Evaluation of hepatitis B viral replication and proteomic analysis of HepG2.2.15 cell line after knockdown of HBx
2011-06-11
Hangzhou,China
Introduction
Hepatitis B virus (HBV) is one of the major pathogens of human liver disease.Globally,there are more than 360 million people chronically infected with HBV,many of whom willfinally develop severe liver diseases,such as cirrhosis and hepatocellular carcinoma (HCC).[1]It has been reported that chronic HBV carriers have a 100-fold higher risk of developing HCC,and among them,over 500 000 die from liver cirrhosis and HCC annually.[2,3]
HBV belongs to the hepadnavirus family and is composed of an outer envelope formed by HBsAg and an inner nucleocapsid that packages the viral genome and polymerase.The HBV genome is a partially doublestranded relaxed circular DNA molecule containing four partially overlapping open-reading frames encoding viral envelope,core,reverse transcriptase-polymerase,and the X gene.[4]HBx,the product of the X gene,promotes viral gene expression and replication by trans-activating the viral promoters and enhancer/promoter complexes.[5,6]It modulates several signaling pathways of protein kinases related to viral replication.For example,HBx stimulates the Src kinase family,[7]including proline-rich tyrosine kinase (Pyk2),[8]and focal adhesion kinase (FAK);[9]activation of these kinases is dependent on cellular calcium signaling and critical in HBV replication.HBx also promotes viral genome replication via the DDB1-dependent pathway and knockdown of DDB1 reduces HBV transcription and replication.[10]In the case of tumorigenesis,HBx stimulates the activity of TGF-β1 and promotes the tumorigenic properties of this wellknown cytokine.[11,12]It also regulates the expression of several matrix metalloproteinases such asfibronectin and integrin beta-1,thereby remodeling the extracellular matrix,which may promote tumor metastasis.[13,14]A technique for target protein identification,twodimensional electrophoresis (2-DE) in combination with image analysis and mass spectrometry (MS) is commonly used in the area of HBV research.[15,16]Thus,to discover novel anti-HBV targets for attenuating the morbidity and mortality of HBV-related disease,we performed global proteomic profiling using 2-DE to identify the downstream functional proteins of HBx.
The HepG2.2.15 cell line was established by transfecting a hepatoblastoma cell line (HepG2) with plasmids containing two head-to-tail dimers of the HBV genome.The HepG2.2.15 cell line releases high levels of HBsAg and HBeAg into the medium during culture.It supports the full replication cycle of HBV,assembly and secretion of HBV DNA and Dane particles.[17,18]Furthermore,HBV virions produced by the cell line also exhibit high endogenous polymerase activity.[18]Thus,HepG2.2.15 is an appropriate tool to identify the molecular events of the viral replication cycle as well as the secretion of HBV particles into the intracellular environment in vitro.
In this study,we first evaluated the effects of HBx knockdown on HBV replication in vitro.To reveal the underlying mechanisms,proteomic techniques were used to analyze the changes of protein expression profile in HepG2.2.15 cells after RNA interference of HBx by small interfering RNA (siRNA) and identify 12 proteins with altered expression that may play critical roles in HBx-related HBV replication.
Methods
Cell culture
HepG2.2.15 cells were cultured in DMEM medium(HyClone,USA) containing 10% fetal bovine serum(FBS) (Gibco,USA) and 200 mg/L G418 (Sigma,USA) at 37 ℃ in a humidified incubator gassed with 5% CO2.
siRNA transfection
The day before transfection,subcon fluent monolayer HepG2.2.15 cells were harvested from the culture dishes with trypsin and seeded in 6-well plates at 20 000 cells/cm2such that they were about 50% con fluent the next day.Then,the cells were treated with siRNA buffer alone for the control group.Transfection with 40 nmol/L of HBx siRNA (sense:5'-GACCUUGAGGCAUACUUCAdTdT-3'and antisense:5'-UGAAGUAUGCCUCAAGGUCdTdT-3′)or mock siRNA (sense:5'-UUCUCCGAACGUGUCAC GUdTdT-3' and antisense:5'-ACGUGACACGUUCG GAGAAdTdT-3') was performed using oligofectamine transfection reagent in OptiMEMI-reduced serum medium (Invitrogen) in the absence of FBS and antibiotics according to the manufacturer's instructions.Six hours after transfection,the medium was removed and replaced with RPMI-1640 containing 200 μg/mL G418.Supernatant was harvested for further analysis 48 hours after siRNA treatment.
Analysis of HBsAg and HBeAg
HBsAg and HBeAg levels in the supernatant were detected by Abbott AxSYM HBsAg and HBeAg QT assay kits (Abbott Diagnostics Division) following the manufacturer's instructions.The suppression rate of replication of HBsAg and HBeAg in the HBx knockdown group was calculated using the following formula:
Suppression rate=[1-A value (HBx knockdown)/A value (negative control)]×100%
Real-time quantitative PCR (qPCR) analysis of hepatitis B virus DNA
Forty-eight hours after treatment with siRNA,supernatant HBV DNA levels were tested using a realtime PCR kit (Bioselex,Hangzhou,China) according to the manufacturer's instructions on an ABI 7500 real-time PCR system (Applied Biosystems,Foster,CA,USA).This kit was approved by the State Food and Drug Administration of China for in vitro diagnosis with a low detection limit.[19]
2-DE
Cells of each sample were harvested and washed three times with PBS,resuspended in 1 mL lysis buffer containing 7 mol/L urea,2 mol/L thiourea,4% CHAPS,1% DTT,2% IPG buffer (pH 4-7,Amersham Biosciences)and protease inhibitor cocktail (Complete tablets,Roche Diagnostics,Germany),then ultrasonically lysed for 4×10 seconds and incubated on ice for 1 hour.After centrifugation at 23 000 g for 30 minutes,the supernatant was harvested.Protein concentration was determined using the Bradford method.
2-DE was carried out following the instructions from Amersham Biosciences.Brie fly,samples containing 200 μg protein were diluted to 450 μL with rehydration solution (8 mol/L urea,2% CHAPS,20 mmol/L DTT,0.5% IPG buffer pH 4-7,0.002% bromophenol blue).After rehydration with 50 V for 12 hours,the sample proteins were subjected on IPG strips to IEF operating the Amersham Biosciences IPGphor as follows:200 V for 1 hour,500 V for 1 hour,1000-4000 V for 1 hour,4000-8000 V for 1 hour,8000 V for 1 hour,then focused at 8000 V for about 8 hours.After IEF,strips were equilibrated for 15 minutes in SDS equilibration buffer(50 mmol/L Tris-HCl,6 mol/L urea,30% glycerol,2%SDS and 0.002% bromophenol blue) containing 1%DTT and shaken at 120 rpm on an orbital shaker,then transferred to the SDS equilibration buffer containing 2.5% iodoacetamide and shaken for another 15 minutes.After equilibration,the strips were loaded onto vertical SDS PAGE (12.5% T constant).The second dimensional SDS electrophoresis was run using an Ettan DALTsix electrophoresis unit (Amersham Biosciences).Experiments with each group (negative control or HBx knockdown) were performed in triplicate.
Silver staining and image analysis
After electrophoresis,silver staining was done to visualize the gel according to the protocol of Yan et al.[20]Protein patterns on silver-stained gels were scanned using a high-resolution scanner (Amersham Biosciences),and analyzed with ImageMaster 2D software (version 6.0) for spot detection,background subtraction,volume normalization and spot matching.The resulting data were exported to Microsoft Excel,and significantly differentially expressed protein spots matching the threshold of 2.0-fold were selected for MALDI-TOF/TOF MS analysis.
In-gel digestion and MALDI-TOF/TOF MS analysis
Protein spots were excised from wet gels,transferred into Eppendorf tubes and destained.After that,the gel pieces were hydrated in 10 μL proteomics grade trypsin(Sigma) solution (20 ng/μL in 25 mmol/L NH4HCO3)and incubated at 37 ℃ for 15 hours.Peptides were extracted with 50% ACN and 2.5% TFA,and then dried in a lyophilizer and reconstituted in 1.5 μL 0.1% TFA Solution (Virtis,Gardiner,NY,USA).
The dried peptide mixtures were mixed with saturated matrix solution (5 mg/mL CHCA (Sigma,USA) in 0.1% TFA and 50% ACN) and spotted onto the MALDI target plate.Mass analysis was performed on an ABI 4700 Proteomics Analyzer MALDI-TOF/TOF(Applied Biosystems,Foster City,CA,USA) operating in result-dependent acquisition mode.Peptide mass maps were acquired in positive ion re flector mode at 20 kV accelerating voltage with 2000 laser shots per spectrum.When there were monoisotopic peak masses within 900-4000 Da with a minimum signalto-noise ratio (S/N) of 10 and a local noise window width of 250 m/z,the firstfive precursor ions showed the highest intensity excluding common trypsin autolysis peaks and matrix ion signals were selected for fragmentation.The instrument was operated in the MS/MS positive ion mode at 2 kV collision energy with default calibration.Monoisotopic peak masses were automatically determined with a minimum S/N of 5 and a local noise window width of 250 m/z.Combined MS and MS/MS spectra were submitted to GPS Explorer(version 3.6,Applied Biosystems) and MASCOT (version 2.1,Matrix Science) searching against the UniprotKB/SwissProt database.The search parameters were as follows:trypsin cleavage,one missed cleavage allowed,carbamidomethylation of cysteine asfixed modification and methionine oxidation as variable modification,peptide mass tolerance to 150 ppm,fragment tolerance to 0.4 Da,and minimum ion score confidence interval for MS/MS data set to 95%.Significance of the identification was evaluated according to the value of probability and sequence coverage.
Antibodies and Western blotting
Western blotting was performed as previously described.[21]HSP70 (human HSPA1A,Santa Cruz,dilution 1∶1000) was detected in total cell extracts.
Statistical analysis
Data of microparticle enzyme immunoassay (MEIA)results and real-time qPCR are shown as mean±SD.One-way ANOVA analysis of Dunnett's test was used to analyze the significance of differences between the negative control and HBx knockdown groups.The statistical significance in terms of the expression profiles of HepG2.2.15 cells with or without HBx knockdown was estimated by Student's t test.All calculations were done with SPSS 11.5 software,and a P value less than 0.05 was considered statistically significant.
Results
Downregulation of HBx inhibits the expression of HBsAg and HBeAg
To investigate the role of HBx in the production of HBsAg and HBeAg,HepG2.2.15 cells were transfected with HBx siRNA or a mock siRNA as a negative control.The supernatant was collected at 48-hour after the treatment with siRNA and the titer of HBsAg and HBeAg was assessed by HBsAg and HBeAg QT assay.We found that after treatment with HBx siRNA,the suppression rates of HBsAg and HBeAg were 55%and 66% respectively when compared with the mock siRNA-transfected group (Fig.1),indicating that the downregulation of HBx inhibited HBsAg and HBeAg production in vitro.
Downregulation of HBx inhibits HBV DNA replication
We also investigated whether the knockdown of HBx in vitro had a direct effect on HBV DNA replication.The HBV DNA in the supernatant was measured at 48 hours after the treatment with siRNA by real-time qPCR analysis.The HBV DNA replication level was lower in the HBx siRNA group than in the mock siRNA-treated group (P=0.005) (Fig.2),indicating that the downregulation of HBx inhibited HBV DNA replication.

Fig.1.Downregulation of HBx inhibited the expression of HBsAg and HBeAg.HepG2.2.15 cells were treated with HBx siRNA,the culture supernatants were collected for MEIA analysis of HBsAg and HBeAg 48 hours later,and suppression rates of HBsAg and HBeAg were calculated [suppression rate=[1-A value (HBx knockdown)/A value (negative control)]×100%].Each test was repeated 3 times.Results indicated that the relative level of HBsAg(*:P<0.001) and HBeAg (#:P=0.007) after downregulation of HBx.
Comparative analysis of 2-DE protein profiles and identification of proteins by MALDI-TOF/TOF
To identify key proteins that may play a critical role in HBV replication,comparative 2-DE analysis of HepG2.2.15 cells treated with mock and HBx siRNA was performed.Protein lysate from each group was resolved by 2-DE and visualized by silver staining,then the gels were digitized prior to computer-based matching and quantitative analysis with image analysis software.MALDI-TOF/TOF analysis was performed in order to identify the protein spots with altered intensity (>2.0-fold) as a result of HBx knockdown.After submitting the combined MS and MS/MS spectra generated by MALDI-TOF/TOF,12 were identified:7 up-regulated proteins and 5 down-regulated proteins.The identified proteins represented a heterogeneous group that included several important molecules relevant to protein folding such as heat shock 70 kDa protein (HSP70) and signal transduction such as calreticulin.The functions of the remaining proteins in HBV pathogenesis are still not clear (Fig.3 and Table ).
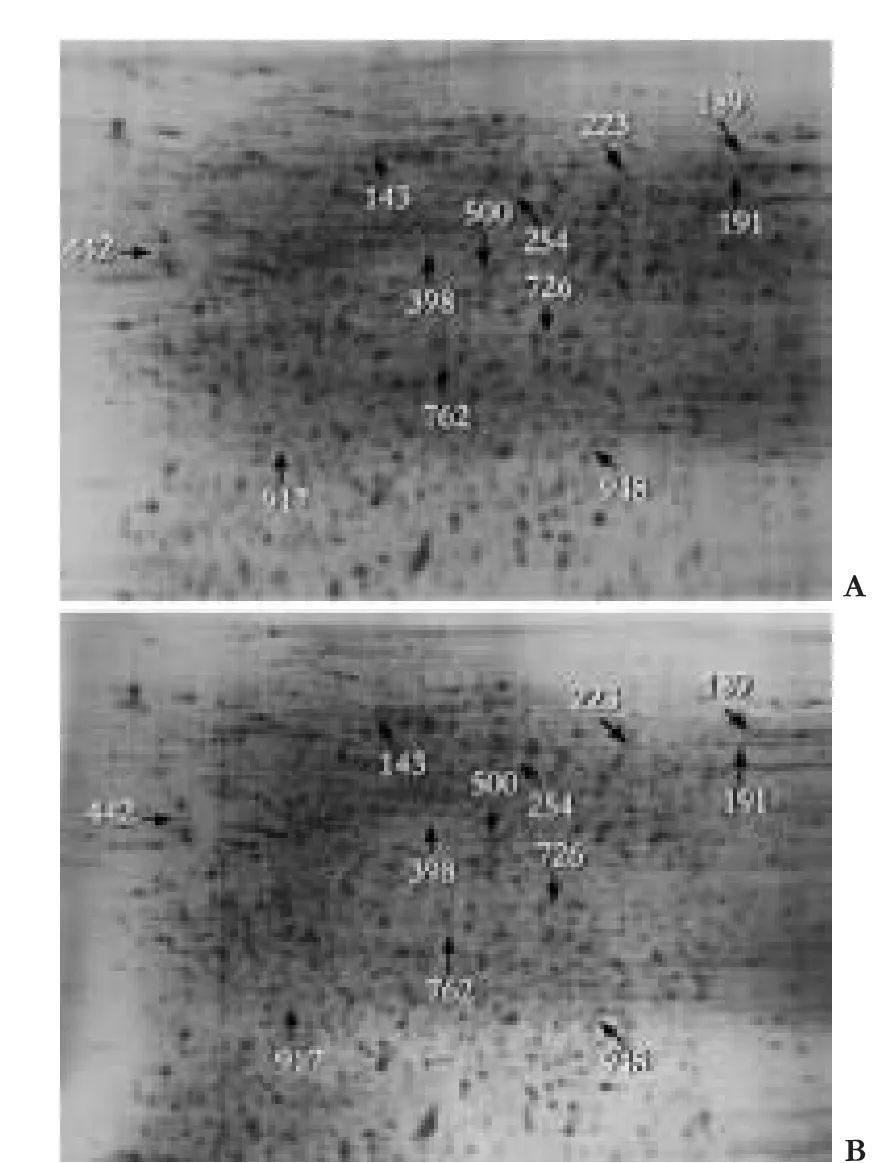
Fig.3.Cells were lysed and 200 μg of total protein lysate was subjected to 2-DE,followed by silver staining and image analysis.Results were quantified from three sets of 2-DE.The 2-DE map indicated protein spots changed in volume after knockdown of HBx in HepG2.2.15 cells.The spots of interest were excised from the gels and digested with trypsin.The digested peptides were used for MALDI-TOF/TOF MS analysis.The identified proteins are listed in Table.A:HBx siRNA treated group; B:Mock siRNA treated group.
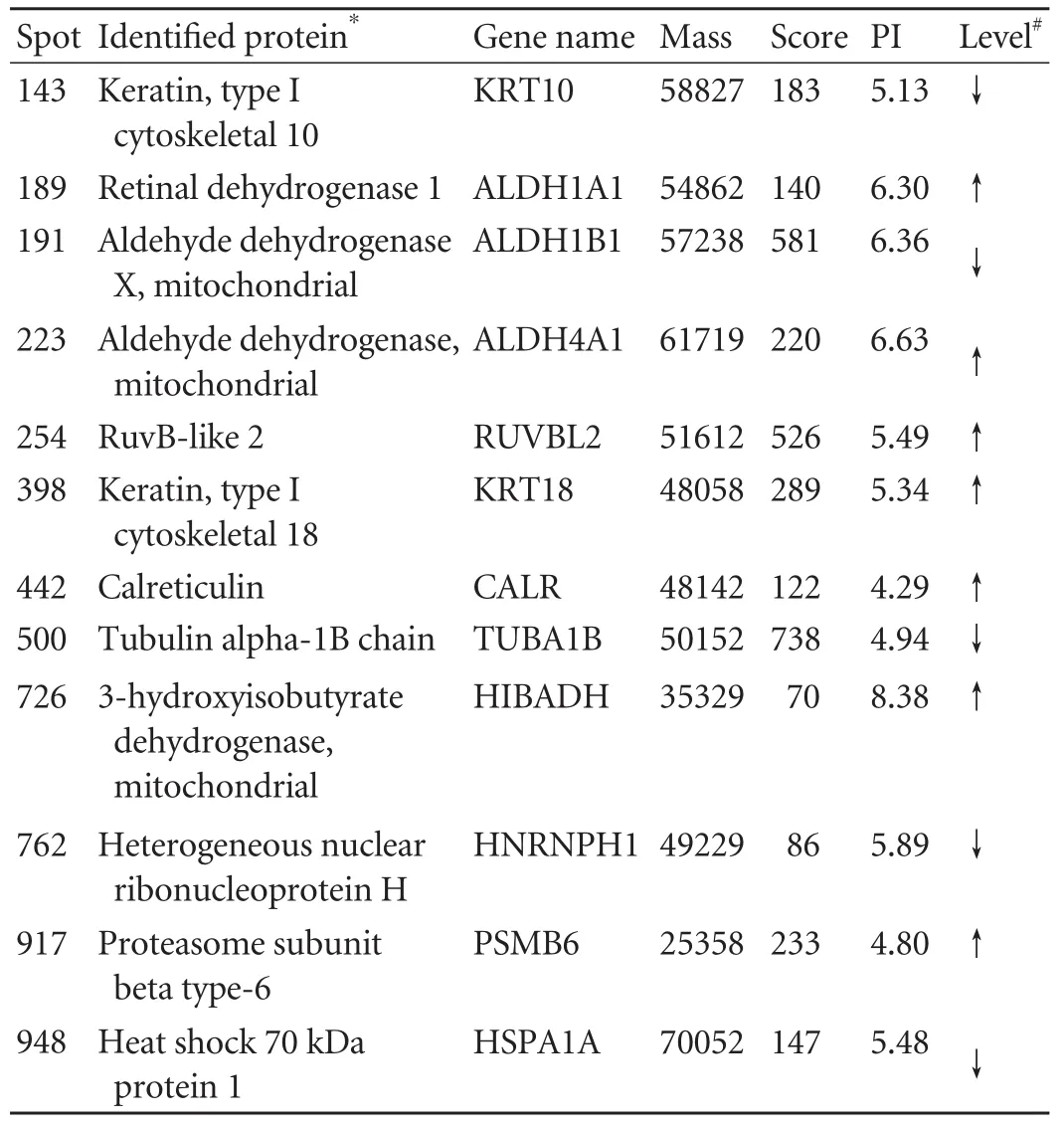
Table.Differentially-expressed proteins identified after knockdown of HBx in HepG2.2.15 cells
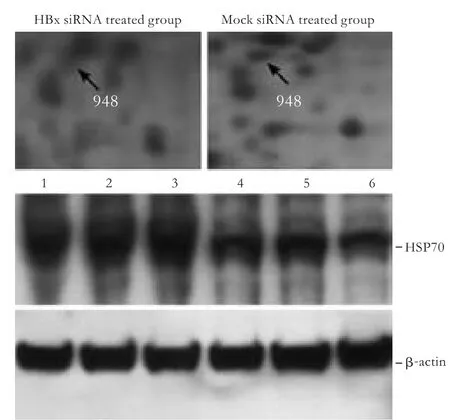
Fig.4.Western blotting analysis of HSP70 (HSPA1A) identified by MS with β-actin as internal reference.Lanes 1,2,3:HepG2.2.15 cells treated with mock siRNA; lanes 4,5,6:HepG2.2.15 treated with siRNA of HBx.The expression level of HSP70 protein was decreased after knockdown of HBx in HepG2.2.15 cells.
Validation of proteins by Western blotting
To validate the 2-DE results and assess the expression changes of several proteins showing differential patterns after treatment with HBx siRNA,Western blotting analysis of protein abundance changes was performed.From the identified candidates,we tested HSP70 by Western blotting analysis using available commercial antibodies.HSP70 protein level was decreased after the knockdown of HBx in HepG2.2.15 cells,which conformed to the 2-DE data (Fig.4).
Discussion
In this study,we first evaluated the effects of knockdown of HBx on HBV replication level by MEIA analysis of HBsAg and HBeAg in the supernatants and real-time qPCR analysis of extracellular HBV DNA.After the knockdown of HBx,the titer of HBsAg and HBeAg as well as the HBV DNA replication level were dramatically decreased,suggesting that HBx plays an important role in HBV replication in this culture system.
The viral HBx protein has been shown to promote transcriptional activation and viral replication in the nucleus and cytoplasm of infected cells.[22]HBx regulates a wide range of genes.It up-regulates the expression of HBV genes by transactivating its own promoters,as well as cellular genes of infected hepatocytes,thereby modifying the environment to facilitate viral replication.[1]Our proteomic screening,which explored global protein changes after the knockdown of HBx,may provide clues to reveal the mechanisms of action of HBx on HBV replication,and possibly identify key factors co-activated with HBx in HBV replication.
In this study,a total of 12 protein spots were identified to exhibit altered expression level after knockdown of HBx with high confidence by peptide massfingerprinting.One of the proteins that decreased after RNA interference of HBx was HSP70,a molecular chaperone involved in the topological reorientation of the HBV large envelope protein.[23]HSP70 plays an important role in viral replication.Gonzalez et al[24]reported that HSP70 coimmunoprecipitates with NS5A,a protein implicated in regulating HCV viral genome replication.[25,26]siRNA-mediated knockdown of HSP70 reduces the augmentation of the HCV internal ribosome entry site,mediates translation induced by NS5A,and modestly suppresses viral protein accumulation.[24]In HBV,HSP70 binds specifically with HBx to form a complex,[27]helping HBx to fold in the right conformation; at the same time,HBx exerts its function by binding to HSP70.[27]It also physically interacts with HSP60 and HSP90 in the HepG2.2.15 cell line to form a multi-chaperone machine that contributes to the HBV life cycle,so down-regulation of HSP70 could significantly inhibit the production of HBV virions.[28]Besides,a carcinogenic role of HSP70 has been reported;[29-31]in the case of HBV-related HCC,expression of HSP70 was increased along with the stepwise progression of hepatocarcinogenesis and there is a positive correlation between HSP70 expression and prognostic factors for HCC.[32]The role of HSP70 in HCC is associated with its function in cell proliferation and apoptosis.HSP70 interacts with p53 as a chaperone,[33]and it has been shown that HSP70 antisense oligomers specifically inhibits tumor cell proliferation by inducing apoptosis.[34]Taken together with these reports,our results strongly suggest that HSP70 plays a pivotal role in HBV replication and HBV-related carcinogenesis.
Calreticulin (CALR),one of the most ubiquitous signaling molecules mainly located in the endoplasmic reticulum (ER),was upregulated after the knockdown of HBx.CALR has important functions in the calcium signaling pathway by homeostatic control of cytosolic and ER calcium levels.[35,36]Ca2+plays critical roles in viral entry,expression of viral genes,posttranslational processing of viral proteins,virion maturation and release.[37]In the case of viral replication,viruses disrupt Ca2+homeostasis and accelerate their replication cycles using Ca2+and cellular Ca2+-binding proteins.[38,39]Modest ER-mitochondrial Ca2+increases activate the Ca2+-dependent Kreb's cycle dehydrogenases to facilitate ATP production,thereby meeting the higher energy demand of viral replication.[40]At the same time,Ca2+overload either in mitochondria or intracellular space has pro-apoptotic effects by several pathways.For example,abnormal increases in ER-mitochondrial Ca2+activates the opening of the permeability transition pore,causes the release of cytochrome C and activation of caspase 9,then triggers apoptosis of the infected cell.[41]Finally,virions are released and viral dissemination is maximized.CALR is also involved in the adaptive immune response; it has been proposed that intracellular viruses subvert this response to pathogeninfected cells through the prevention of cell surface exposure of CALR.[42]In this study,we unveiled a direct link between CALR and HBx,providing a mechanism by which HBx might contribute to HBV replication.
RuvB-like 2 (RUVBL2),which belongs to the ATPase associated with diverse cellular activities family of helicases,was also overexpressed in HepG2.2.15 cells after treatment with HBx siRNA in this study and participated in cellular processes such as cellular transformation,cell signaling,apoptosis,and the DNA damage response.It was reported that RUVBL2 interacts with proteins like β-catenin,c-Myc,and ATF2,[43-45]the former two are believed to contribute to HBV- or HCV-induced liver pathogenesis,including liver cell damage,viral replication and carcinogenesis.[46-49]The mechanism of action of RUVB2 on virus replication remains unclear.As to in fluenza A virus,Kakugawa et al[50]reported that RUVBL2 interacts with the viral nucleoprotein and interrupts its oligomerization,a critical step in the assembly of viral replication complexes,thereby suppressing viral replication in embryonic kidney cells.In this study,our novel finding reveals the association between RUVBL2 and HBx,which may have important anti-HBV functions.RUVBL2 might play this role through the same mechanism.It acts in the in fluenza A virus,but the underlying mechanism is still unclear and needs further investigation.
In summary,we studied the biological functions of HBx protein,and found that HBV replication was significantly inhibited by the knockdown of HBx in vitro.Using a global quantitative proteomic approach,we identified 12 proteins with altered expression after treatment with HBx siRNA.These findings extend our knowledge and deepen understanding of the molecular events of HBV replication mediated by HBx.This is only the first step of our study; more work is needed to reveal the underlying mechanisms of particular proteins in HBV pathogenesis,and identify novel targets for anti-HBV therapeutic intervention.
Funding:This study was supported by grants from the Zhejiang Provincial Natural Science Foundation (Y207465) and the National Basic Research Program of China (973 Program) (2009CB522403).
Ethical approval:Not needed.
Contributors:XHY and ZSS proposed the study.XHY and XCY wrote the first draft.All authors contributed to the design and interpretation of the study and to further drafts.ZSS is the guarantor.
Competing interest:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Nguyen DH,Ludgate L,Hu J.Hepatitis B virus-cell interactions and pathogenesis.J Cell Physiol 2008;216:289-294.
2 Parkin DM,Bray F,Ferlay J,Pisani P.Estimating the world cancer burden:Globocan 2000.Int J Cancer 2001;94:153-156.
3 Parkin DM,Bray F,Ferlay J,Pisani P.Global cancer statistics,2002.CA Cancer J Clin 2005;55:74-108.
4 Tsai WL,Chung RT.Viral hepatocarcinogenesis.Oncogene 2010;29:2309-2324.
5 Keasler VV,Hodgson AJ,Madden CR,Slagle BL.Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo.J Virol 2007;81:2656-2662.
6 Tang H,Oishi N,Kaneko S,Murakami S.Molecular functions and biological roles of hepatitis B virus x protein.Cancer Sci 2006;97:977-983.
7 Klein NP,Schneider RJ.Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras.Mol Cell Biol 1997;17:6427-6436.
8 Bouchard MJ,Wang LH,Schneider RJ.Calcium signaling by HBx protein in hepatitis B virus DNA replication.Science 2001;294:2376-2378.
9 Bouchard MJ,Wang L,Schneider RJ.Activation of focal adhesion kinase by hepatitis B virus HBx protein:multiple functions in viral replication.J Virol 2006;80:4406-4414.
10 Leupin O,Bontron S,Schaeffer C,Strubin M.Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death.J Virol 2005;79:4238-4245.
11 Oshikawa O,Tamura S,Kawata S,Ito N,Tsushima H,Kiso S,et al.The effect of hepatitis B virus X gene expression on response to growth inhibition by transforming growth factorbeta 1.Biochem Biophys Res Commun 1996;222:770-773.
12 Yoo YD,Ueda H,Park K,Flanders KC,Lee YI,Jay G,et al.Regulation of transforming growth factor-beta 1 expression by the hepatitis B virus (HBV) X transactivator.Role in HBV pathogenesis.J Clin Invest 1996;97:388-395.
13 Norton PA,Reis HM,Prince S,Larkin J,Pan J,Liu J,et al.Activation offibronectin gene expression by hepatitis B virus x antigen.J Viral Hepat 2004;11:332-341.
14 Lara-Pezzi E,Majano PL,Yánez-Mó M,Gómez-Gonzalo M,Carretero M,Moreno-Otero R,et al.Effect of the hepatitis B virus HBx protein on integrin-mediated adhesion to and migration on extracellular matrix.J Hepatol 2001;34:409-415.
15 Tong A,Gou L,Lau QC,Chen B,Zhao X,Li J,et al.Proteomic profiling identifies aberrant epigenetic modifications induced by hepatitis B virus X protein.J Proteome Res 2009;8:1037-1046.
16 Wang J,Jiang D,Zhang H,Lv S,Rao H,Fei R,et al.Proteome responses to stable hepatitis B virus transfection and following interferon alpha treatment in human liver cell line HepG2.Proteomics 2009;9:1672-1682.
17 Sells MA,Chen ML,Acs G.Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA.Proc Natl Acad Sci U S A 1987;84:1005-1009.
18 Sells MA,Zelent AZ,Shvartsman M,Acs G.Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions.J Virol 1988;62:2836-2844.
19 Shi M,Zhang Y,Zhu YH,Zhang J,Xu WJ.Comparison of real-time polymerase chain reaction with the COBAS Amplicor test for quantitation of hepatitis B virus DNA in serum samples.World J Gastroenterol 2008;14:479-483.
20 Yan JX,Wait R,Berkelman T,Harry RA,Westbrook JA,Wheeler CH,et al.A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry.Electrophoresis 2000;21:3666-3672.
21 Cheng J,Zhou L,Xie QF,Xie HY,Wei XY,Gao F,et al.The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells.Proteomics 2010;10:1557-1572.
22 Henkler F,Hoare J,Waseem N,Goldin RD,McGarvey MJ,Koshy R,et al.Intracellular localization of the hepatitis B virus HBx protein.J Gen Virol 2001;82:871-882.
23 Lambert C,Prange R.Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein:Implications for translocational regulation.Proc Natl Acad Sci U S A 2003;100:5199-5204.
24 Gonzalez O,Fontanes V,Raychaudhuri S,Loo R,Loo J,Arumugaswami V,et al.The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production.Hepatology 2009;50:1756-1764.
25 Girard S,Vossman E,Misek DE,Podevin P,Hanash S,Bréchot C,et al.Hepatitis C virus NS5A-regulated gene expression and signaling revealed via microarray and comparative promoter analyses.Hepatology 2004;40:708-718.
26 Huang Y,Staschke K,De Francesco R,Tan SL.Phosphorylation of hepatitis C virus NS5A nonstructural protein:a new paradigm for phosphorylation-dependent viral RNA replication?Virology 2007;364:1-9.
27 Zhang SM,Sun DC,Lou S,Bo XC,Lu Z,Qian XH,et al.HBx protein of hepatitis B virus (HBV) can form complex with mitochondrial HSP60 and HSP70.Arch Virol 2005;150:1579-1590.
28 Liu K,Qian L,Wang J,Li W,Deng X,Chen X,et al.Twodimensional blue native/SDS-PAGE analysis reveals heat shock protein chaperone machinery involved in hepatitis B virus production in HepG2.2.15 cells.Mol Cell Proteomics 2009;8:495-505.
29 Ciocca DR,Clark GM,Tandon AK,Fuqua SA,Welch WJ,McGuire WL.Heat shock protein hsp70 in patients with axillary lymph node-negative breast cancer:prognostic implications.J Natl Cancer Inst 1993;85:570-574.
30 Malusecka E,Zborek A,Krzyzowska-Gruca S,Krawczyk Z.Expression of heat shock proteins HSP70 and HSP27 in primary non-small cell lung carcinomas.An immunohistochemical study.Anticancer Res 2001;21:1015-1021.
31 Kaur J,Ralhan R.Differential expression of 70-kDa heat shock-protein in human oral tumorigenesis.Int J Cancer 1995;63:774-779.
32 Lim SO,Park SG,Yoo JH,Park YM,Kim HJ,Jang KT,et al.Expression of heat shock proteins (HSP27,HSP60,HSP70,HSP90,GRP78,GRP94) in hepatitis B virus-related hepatocellular carcinomas and dysplastic nodules.World J Gastroenterol 2005;11:2072-2079.
33 Zylicz M,King FW,Wawrzynow A.Hsp70 interactions with the p53 tumour suppressor protein.EMBO J 2001;20:4634-4638.
34 Zhao ZG,Shen WL.Heat shock protein 70 antisense oligonucleotide inhibits cell growth and induces apoptosis in human gastric cancer cell line SGC-7901.World J Gastroenterol 2005;11:73-78.
35 Bedard K,Szabo E,Michalak M,Opas M.Cellular functions of endoplasmic reticulum chaperones calreticulin,calnexin,and ERp57.Int Rev Cytol 2005;245:91-121.
36 Michalak M,Groenendyk J,Szabo E,Gold LI,Opas M.Calreticulin,a multi-process calcium-buffering chaperone of the endoplasmic reticulum.Biochem J 2009;417:651-666.
37 Zhou Y,Frey TK,Yang JJ.Viral calciomics:interplays between Ca2+and virus.Cell Calcium 2009;46:1-17.
38 Bergqvist A,Rice CM.Transcriptional activation of the interleukin-2 promoter by hepatitis C virus core protein.J Virol 2001;75:772-781.
39 Kinoshita S,Su L,Amano M,Timmerman LA,Kaneshima H,Nolan GP.The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells.Immunity 1997;6:235-244.
40 Li Y,Boehning DF,Qian T,Popov VL,Weinman SA.Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+uniporter activity.FASEB J 2007;21:2474-2485.
41 D'Agostino DM,Bernardi P,Chieco-Bianchi L,Ciminale V.Mitochondria as functional targets of proteins coded by human tumor viruses.Adv Cancer Res 2005;94:87-142.
42 Kepp O,Senovilla L,Galluzzi L,Panaretakis T,Tesniere A,Schlemmer F,et al.Viral subversion of immunogenic cell death.Cell Cycle 2009;8:860-869.
43 Bauer A,Chauvet S,Huber O,Usseglio F,Rothbacher U,Aragnol D,et al.Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity.EMBO J 2000;19:6121-6130.
44 Cho SG,Bhoumik A,Broday L,Ivanov V,Rosenstein B,Ronai Z.TIP49b,a regulator of activating transcription factor 2 response to stress and DNA damage.Mol Cell Biol 2001;21:8398-8413.
45 Wood MA,McMahon SB,Cole MD.An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc.Mol Cell 2000;5:321-330.
46 Kim CW,Yoon SK,Jung ES,Jung CK,Jang JW,Kim MS,et al.Correlation of hepatitis B core antigen and beta-catenin expression on hepatocytes in chronic hepatitis B virus infection:relevance to the severity of liver damage and viral replication.J Gastroenterol Hepatol 2007;22:1534-1542.
47 Su F,Theodosis CN,Schneider RJ.Role of NF-kappaB and myc proteins in apoptosis induced by hepatitis B virus HBx protein.J Virol 2001;75:215-225.
48 Tien LT,Ito M,Nakao M,Niino D,Serik M,Nakashima M,et al.Expression of beta-catenin in hepatocellular carcinoma.World J Gastroenterol 2005;11:2398-2401.
49 Zhang Z,Harris D,Pandey VN.The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus.J Virol 2008;82:5761-5773.
50 Kakugawa S,Shimojima M,Neumann G,Goto H,Kawaoka Y.RuvB-like protein 2 is a suppressor of in fluenza A virus polymerases.J Virol 2009;83:6429-6434.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Predictors of patient survival following living donor liver transplantation
- Hepatocellular carcinoma HepG2 cell apoptosis and caspase-8 and Bcl-2 expression induced by injectable seed extract of Coix lacryma-jobi
- A new scoring system for assessment of liver function after successful hepatectomy in patients with hepatocellular carcinoma
- Relationship between alcohol consumption and clinical manifestation of patients with fatty liver:a single-center study
- Oncofetal antigen glypican-3 as a promising early diagnostic marker for hepatocellular carcinoma
- Necessity and indications of invasive treatment for Budd-Chiari syndrome
