of delayed gastric emptying and the effects of various surgical modif i cations on the occurrence of delayed gastric emptying after pancreatoduodenectomy
- Hepatopancreatoduodenectomy for advanced hepatobiliary malignancies: a single-center experience
- Bilioenteric anastomotic stricture in patients with benign and malignant tumors: prevalence, risk factors and treatment
- Effects of multimodal fast-track surgery on liver transplantation outcomes
