ARTICLE In situ Detection of Amide A Bands of Proteins in Water by Raman Ratio Spectrum†
2016-04-08ChengqinTngKeLinXioguoZhouShilinLiuHefeiNtionlLortoryforPhysiclSciencestheMicroscleDeprtmentofChemiclPhysicsUniversityofSciencendTechnologyofChinHefei230026ChinSchoolofPhysicsndOptoelectronicEngineeringXidinUniversi
Cheng-qin Tng,Ke Lin∗,Xio-guo Zhou,c,Shi-lin Liu∗.Hefei Ntionl Lortory for Physicl Sciences t the Microscle,Deprtment of Chemicl Physics, University of Science nd Technology of Chin,Hefei 230026,Chin.School of Physics nd Optoelectronic Engineering,Xidin University,Xi’n 710071,Chinc.Synergetic Innovtion Center of Quntum Informtion&Quntum Physics,University of Science nd Technology of Chin,Hefei 230026,Chin(Dted:Received on Novemer 30,2015;Accepted on Jnury 27,2016)
ARTICLE In situ Detection of Amide A Bands of Proteins in Water by Raman Ratio Spectrum†
Cheng-qian Tanga,Ke Linb∗,Xiao-guo Zhoua,c,Shi-lin Liua∗
a.Hefei National Laboratory for Physical Sciences at the Microscale,Department of Chemical Physics, University of Science and Technology of China,Hefei 230026,China
b.School of Physics and Optoelectronic Engineering,Xidian University,Xi’an 710071,China
c.Synergetic Innovation Center of Quantum Information&Quantum Physics,University of Science and Technology of China,Hefei 230026,China
(Dated:Received on November 30,2015;Accepted on January 27,2016)
The amide A band of protein is sensitive to the hydrogen bands of amide groups of proteins. However,it is hard to distinguish the amide A band of aqueous protein in situ directly, since it overlaps with O−H stretching vibration of water.In this work,we presented a new analytical method of Raman ratio spectrum,which can extract the amide A band of proteins in water.To obtain the Raman ratio spectrum,the Raman spectrum of aqueous protein was divided by that of pure water.A mathematical simulation was employed to examine whether Raman ratio spectrum is e ff ective.Two kinds of protein,lysozyme and α-chymotrypsin were employed.The amide A bands of them in water were extracted from Raman ratio spectra. Additionally,the process of thermal denaturation of lysozyme was detected from Raman ratio spectrum.These results demonstrated the Raman ratio spectra could be employed to study the amide A modes of proteins in water.
Key words:Raman ratio spectrum,Amide A band,In situ,Protein,Water
†Part of the special issue for“the Chinese Chemical Society’s 14th National Chemical Dynamics Symposium”.
∗Authors to whom correspondence should be addressed.E-mail: klin@xidian.edu.cn,Klin@ustc.edu.cn,slliu@ustc.edu.cn
I.INTRODUCTION
Protein molecules are important to life activities.For example,enzyme could accelerate the chemical reactions in body,hemoglobin could transport oxygen in red blood cells,insulin could regulate the metabolism of fats and carbohydrates,and inhibit the production of glucose in the liver.The functions of proteins are determined by their structures.The structures have been studied by plenty of technologies.One of the popular technologies is the vibrational spectroscopy.The vibrational spectra of proteins in amide region could identify their secondary structure.In the amide I spectral region,the vibrational band locates in 1650−1660 and 1670−1680 cm−1for α-helix and β-sheet structures of proteins,respectively[1–4].The amide I bands of different secondary structure overlap seriously with each other.Thus,the overlapping amide I band locates in a higher wavenumber for the protein with more β-sheet structure[5].In the amide II mode spectral region,the band width of α-helix structure is larger than that of βsheet[6].Additionally,in the amide III spectral region, the vibrational band locates in the 1299 and 1235 cm−1 for α-helix and β-sheet structures of protein,respectively[7,8].
Besides the amide I,II,III bands,the amide A(N−H stretching vibration)band was used to analyze qualitatively the secondary structure of protein.For example,the amide A modes of some chiral proteins were recorded through the vibrational sum frequency generation spectroscopy to study their structures on the surface of aqueous solutions[9].It was found that the amide A vibrational band located in 3268−3274, 3278−3304,and 3355 cm−1for β-sheet,α-helix,and 310-helix,respectively.Di ff erent from the surface,in the bulk of aqueous proteins,it is di ffi cult to measure in situ the vibrational spectrum of amide A mode.This is because the O−H stretching band of H2O overlaps seriously with the amide A band.Previously,D2O was used to replace H2O to avoid the overlap[10].However,D2O increases the rigidity of the native structure of protein[11].Furthermore,the atoms D of D2O exchange with H atoms of amide groups and OH groups of proteins,which also a ff ects the properties of proteins. Di ff erent from the proteins in water,the amide A band could be easily recorded for the proteins in solid state, in organic solvents[12],and the peptides in gas phase [13–15].Since proteins only present biological activity in water,it is signi fi cant to record in situ the spectra of proteins in water.To our best knowledge,the amide A bands of proteins in water were not recorded in situ previously.
In this study,a new analytical method of Raman ratio spectrum was proposed to measure the amide A modes of lysozyme and α-chymotrypsin in H2O.The amide A bands of proteins in water were recorded directly through dividing the spectra of aqueous proteins to that of water.It was found that the amide A band of protein with more β-sheet located in a lower wavenumber. Furthermore,the Raman ratio spectrum was employed to record in situ the thermal denaturation of lysozyme in water.
II.EXPERIMENTS
Lysozyme was purchased from Life Science Products and Services.α-Chymotrypsin was purchased from Sinopharm Chemical Reagent Co.Water was puri fi ed by an ultrapure water system(Milli-Q,Millipore Co). All the aqueous proteins were prepared to be about 10% mass fraction.A drop of aqueous lysozyme at 84◦C was dried on the surface of a glass slide at the same temperature.The temperature was controlled with a thermostatic bath(THD-2006,Ningbo).
Raman spectra of these aqueous proteins were recorded with an instrument similar to that used previously[16–18].A CW laser(Coherent,Verdi-V5, 532 nm)was employed to excite the aqueous solutions, and the power of the laser was set to 1 W.The polarization of the laser was puri fi ed with a Glan-laser prism,and then controlled with a halfwave plate.The Raman signal was dispersed with a triple monochromator(Acton Research,TriplePro)and then recorded with a liquid nitrogen cooled CCD(Princeton Instruments,Spec-10:100B).During the measurements,the aqueous solutions were put into a cuboid SiO2cuvette. For solid proteins,a commercial confocal Raman spectrometer(WITec)was employed.A CW laser(532 nm, 50 mW)was focused on the surface of proteins through a objective(100∗,Olympus).The Raman spectra were recorded with an EMCCD(Andor).The wavelengths of the Raman signals were calibrated with the spectrum of mercury lamp.All the data were analyzed in the Igor pro software.
III.RESULTS AND DISCUSSION
A.Raman spectra of protein

FIG.1 Raman spectra of aqueous lysozyme(solid line)and pure water(dashed line)in the N−H(amide A)and O−H stretching region.
Figure 1 shows the Raman spectra of water and aqueous lysozyme in the region of 3000−3700 cm−1.The two spectra are similar obviously.In the Raman spectrum of aqueous lysozyme,the small peak at about 3070 cm−1 was assigned to the C−H stretching vibration of benzene ring group of lysozyme[4].As the amide A band of proteins locates in the region of 3250−3350 cm−1[9–15,19],it overlaps seriously with the Raman spectrum of water in the O−H stretching vibration region.Because of the small intensity of amide A band and the low concentration of lysozyme,it is di ffi cult to distinguish the amide A band from the Raman spectrum of aqueous lysozyme.Previously,the proteins were usually studied through the spectra in the amide I,II and III region[1–8],as these modes could be easily distinguished from the spectrum of water.However,if the vibrational bands of some particular solvents overlapped with the amide I,II or III bands,the amide A band should be a candidate to be employed to study the structure of proteins.In the following section,we will introduce a new method to distinguish the amide A band from the O−H stretching band of water.
B.Raman ratio spectrum
Raman ratio spectrum was proposed fi rstly to extract the amide A band of proteins in the spectra of aqueous proteins.It was de fi ned with the equation,
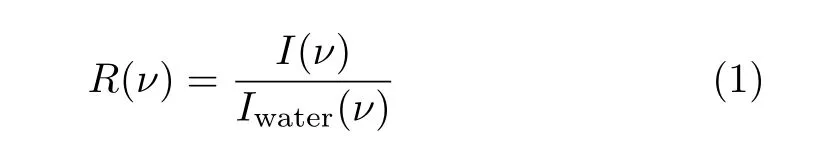
where R(ν)is the Raman ratio spectrum,I(ν)and Iwater(ν)are the Raman spectra of aqueous protein and pure water,respectively.Aqueous protein contains bulk water,protein and the hydration shell of protein,thus the spectrum of the aqueous protein could be written as:
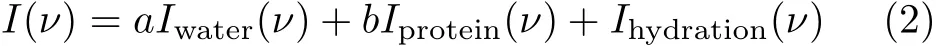
where Iprotein(ν)and Ihydration(ν)are the Raman spectra of protein and their hydration,respectively,a and b are coe ffi cients.For large protein,its hydration is much less than that of the protein and bulk water,the spectrum of the hydration shell could be ignored in Eq.(2). Hence the Raman ratio spectrum could be expressed as the following equation,Around 3300 cm−1region the intensity of water is approximately constant,thus amide A band of protein at ~3300 cm−1could be directly distinguished in the Raman ratio spectrum.

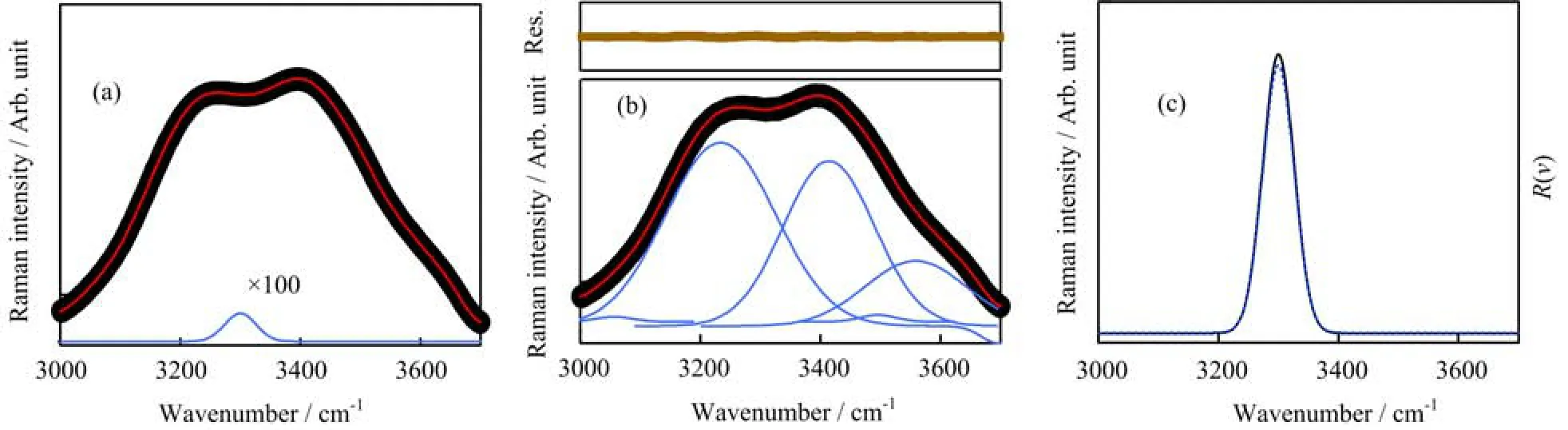
FIG.2(a)The simulated Raman spectrum of water(red solid line),amide A band(blue solid line,100 times ampli fi ed)and aqueous protein(black solid line);(b)Experimental Raman spectrum of pure water(black solid line)at room temperature and the corresponding multi-peak fi tting;(c)The simulated amide A band(blue dashed line)and the Raman ratio spectrum (black solid line).

FIG.3 The simulated Gaussian bands(blue dashed line)and the corresponding Raman ratio spectra(black solid line)at low(a)and high(b)wavenumber in the OH stretching region.(c)Comparison of the wavenumbers of the Gaussian bands and these of the corresponding Raman ratio spectra.
To examine whether amide A band can be extracted from Raman ratio spectrum,a mathematical simulation was employed.As shown in Fig.2(a),the simulated spectrum of water added a weak Gaussian peak to simulate the spectrum of aqueous protein.The simulated spectrum of water was obtained by the multi-peak fi tting of the experimental Raman spectrum of pure water (Fig.2(b)).The fi tting procedure only o ff ered the mathematic function of the experimental spectrum,it did not present any real physical meaning.A Gaussian peak at 3300 cm−1was employed to simulate the spectrum of amide A band.The intensity of this peak was set to be about 1/1000 of that of water at maximum.The full width at half maximum(FWHM)of this peak was set to be 62 cm−1,which was similar to the FWHM of the experimental amide A band of protein on surface[9]. The simulated amide A band was very small,thus the simulated spectrum of aqueous protein was very similar to that of water,and the amide A band cannot be distinguished.The Raman ratio spectrum was calculated, and was shown in Fig.2(c).A single band was observed in the ratio spectrum.This band are almost the same as the simulated amide A band.Both the Raman shift and the FWHM of this band agreed well with these of the simulated band.Consequently,Raman ratio spectrum could be employed to distinguish amide A band from the spectrum of aqueous protein.
The general applicability of Raman ratio spectrum was checked in the whole O−H stretching regions.Two Gaussian bands at the lowest(Fig.3(a))and highest wavenumber(Fig.3(b))in this region were employed to simulate the amide A band and the spectrum of aqueous solutions.Similar to the above simulation,the simulated Raman ratio spectra were calculated(Fig.3 (a)and(b)).The bands in the ratio spectra matched well with the Gaussian bands.In addition,the Raman shifts of the bands in Raman ratio spectra were compared with those of the Gaussian bands(Fig.3(c)). They matched well with each other.Consequently,the Raman ratio spectra could be employed to extract a small band from the Raman spectrum of water mixtures in the whole O−H stretching region.Furthermore,the di ff erence between the Raman shift of the bands in ratio spectra and that of the Gaussian bands was the small-est at~3300 cm−1,thus Raman ratio spectra were the most exact to determine the band in the this region. Luckily,the amide A band locates in this region.
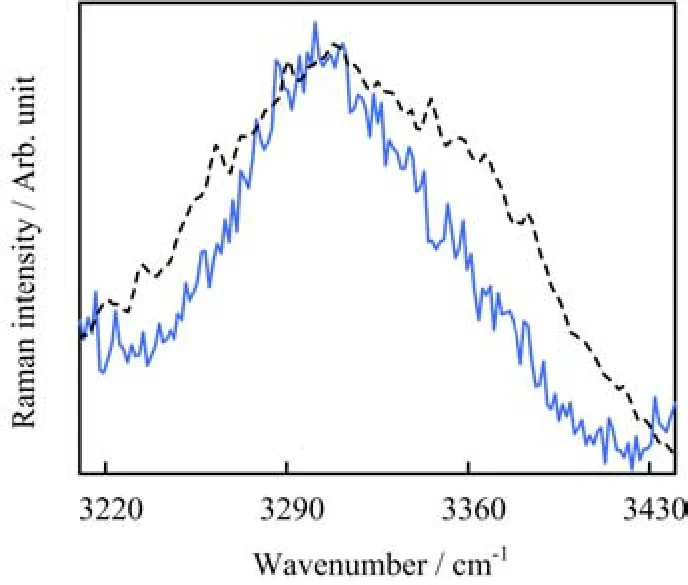
FIG.4The Raman ratio spectrum R(ν)of aqueous lysozyme(solid line)and the Raman spectrum of solid lysozyme(dash-dotted line)at room temperature.
The experimental spectra were employed to check whether the Raman ratio spectrum could be employed to extract the amide A band of protein in water.Using the experimental Raman spectrum of aqueous lysozyme and that of water,the Raman ratio spectrum was obtained,and it was shown in Fig.4.A single band was observed in this spectrum.The band located at about 3300 cm−1,which agreed well with the amide A band of protein on the surface[9].The amide A band of solid lysozyme was directly recorded in its Raman spectrum (Fig.4).It was found that the amide A band of solid lysozyme matched well with the band in the Raman ratio spectrum of aqueous lysozyme.Consequently,the band in the Raman ratio spectrum was assigned to amide A mode.
Previously the secondary structures of protein were widely studied through amide I band in Raman spectra[1–5],and the amide A band in the sum frequency generation spectra[9].Here we examined whether the amide A band in Raman ratio spectrum can be employed to identify qualitatively the secondary structure of protein in water.Lysozyme[20](α-helix 40%, β-sheet 10%)and α-chymotrypsin(α-helix 14%,β-sheet 32%)[21]were used,since their secondary structures are much di ff erent.The secondary structures of the solid proteins were recorded from the X-ray di ff raction of solid lysozyme and α-chymotrypsin.In the solid state, the Raman spectra in both amide I and amide A region could be easily recorded.The spectra of both proteins were plotted in Fig.5(a)and(b).It was widely accepted that the amide I band of α-helix located in a lower wavenumber than that of β-sheet[1–4],and the amide A band of α-helix located in a higher wavenumber than that of β-sheet[9].The Raman spectra of both solid proteins in amide I(Fig.5(a))and amide A (Fig.5(b))region supported the relationship.This relationship was also observed in the Raman spectra and Raman ratio spectra of proteins in water.The spectra in amide I region of both aqueous proteins were measured and plotted in Fig.5(c).The amide I band of lysozyme located at a lower wavenumber than that of α-chymotrypsin,which demonstrated α-helix structure of lysozyme in water were much more than that of αchymotrypsin in water.The amide A bands of both proteins in water were obtained through the Raman ratio spectra(Fig.5(d)).It was found that the frequency of the amide A band of the lysozyme with more α-helix was larger than that of the α-chymotrypsin with more β-sheet.As a consequence,Raman ratio spectrum in amide A region can be used to identify qualitatively in situ the secondary structure of protein in water.

FIG.5 Raman spectra of solid lysozyme(solid line)and αchymotrypsin(dashed line)in amide I(a)and amide A(b) region.Raman spectra(c)and Raman ratio spectra(d)of lysozyme(solid line)and α-chymotrypsin(dashed line)in water,respectively.
C.Thermal denaturation of lysozyme

FIG.6(a)Raman spectra of solid lysozyme at 25◦C (dashed line)and at 84◦C(solid line).(b)Raman ratio spectra of aqueous lysozyme at 25◦C(dashed line)and at 84◦C(solid line).
As the Raman ratio spectrum of amide A band can identify qualitatively the secondary structure of proteins,it could be employed to study the denaturation of proteins.Previously,the thermal denaturation of protein was usually monitored through amide I band[22–24].It was found the fraction of α-helix decreased when thermal denaturation occurred[25].The Raman spectrum of solid native lysozyme at room temperature was compared with that of the solid denaturated lysozyme at 84◦C.The solid lysozyme at 84◦C was dried up from its aqueous solution.The amide A band was directly recorded for both solid lysozyme,as shown in Fig.6(a).The band located at 3304.1 and 3312.5 cm−1 for solid lysozyme at 84 and 25◦C respectively.The red shift of 8.4 cm−1indicates that the fraction of αhelix decreased during the thermal denaturation.To detect in situ the change of the secondary structure of lysozyme in water during the thermal denaturation process,the Raman ratio spectra of aqueous lysozyme at 25 and 84◦C were recorded.The raw Raman ratio spectra were fi tted with multi-peaks to remove the spectral contribution of the strong alkyl stretching band.The fi nal Raman ratio spectra at both temperatures were plotted in Fig.6(b).The amide A band of lysozyme in water at 84 and 25◦C located at 3299.6 and 3308 cm−1 respectively.The red shift of 8.4 cm−1was obviously observed.It agreed very well with that from the Raman spectra of the solid lysozymes.Consequently,the Raman ratio spectrum could be employed to study qualitatively in situ the secondary structure of protein in water with high accuracy.
Furthermore,the process of thermal denaturation of aqueous lysozyme can be monitored in situ by amide A band in the Raman ratio spectrum.The Raman spectra were recorded in the O−H stretching and amide A region for aqueous lysozyme from 60◦C to 84◦C.The Raman ratio spectra of amide A band at these temperatures were obtained and plotted in Fig.7(a).The frequencies of the amide A mode were listed in Fig.7(b). These temperature dependent frequencies were fi tted with a sigmoid function.The denaturation temperature ww ai t sh dtehtae rt m fri o nme d ottoh b ere t~ec7h5n ◦o C lo. gTi ehse[ 2t e6 m ].p P erreavt u ior u es a lyg , r et eh de thermal denaturation was also detected with the vibrational spectra of the amide I band[22–24].Here the Raman spectra of amide I band were also recorded, they were analyzed to compare with the results from the amide A band in the Raman ratio spectra.The spectra of amide I band at the temperatures were plotted in Fig.7(c).The frequencies of this mode were plotted in Fig.7(d).These temperature dependent frequencies were also fi tted with a sigmoid function.The denaturt a emtio pne r t a etmu r pe e ra ag true re ed ww ae s ll d w eittehr m thinae td fr t o om b te h~e 7a7m ◦idC e .AT hi ne the Raman ratio spectra.Thus,the thermal denaturation of lysozyme in water indeed could be monitored in situ by Raman ratio spectrum.The amide A band provides a candidate method to study qualitatively the secondary structure of proteins in water when amide I band was overlapped with other bands.

FIG.7(a)Raman ratio spectra of aqueous lysozyme in the amide A region at temperatures of 60,63,66,69,71,73,75, 77,79,81,and 84◦C(from bottom to top,respectively). (b)Temperature dependent frequencies of amide A mode from Raman ratio spectra.(c)Raman spectra of aqueous lysozyme in the amide I region at the same temperatures as in(a).(d)Temperature dependent frequencies of amide I mode from Raman spectra.
Besides Raman spectroscopy,IR spectroscopy is also widely used to study the secondary structure of protein [27–30].Here,the IR spectrum of aqueous lysozyme was measured,and the IR ratio spectrum was also obtained.However,the amide A band was not observed in the IR ratio spectrum.One of the reason may be that IR absorption coe ffi cient of O−H stretching mode of water is extremely larger than that of amide A mode of protein.Another reason may be the small signal noise ratio in our IR spectrum.
IV.CONCLUSION
Because of the strong overlapping with O−H stretching vibration of water,the weak amide A band of protein in water was not obtained previously.In this work, we presented a new analytical method of Raman ratio spectrum to record in situ the amide A band of protein in water.The viability of this method was proven by a mathematical simulation and the experimental spectrum of proteins in water.This new method was supported by comparing the Raman spectra in amide I region and Raman ratio spectra in amide A region of lysozyme and α-chymotrypsin in water.The Raman ra-tio spectrum was employed to study in situ the thermal denaturation of lysozyme in water.The Raman ratio spectrum is a new approach to study qualitatively the secondary structure of protein in water.
V.ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China(No.91127042,No.21103158, No.21273211,No.21473171),the National Key Basic Research Special Foundation(No.2013CB834602 and No.2010CB923300),the Fundamental Research Funds for the Central Universities(No.7215623603),and the Hua-shan Mountain Scholar Program.We also thank Doctor Kang-zhen Tian and Professor Shu-ji Ye for the measurement of IR spectra of aqueous lysozyme.
[1]T.G.Spiro and B.P.Gaber,Annu.Rev.Biochem.46, 553(1977).
在推进地下水超采治理试点工作中,始终牢牢把握五条基本原则:一是政府引导、全民行动,发挥好政府和群众两个积极性;二是规划统领、科学治理,年度实施方案与中长期规划有机衔接,集中连片规模实施,务求治理一片、见效一片、巩固一片;三是创新机制、示范带动,探索建立地下水超采治理的有效途径,力求取得可示范、可复制、可推广的经验;四是因地制宜、积极稳妥,根据实际情况,科学确定治理模式和工程规模;五是竞争立项、绩效考核,依据项目前期工作和压采效果择优实施,严格把关、严格奖惩。
[2]D.I.Ellis,D.P.Cowcher,L.Ashton,S.O’Hagan,and R.Goodacre,The Analyst 138,3871(2013).
[3]V.Kocherbitov,J.Latynis,A.Misi¯unas,J.Barauskas, and G.Niaura,J.Phys.Chem.B 117,4981(2013).
[4]A.Rygula,K.Majzner,K.M.Marzec,A.Kaczor,M. Pilarczyk,and M.Baranska,J.Raman Spectrosc.44, 1061(2013).
[5]A.J.P.Alix,G.Pedanou,and M.Berjot,J.Mol. Struct.174,159(1988).
[6]F.Dousseau and M.Pezolet,Biochemistry(Mosc.)29, 8771(1990).
[7]S.A.Oladepo,K.Xiong,Z.M.Hong,S.A.Asher, J.Handen,and I.K.Lednev,Chem.Rev.112,2604 (2012).
[8]S.J.Ye,H.C.Li,W.L.Yang,and Y.Luo,J.Am. Chem.Soc.136,1206(2014).
[10]P.K.Johansson and P.Koelsch,J.Am.Chem.Soc. 136,13598(2014).
[11]P.Cioni and G.B.Strambini,Biophys.J.82,3246 (2002).
[12]A.Fulara and W.Dzwolak,J.Phys.Chem.B 114,8278 (2010).
[13]E.E.Baquero,W.H.James,S.H.Choi,S.H.Gellman, and T.S.Zwier,J.Am.Chem.Soc.130,4784(2008).
[14]E.E.Baquero,W.H.James,S.H.Choi,S.H.Gellman, and T.S.Zwier,J.Am.Chem.Soc.130,4795(2008).
[15]H.S.Biswal,E.Gloaguen,Y.Loquais,B.Tardivel,and M.Mons,J.Phys.Chem.Letters 3,755(2012).
[16]K.Lin,X.G.Zhou,Y.Luo,and S.L.Liu,J.Phys. Chem.B 114,3567(2010).
[17]K.Lin,N.Y.Hu,X.G.Zhou,S.L.Liu,and Y.Luo, J.Raman Spectrosc.43,82(2012).
[18]L.Chen,W.D.Zhu,K.Lin,N.Y.Hu,Y.Q.Yu,X. G.Zhou,L.F.Yuan,S.M.Hu,and Y.Luo,J.Phys. Chem.A 119,3209(2015).
[19]J.Kong and S.Yu,Acta Biochim.Biophys.Sin.39, 549(2007).
[20]Z.M.Wang,G.Y.Zhu,Q.C.Huang,M.X.Qian, M.C.Shao,Y.S.Jia,and Y.Q.Tang,Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology 1384,335(1998).
[21]W.Kabsch and C.Sander,Biopolymers 22,2577 (1983).
[22]A.Giugliarelli,P.Sassi,M.Paolantoni,A.Morresi, R.Dukor,and L.Na fi e,J.Phys.Chem.B 117,2645 (2013).
[23]G.Bellavia,L.Paccou,S.Achir,Y.Guinet,J.Siepmann,and A.Hedoux,Food Biophysics 8,170(2013).
[24]A.Hedoux,R.Ionov,J.F.Willart,A.Lerbret,F.Affouard,Y.Guinet,M.Descamps,D.Prevost,L.Paccou,and F.Danede,J.Chem.Phys.124,(2006).
[25]Y.Moriyama,N.Kondo,and K.Takeda,Langmuir 28, 16268(2012).
[26]T.Yamamoto,N.Fukui,A.Hori,and Y.Matsui,J. Mol.Struct.782,60(2006).
[27]H.Baltacioglu,A.Bayindirli,M.Severcan,and F.Severcan,Food Chem.187,263(2015).
[28]A.Saeed,G.A.Raouf,S.S.Nafee,S.A.Shaheen,and Y.Al-Hadeethi,PLoS ONE 10,(2015).
[29]J.V.Coe,S.V.Nystrom,Z.M.Chen,R.Li,D.Verreault,C.L.Hitchcock,E.W.Martin,and H.C.Allen, J.Phys.Chem.B 119,13079(2015).
[30]H.Y.Yang,S.N.Yang,J.L.Kong,A.C.Dong,and S.N.Yu,Nat.Protoc.10,(2015).
猜你喜欢
杂志排行
CHINESE JOURNAL OF CHEMICAL PHYSICS的其它文章
- ARTICLE E ffi cient Separation of Ar and Kr from Environmental Samples for Trace Radioactive Noble Gas Detection†
- ARTICLE Spectrum Correction in Study of Solvation Dynamics by Fluorescence Non-collinear Optical Parametric Ampli fi cation Spectroscopy†
- I.INTRODUCTION
- ARTICLE High-Resolution Experimental Study on Photodissocaition of N2O†
- ARTICLE Photoelectron Spectroscopy and Density Functional Calculations of TiGen−(n=7−12)Clusters†
- ARTICLE Hermiticity of Hamiltonian Matrix using the Fourier Basis Sets in Bond-Bond-Angle and Radau Coordinates†
