Methylprednisolone inhibits activated CD4+ T cell survival promoted by toll-like receptor ligands
2010-12-14YouShengLuLiYongPuXiangChengLiandXueHaoWang
You-Sheng Lu, Li-Yong Pu, Xiang-Cheng Li and Xue-Hao Wang
Nanjing, China
Methylprednisolone inhibits activated CD4+T cell survival promoted by toll-like receptor ligands
You-Sheng Lu, Li-Yong Pu, Xiang-Cheng Li and Xue-Hao Wang
Nanjing, China
(Hepatobiliary Pancreat Dis Int 2010; 9: 376-383)
methylprednisolone;toll-like receptor;CD4+T lymphocytes;NF-κB
Introduction
Methylprednisolone (MP) has been widely used as an anti-in fl ammatory and immunosuppressive agent for a long time in clinical applications,especially in the fi eld of organ transplantation.[1]Its effects have mainly been attributed to the suppression of T cells through at least two pathways: one regulating the differentiation of CD4+T cells and the other regulating the survival or proliferation rate of CD4+T cells. Still,several issues concerning the mechanism of MP remain unclear.[2]
Toll-like receptors (TLRs) mediate the recognition of pathogen-associated molecular patterns (PAMPs) by cells of the innate immune system, allowing the detection of infection and in fl ammation. These PAMPs can promote the survival of activated CD4+T cells indirectly by initiating maturation responses in antigen presenting cells(APCs). For example, T cell receptor (TCR) engagement activates nuclear factor-kappa B (NF-κB), a transcription factor that mediates many in fl ammatory responses and is important in maintaining the survival of activated CD4+T cells. Interestingly, CD4+T cells also express TLRs, suggesting that PAMPs may directly induce the survival of activated CD4+T cells. The function of TLRs on CD4+T cells, however, remains poorly understood.Whether PAMPs are capable of inducing direct functional responses in activated, non-regulatory CD4+T cells or whether TLR-mediated responses in CD4+T cells use the same signaling pathways that have previously been described in APCs is not known.[3,4]
TLR signaling can be initiated through at least two pathways: one that is dependent on the adaptor molecule myeloid differentiation factor 88 (MyD88)and the other that is MyD88-independent. Nevertheless,both pathways lead to the activation of NF-κB andactivator protein 1 (AP-1). The MyD88-dependent and MyD88-independent pathways involve TLR-3 and TLR-9, respectively. In this study, therefore, we used polyinosinic-polycytidylic acid (poly I:C) and CpG oligodeoxynucleotides (CpG DNA) as ligands for TLR-3 and TLR-9, respectively.[5,6]
In this study, activated CD4+T cells express TLR-3,TLR-5, and TLR-9 mRNAs. To assess the functional signi fi cance of this pattern of TLR expression, we added TLR ligands to highly puri fi ed preparations of activated CD4+T cells. Both poly I:C and CpG DNA, ligands for TLR-3 and TLR-9, respectively, directly enhanced the survival of activated CD4+T cells in vitro, but this effect could be inhibited by MP. Both CpG DNA and poly I:C were dependent on the activation of NF-κB, but not AP-1,to mediate the survival of activated CD4+T cells, but this effect could be inhibited by MP. Both CpG DNA and poly I:C upregulated the expression of NF-κB, but not AP-1, and this effect could be inhibited by MP.Neither CpG DNA nor poly I:C had any effect on the proliferation of activated CD4+T cells. These data show that PAMPs directly promote the survival of activated CD4+T cells and that this effect can be inhibited by MP.
MethodsAnimals
Eight- to 10-week-old male inbred BALB/c mice were purchased from Vital River Experimental Animal Co.,Beijing, China. All experiments were conducted in accordance with the guidelines approved by the Chinese Association of Laboratory Animal Care and the standards for animal use and care set by the Institutional Animal Care Committee.
CD4+ T cell puri fi cation
In experiments using BALB/c CD4+T cells,splenocytes and lymph node cells were pooled,erythrocyte-depleted by hypotonic lysis, and incubated with mouse monoclonal antibodies (mAb) against OX-7(CD90.1, IgG1), OX-8 (CD8a, IgG1), OX-42 (CD11b/c,IgG2a), OX-33 (CD45RA, IgG1), and 10/78 (NKR-P1A,IgG1) (BD Pharmingen, San Diego, CA) for 10 minutes on ice. The cells were then washed with phosphate-buffered saline (PBS) before undergoing two consecutive rounds of depletion with goat anti-mouse IgG (H+L) MicroBeads(Miltenyi Biotec, Bergisch Gladbach, Germany). The mouse monoclonal antibodies (IgG) and anti-mouse IgG MicroBeads were used to deplete CD4-lymphocytes such as CD8+T lymphocytes, natural killer (NK) cells,B lymphocytes, monocytes, granulocytes, macrophages,and dendritic cells, and the remained cells were then labeled with a CD4-FITC mAb (GK1.5; BD Biosciences,Mountain View, CA) and a CD25-PE mAb (PC61; BD Biosciences, Mountain View, CA). The labeled cells were sorted into CD25-CD4+populations using a FACSVantage high-speed sorter (BD Biosciences).[6]
CD4+ T cell activation
All CD4+T cell activation was conducted in complete culture medium composed of RPMI 1640(BD Pharmingen, San Diego, CA), 1.5 μmol/L 2-ME(Sigma-Aldrich, St. Louis, MO), 50 μg/ml gentamicin(Life Technologies, Grand Island, NY), and 10% FCS(Shenergy Biocolor, Shanghai, China) at 37 ℃ in 5%CO2. Puri fi ed CD4+T cells were activated in 24-well plates (Pierce Biotechnology, USA) coated with 1.0 μg/ml CD3 mAb (2C11; BD Biosciences) and 1.0 μg/ml CD28 mAb (37.51; BD Biosciences) for 24 hours.[7]
Semiquantitative RT-PCR
Total RNA from activated CD4+T cells was prepared by lysis with RLT buffer (BD Biosciences Pharmingen,USA) and with buffers and columns supplied in the RNeasy Kit with DNaseⅠ (BD Biosciences Pharmingen,USA) in accordance with the manufacturer's instructions.The puri fi ed RNA was then reversed transcribed and ampli fi ed using the TITANIUM One Step RT-PCR Kit(Roche, USA) under nonsaturating conditions. The following PCR cycling conditions were used: one cycle at 95 ℃ for 3 minutes followed by 25-28 cycles at 94.5 ℃for 30 seconds and 60 ℃ for 1 minute and a fi nal cycle at 72 ℃ for 20 minutes. The speci fi c primer sequences for each gene were as follows: TLR-3, 5'-CCC CTC GCT CTT TTT ATG GAC, 3'-CCT GGC CGC TGA GTT TTT GTT C; TLR-5, 5'-CAG CCC CGT GTT GGT AAT A, 3'-CCC GGA ATG AAG AAT GGA G; TLR-9, 5'-CTA TAC AGC CTG CGC GTTCTC TTC, 3'-AGC TTG CGC AGG CGG GTT AGG TTC; G3PDH, 5'-ACC ACA GTC CAT GCC ATC AC, 3'-TCC ACC ACC CTG TTG CTG TA. The PCR products were resolved by 2% agarose gel electrophoresis,stained with ethidium bromide, and imaged with a Gel Doc analyzer (Bio-Rad, Hercules, CA).[8,9]
Expression analysis of NF-κB and AP-1 by Western blotting
Nuclear proteins were extracted from CD4+T cells using a commercial extraction kit (Pierce Biotechnology,USA), and the protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit(Shenergy Biocolor, Shanghai, China). These samples were then used to evaluate the expression of NF-κB and AP-1. Equal amounts of the protein samples (30 μg of nuclear proteins) were subjected to 10% SDS-PAGE,followed by electrophoretic transfer to polyvinylidene di fl uoride (PVDF) membranes (Roche, USA). The membranes were then probed with primary antibodies against NF-κB (Cell Signaling Technology, USA) and AP-1 (BD Biosciences Pharmingen, USA), followed by a horseradish peroxidase-conjugated secondary antibody.Signals were visualized by enhanced chemiluminescence(Pierce Biotechnology, USA).[10,11]
TLR ligands and inhibitors
The CpG oligonucleotide TCC ATG ACG TTC CTG ACG TT (CpG DNA) and poly I:C were purchased from Amersham Biosciences (Arlington Heights, IL). In all experiments, the TLR ligands were dissolved in PBS.
Survival and proliferation analysis
Following activation, puri fi ed CD4+T cells were washed twice in PBS and replated in culture medium at 106/ml. The cultures were left untreated or treated with TLR ligands, MP, inhibitors of NF-κB and AP-1 for the indicated times and at the indicated concentrations.Following incubation, CD4+T cells were washed twice in phosphate-buffered saline (PBS)/2% foetal bovine serum (FBS) and stained with a CD4-allophycocyanin mAb and 7-aminoactinomycin D (7-AAD; BD Pharmingen) or annexin (BD Pharmingen). Survival was assessed by exclusion of either of these two stains. For absolute live cell counts 50 000 CD45.1+splenocytes labeled with anti-CD45.1-PE mAb (A20; BD Biosciences,Mountain View, CA) were also added to stained CD4+T cell sample FACS tubes before FACS analysis. Live CD4+T cell counts were calculated by taking the ratio of the number of CD4+CD45.1-7-ADD-events collected to the number CD45.1+events collected and multiplying by 50 000. Proliferation was measured by carboxy fl uorescein succinimidyl ester (CFSE) dye dilution. At the time of harvest, CFSE-labeled CD4+T cells were washed in cold PBS containing 2% FCS and 0.01% sodium azide,and at least 2×106cells per sample were stained with a combination of a PE-conjugated monoclonal antibody speci fi c for Thy1.2 and a biotin-conjugated monoclonal antibody speci fi c for CD25, followed by Cy-Chromeconjugated streptavidin (Pierce Biotechnology, USA). The vital dye TOPRO-3 (Molecular Probes, Inc.) was added to each sample (1 nmol/L fi nal concentration) before acquisition to distinguish between live and dead cells.Four-color fl ow cytometry was performed on a Becton Dickinson Immunocytometry Systems (San Jose, CA)FACSCalibur dual-laser cytometer using standard Cell QuestTMacquisition/analysis and ModFitTMsoftware,and fl uorescence compensation was achieved using the appropriate single fl uorochrome-labeled samples. We collected 20 000 events, and absolute cell counts were determined by dividing the total number of events by the product of the fl ow rate (0.001 ml/s) and the elapsed time (in seconds). Analysis of CD25 expression and cell division (CFSE- fl uorescence pro fi le) was restricted to the Thy1.2 subset of CFSE-labeled CD4+T cells.[12,13]
Results CD4+ T cells modulate TLR expression in response to TCR stimulation
In order to examine TLR expression patterns in activated CD4+T cells, highly puri fi ed, naive CD4+CD25-T cells were either left to rest for 24 hours or activated for 24 hours with plate-bound anti-CD3 and anti-CD28 mAbs. We then performed RT-PCR under nonsaturating conditions for TLRs known to have naturally occurring ligands. After stimulation, TLR-3, TLR-5 and TLR-9 mRNA levels were upregulated, but poly I:C had no effect on the expression of TLR-3 RNA (Fig. 1A).
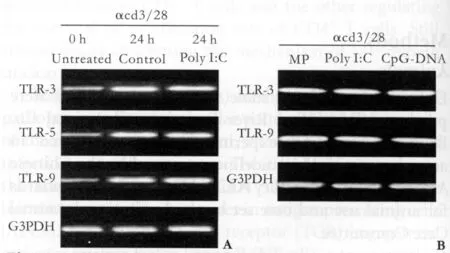
Fig. 1. A: TLR mRNA expression patterns in activated CD4+T cells. T cell-depleted, pooled BALB/c splenocytes and lymph node cells (APC), BALB/c CD4+CD25- T cells, and BALB/c CD4+CD25- T cells were activated for 24 hours with plate-bound anti-CD3 and anti-CD28 mAbs, harvested and lysed to collect total RNA. This RNA was then reverse transcribed and ampli fi ed under nonsaturating conditions using TLR-specific primers or G3PDH primers. The PCR products were resolved by agarose gel electrophoresis and stained with ethidium bromide. The results shown are representative of three independent experiments. B:TLR mRNA expression patterns in activated CD4+ T cells after treatment with poly I:C, CpG DNA, or MP. Activated CD4+ T cells (prepared as in Fig. 1A) were cultured in either poly I: C (90 μg/ml), MP (1 μg/ml) or CpG DNA (30 μmol/L) for 24 hours and then lysed to collect total RNA. The RNA was then reverse transcribed and amplified under nonsaturating conditions with TLR-speci fi c primers or G3PDH primers. The PCR products were resolved by agarose gel electrophoresis and stained with ethidium bromide. The results shown are representative of three independent experiments.
To examine changes in the TLR expression patterns in activated CD4+T cells following treatment with poly I:C, MP or CpG DNA, highly puri fi ed, naive CD4+CD25-T cells were activated for 24 hours with plate-bound anti-CD3 and anti-CD28 mAbs and treated with poly I:C, CpG DNA, or MP for 24 hours or left untreated for 0 hour. We then performed RT-PCR under nonsaturating conditions for TLRs known to have naturally occurring ligands. The mRNA levels of TLR-3, TLR-5 and TLR-9 were unchanged after treatment (Fig. 1B).
Effect of MP on the enhanced survival of activated CD4+ T cells treated with poly I:C or CpG DNA
TLR ligands can directly promote the survival of several cell types, including neutrophils, dendritic cells(DCs), and B cells.[14]Given that our activated CD4+T cells also expressed TLRs and generated signals in response to TLR ligands, we asked whether TLR ligands could directly enhance the survival of these cells. BALB/c CD4+T cells were activated with plate-bound anti-CD3 and anti-CD28 mAbs for 24 hours, puri fi ed by fl ow cytometry to remove all non-CD4+T cells, and replated in unsupplemented culture medium in the absence or presence of TLR ligands and MP. Survival was then assessed at 72 hours following activation. The survival of activated CD4+T cells was increased by poly I:C and CpG DNA from 40.32% to 76.04% and 72.45%(Fig. 2A-C), respectively. In contrast, MP decreased the survival of activated CD4+T cells from 40.32% to 36.08% (Fig. 2C, F).
In order to examine the role of MP in TLR ligandmediated survival responses in activated CD4+T cells,we treated activated CD4+T cells with CpG DNA and poly (I:C) co-cultured with MP and assessed survival.We observed that the increased survival of activated CD4+T cells induced by poly I:C or CpG DNA could be reduced by MP from 76.04% to 35.72% (Fig. 2D) and from 72.45% to 34.80% (Fig. 2E), respectively.
Effect of poly I:C and CpG DNA on the survival of activated CD4+ T cells
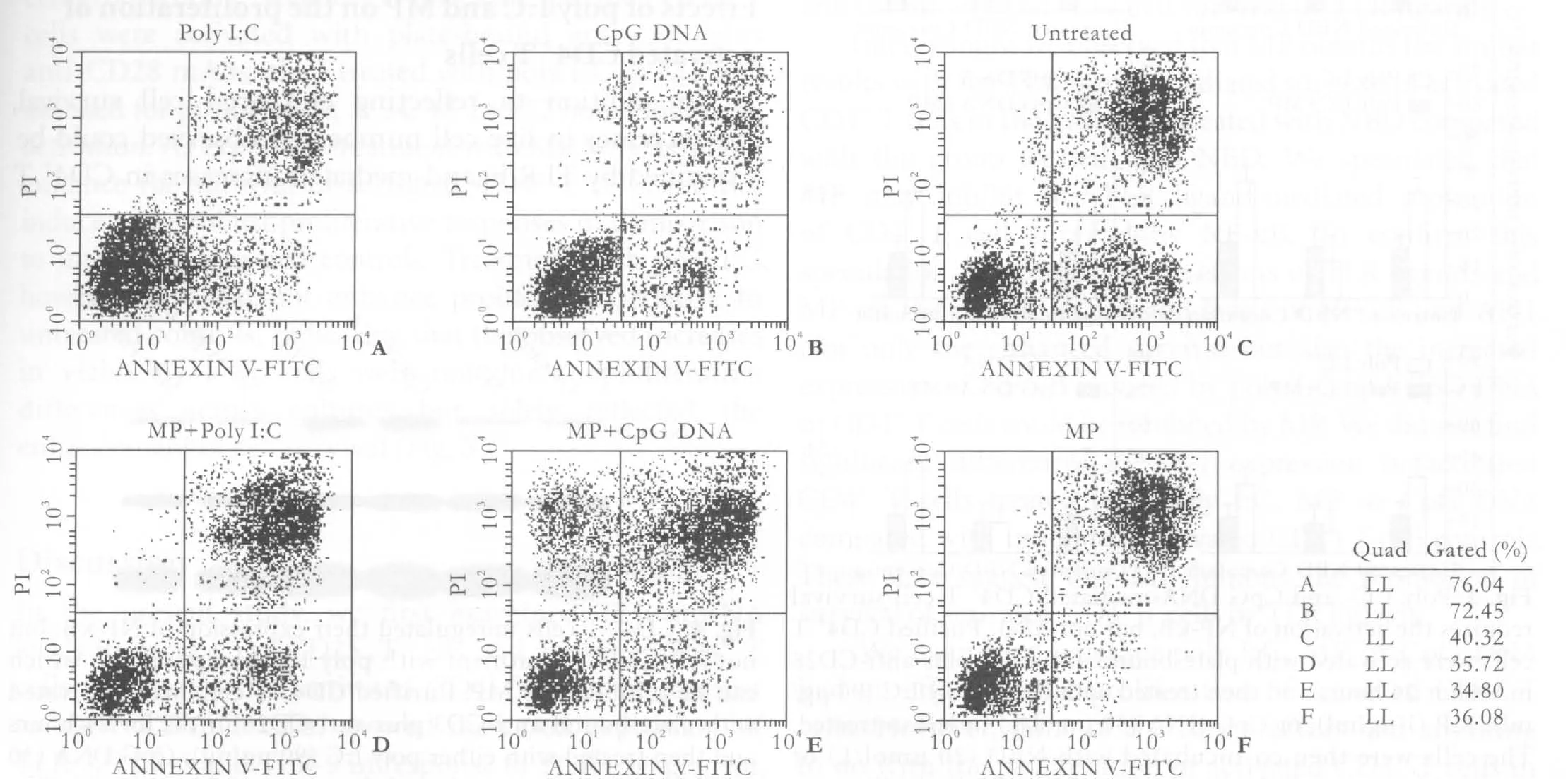
Fig. 2. MP inhibited poly I:C- and CpG DNA-enhanced survival of activated CD4+ T cells. Puri fi ed CD4+ T cells were activated with plate-bound anti-CD3 plus anti-CD28 mAbs for 24 hours and then treated with either poly I:C (90 μg/ml), CpG DNA (30 μmol/L)or MP (1 μg/ml) or left untreated for 72 hours. The cells were stained with annexin and PI and then sorted using a FACSVantage high-speed sorter. The results are representative of three independent experiments.
The signaling pathways of TLRs can be divided into two categories, MyD88-dependent and MyD88-independent, although both induce activation of the transcription factors NF-κB and AP-1.[4,15,16]This activation is known to be associated with survival responses in activated CD4+T cells. Given that poly I:C and CpG DNA were able to enhance the survival of activated CD4+T cells, we asked whether this enhancement was dependent on NF-κB or AP-1. We inhibited NF-κB using the lipid-soluble peptide NBD,which selectively binds to the NF-κB essential modi fi er(NEMO) and blocks its association with the IκB kinases (IKK) IKKα and IKKβ (IKKαβ). The NEMOIKKαβ interaction is necessary for IκB signal-induced phosphorylation; therefore, inhibiting this interaction prevents subsequent IκB degradation and NF-κB translocation to the nucleus.[17]Meanwhile, we inhibited AP-1 using crucumin.[18]Blocking NF-κB activation with NBD inhibited the ability of both poly I:C and CpG DNA to enhance the survival of activated CD4+T cells, but crucumin did not have the same effect. The effects of NBD were dose-dependent.[17]At 20 μmol/L NBD, the effects of the TLR ligands on the survival of activated CD4+T cell were nearly completely reversed.Thus, activation of NF-κB, but not AP-1, is required to mediate the TLR ligand-induced survival of activated CD4+T cells.
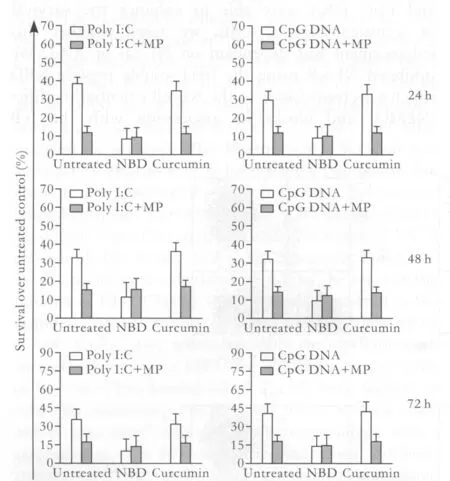
Fig. 3. Poly I:C- and CpG DNA-mediated CD4+ T cell survival requires the activation of NF-κB, but not AP-1. Puri fi ed CD4+ T cells were activated with plate-bound anti-CD3 plus anti-CD28 mAbs for 24 hours and then treated with either poly I:C (90 μg/ml), MP (1 μg/ml), or CpG DNA (30 μmol/L) or left untreated.The cells were then co-incubated with NBD (20 μmol/L) or curcumin (25 μg/ml) for 24, 48 or 72 hours. Cell viability was expressed as the percentage difference in 7-AAD-excluding cells between TLR ligand-treated and untreated cells. The results shown are from four independent experiments (±SEM).
Since TLR ligand-mediated survival in CD4+T cells types was NF-κB dependent, MP could reduce the survival of activated CD4+T cells induced by poly I:C or CpG DNA. We studied whether MP achieve the effect by NF-κB. So, as controls, MP was cultured together with one sample in each group of activated CD4+T cells.We found that, after co-culture of MP, the survival of activated CD4+T cells in the group of untreated and the group treated with crucumin was similar to that in the group treated with NBD-reduced in activated CD4+T cells. Therefore, our results demonstrated that,by NF-κB, MP inhibited the promotion of CD4+T cell survival enhanced by poly I:C and CpG DNA (Fig. 3).
Poly I:C and CpG DNA treatment of activated CD4+ T cells
Given that blocking the activation of NF-κB, but not AP-1, inhibited the ability of both poly I:C and CpG DNA to enhance the survival of activated CD4+T cells, we measured levels of each of these transcription factors following the treatment of activated CD4+T cells with TLR ligands. The protein levels of AP-1 were unchanged by treatment with the TLR ligands relative to every activated CD4+T cell control; however, we observed signi fi cant increases in NF-κB protein levels in activated CD4+T cells treated with CpG DNA and poly I:C compared to control CD4+T cells. Thus, the enhancement of activated CD4+T cell survival mediated by poly I:C and CpG DNA is associated with the speci fi c upregulation of NF-κB, but not AP-1. Furthermore, MP inhibited the upregulation of NF-κB induced by poly I:C and CpG DNA in activated CD4+T cells (Fig. 4).
Effects of poly I:C and MP on the proliferation of activated CD4+ T cells
In addition to re fl ecting enhanced cell survival,the increases in live cell number we observed could be explained by TLR ligand-mediated increases in CD4+Tcell proliferation. To address this issue, puri fi ed CD4+T cells were activated with plate-bound anti-CD3 plus anti-CD28 mAbs, then treated with poly I:C or MP and assessed for proliferation at 24, 48 and 72 hours following activation. As expected, treatment with MP, which did not enhance the survival of activated CD4+T cells, did not induce more robust proliferative responses in comparison to untreated, activated controls. Treatment with poly I:C,however, also did not enhance proliferation relative to untreated controls, indicating that the observed increases in viable CD4+T cells were not due to proliferation differences across cultures but solely re fl ected the enhancement of cell survival (Fig. 5).
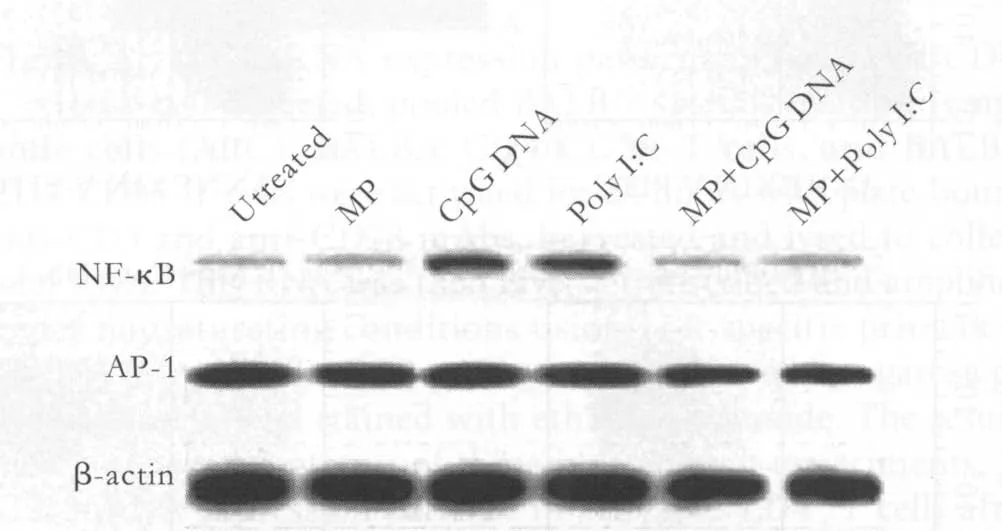
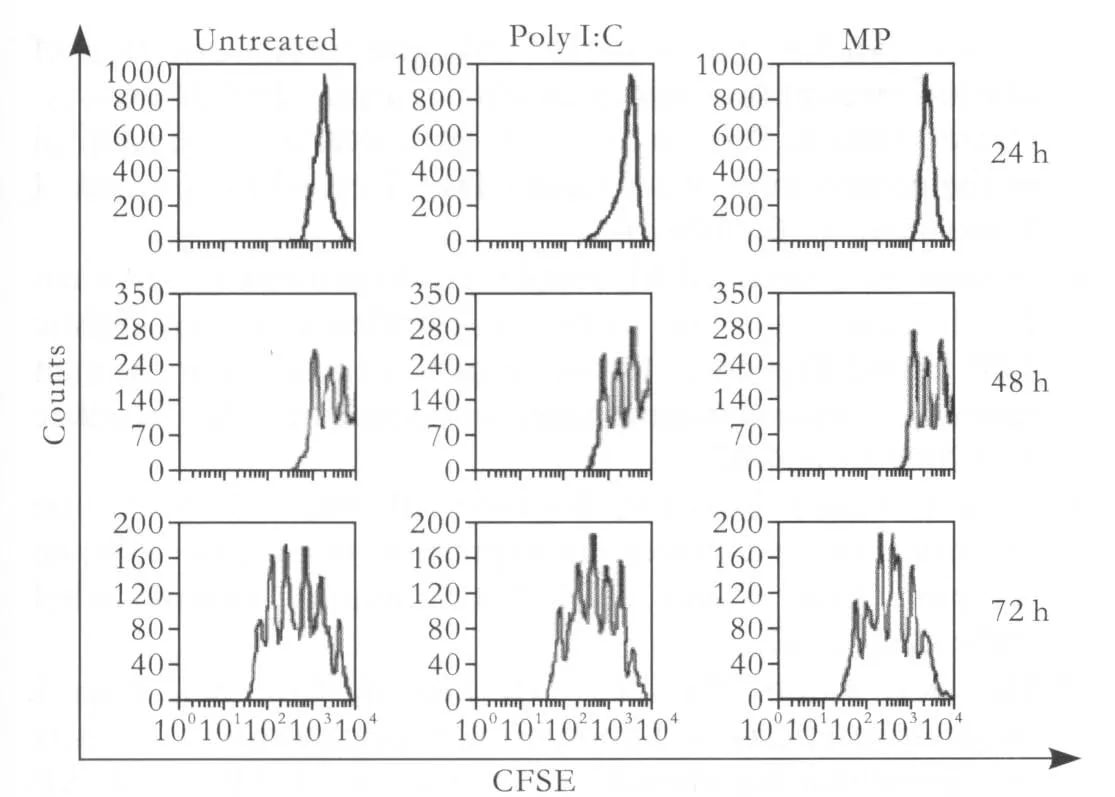
Fig. 5. Poly I:C and MP had no effects on the proliferation of activated CD4+ T cells. Puri fi ed CD4+ T cells labeled with CFSE were activated with plate-bound anti-CD3 plus anti-CD28 mAbs for 24 hours and then cultured in either poly I:C (90 μg/ml) or MP (1 μg/ml) or left untreated for 24, 48, or 72 hours. The cells were assessed for proliferation by CFSE dilution. The histograms showed the CFSE fl uorescence pro fi le of the live Thy1.21 subset of the CFSE-labeled CD4+ T cells. The numbers appearing above each histogram denote each division population, with the undivided T cells residing in the rightmost peak and the T cells having divided eight times residing in the leftmost peak.The experiment shown here is representative of more than 20 separate experiments.
Discussion
In the present study, we fi rst examined TLR mRNA levels in naive CD4+CD25-T cells and activated CD4+T cells and found that activated CD4+T cells, in contrast to naive CD4+T cells, showed increased expression of TLR-3, TLR-5 and TLR-9 in response to TCR engagement.These mRNA levels were used as a proxy for expression levels due to the lack of Abs recognizing the mouse TLRs.Recent studies[19,20]have shown an incomplete picture about TLRs expression in T cells, and several studies[21,22]have obtained different results. In this study, CD4+T cells upregulated their expression of TLR-3, TLR-5 and TLR-9 following activation, and poly I:C, MP and CpG DNA had no effects on the expression of these TLRs.
Since TLR ligands mediated survival in several cell types, we asked whether the same were true in activated CD4+T cells. We chose to treat activated CD4+T cells with poly I:C or CpG DNA. Our data indicate that both poly I:C and CpG DNA can enhance the survival of activated CD4+T cells compared to the untreated,activated CD4+T cell controls. In order to examine the role of MP in TLR ligand-mediated survival responses in activated CD4+T cells, MP was co-cultured in some groups as controls. We observed that TLR ligandenhanced survival in activated CD4+T cells can be inhibited by MP.
Given that TLR ligand-mediated survival in several cell types is dependent on NF-κB or AP-1, we asked whether the same would also be true for activated CD4+T cells. We chose to inhibit NF-κB activation using NBD, a peptide that prevents IκB phosphorylation by selectively preventing the association of IKKαβ with its regulatory protein NEMO.[17]Meanwhile, we inhibited AP-1 activation using crucumin. Treatment with NBD promoted TLR ligand-mediated NF-κB activation to enhance the survival of activated CD4+T cells. In contrast, AP-1 activation was not necessary for the enhancement of CD4+T cell survival by TLR ligands.
Interestingly, we observed that MP obtains the similar results with the TLR ligand-mediated survival of activated CD4+T cells in the groups untreated with NBD compared with the group treated with NBD. We speculated that MP may inhibit the TLR ligand-mediated promotion of CD4+T cell survival by NF-κB. To con fi rm this speculation, we examined the effects of TLR ligands and MP on the protein expression levels of NF-κB and AP-1.Not only the enhanced survival but also the increased expression of NF-κB induced by poly I:C and CpG DNA in CD4+T cells could be inhibited by MP. We did not fi nd signi fi cant differences in AP-1 expression in activated CD4+T cells treated with poly I:C, MP or CpG DNA compared with untreated, activated CD4+T cell controls.These data suggest that MP inhibits the promotion of survival of activated CD4+T cells by TLR ligands.
We examined whether poly I:C, MP or CpG DNA had an effect on the proliferation of activated CD4+T cells. However, none of the three factors had anything to do with the proliferation of activated CD4+T cells in our assay.
In conclusion, MP can inhibit the promotion of survival of activated CD4+T cells induced by TLR ligands. Although MP and TLR ligands have no effects on the expression of activated CD4+T cells TLR mRNA, TLR-mediated survival may increase the expansion and lower the contraction rates of activated CD4+T cells without accentuating proliferation, and MP reduces the TLR-mediated promotion of the survival of activated CD4+T cells while inhibiting the activation of proin fl ammatory cytokines NF-κB. As reported, TLRs can control adaptive immunity and organ transplantation by DCs,[23,25]and TLR ligands directly enhancing the survival of activated CD4+T cells will enhance our knowledge about the role of TLRs in adaptive immunity and organ transplantation. By inhibiting the activation of proin fl ammatory cytokines NF-κB, MP not only directly reduces the survival of activated CD4+T cells, but also suppresses the activity of TLR ligands that enhance the survival of activated CD4+T cells. Taking into account that antigen-speci fi c CD4+T cells or TLRs play an important role in adaptive immunity and organ transplantation, inhibiting the promotion of survival of activated CD4+T cells induced by TLR ligands may be one of important mechanisms of anti-in fl ammatory and anti-rejection effects of MP.
Funding: None.
Ethical approval: Not needed.
Contributors: WXH proposed the study. LYS wrote the fi rst draft and analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. WXH is the guarantor.
Competing interest: No bene fi ts in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Kotsch K, Ulrich F, Reutzel-Selke A, Pascher A, Faber W,Warnick P, et al. Methylprednisolone therapy in deceased donors reduces in fl ammation in the donor liver and improves outcome after liver transplantation: a prospective randomized controlled trial. Ann Surg 2008;248:1042-1050.
2 D'Acquisto F, Paschalidis N, Raza K, Buckley CD, Flower RJ, Perretti M. Glucocorticoid treatment inhibits annexin-1 expression in rheumatoid arthritis CD4+T cells. Rheumatology(Oxford) 2008;47:636-639.
3 Essakalli M, Atouf O, Bennani N, Benseffaj N, Ouadghiri S,Brick C. Toll-like receptors. Pathol Biol (Paris) 2009;57:430-438.
4 Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373-384.
5 Atkinson TJ. Toll-like receptors, transduction-effector pathways,and disease diversity: evidence of an immunobiological paradigm explaining all human illness? Int Rev Immunol 2008;27:255-281.
6 Pu LY, Wang XH, Zhang F, Li XC, Yao AH, Yu Y, et al.Adoptive transfusion of ex vivo donor alloantigen-stimulated CD4(+)CD25(+) regulatory T cells ameliorates rejection of DA-to-Lewis rat liver transplantation. Surgery 2007;142:67-73.
7 Oviedo-Orta E, Perreau M, Evans WH, Potolicchio I. Control of the proliferation of activated CD4+ T cells by connexins. J Leukoc Biol 2010;88:79-86.
8 Mäkelä SM, Strengell M, Pietilä TE, Osterlund P, Julkunen I. Multiple signaling pathways contribute to synergistic TLR ligand-dependent cytokine gene expression in human monocyte-derived macrophages and dendritic cells. J Leukoc Biol 2009;85:664-672.
9 Sang Y, Yang J, Ross CR, Rowland RR, Blecha F. Molecular identi fi cation and functional expression of porcine Toll-like receptor (TLR) 3 and TLR7. Vet Immunol Immunopathol 2008;125:162-167.
10 Han KY, Kwon TH, Lee TH, Lee SJ, Kim SH, Kim J.Suppressive effects of Lithospermum erythrorhizon extracts on lipopolysaccharide-induced activation of AP-1 and NF-kappaB via mitogen-activated protein kinase pathways in mouse macrophage cells. BMB Rep 2008;41:328-333.
11 Valera FC, Queiroz R, Scrideli C, Tone LG, Anselmo-Lima WT.Expression of transcription factors NF-kappaB and AP-1 in nasal polyposis. Clin Exp Allergy 2008;38:579-585.
12 Kondadasula SV, Varker KA, Lesinski GB, Benson DM Jr,Lehman A, Olencki T, et al. Activation of extracellular signaling regulated kinase in natural killer cells and monocytes following IL-2 stimulation in vitro and in patients undergoing IL-2 immunotherapy: analysis via dual parameter fl ow-cytometric assay. Cancer Immunol Immunother 2008;57:1137-1149.
13 Lv XH, Zhou LP, Liu DP, Wang Y, Wang BY, Fu BY, et al.Traditional Chinese medicine Kang Xian Fu Fang I is effective for prophylaxis and treatment of alcoholic liver disease in rats.Hepatobiliary Pancreat Dis Int 2007;6:182-187.
14 Grillot DA, Merino R, Pena JC, Fanslow WC, Finkelman FD,Thompson CB, et al. bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med 1996;183:381-391.
15 Akira S, Yamamoto M, Takeda K. Role of adapters in Toll-like receptor signalling. Biochem Soc Trans 2003;31:637-642.
16 Takeuchi O, Akira S. MyD88 as a bottle neck in Toll/IL-1 signaling. Curr Top Microbiol Immunol 2002;270:155-167.
17 May MJ, D'Acquisto F, Madge LA, Glöckner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science 2000;289:1550-554.
18 Lee SS, Tsai CH, Yang SF, Ho YC, Chang YC. Hypoxia inducible factor-1alpha expression in areca quid chewing-associated oral squamous cell carcinomas. Oral Dis 2010 May 30;[Epub ahead of print].
19 Zarember KA, Godowski PJ. Tissue expression of human Tolllike receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products,and cytokines. J Immunol 2002;168:554-561.
20 Applequist SE, Wallin RP, Ljunggren HG. Variable expression of Toll-like receptor in murine innate and adaptive immune cell lines. Int Immunol 2002;14:1065-1074.
21 Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T, Endres S, Hartmann G. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol 2002;168:4531-4537.
22 Caramalho I, Lopes-Carvalho T, Ostler D, Zelenay S, Haury M, Demengeot J. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. J Exp Med 2003;197:403-411.
23 Rozkova D, Horvath R, Bartunkova J, Spisek R. Glucocorticoids severely impair differentiation and antigen presenting function of dendritic cells despite upregulation of Toll-like receptors.Clin Immunol 2006;120:260-271.
24 Tong HS, Zhang Y, Yuan K, Fu XW. HBsAg loading on dendritic cells in patients with chronic hepatitis B: expressions of phenotypic molecules. Hepatobiliary Pancreat Dis Int 2006;5:56-59.
25 Zhang H, Zheng SS, Jiang GP, Wu LH, Zhu F, Yang ZL.Antitumor effect of immunization with fusion of dendritic cells and hepatocellular carcinoma cells in mice. Hepatobiliary Pancreat Dis Int 2004;3:235-240.
BACKGROUND: Methylprednisolone (MP) can affect the survival of CD4+T lymphocytes and plays an important role in adaptive immune responses; however, its mechanism of action is not clear. Recent studies have shown that tolllike receptors (TLRs) on CD4+T cells can directly modulate adaptive immune responses by affecting the survival and proliferation of activated CD4+T cells. This study aimed to investigate the relationship between MP, TLRs and activated CD4+T cells.
METHODS: We separated and puri fi ed CD4+T cells from mice, activated them in vitro, and co-cultured them with TLR ligands, MP or inhibitors of nuclear factor-kappa B (NF-κB) and activator protein 1 (AP-1). We then assessed CD4+T cell survival
and proliferation and the expression of NF-κB and AP-1.
RESULTS: Activated CD4+T cells showed increased TLR-3 and TLR-9 mRNA expression, but polyinosinic-polycytidylic acid (poly I:C) and MP had no effect on the expression of these mRNAs. Still, poly I:C and CpG oligodeoxynucleotides
(CpG DNA) increased the survival of activated CD4+T cells,whereas MP reduced the survival of activated CD4+T cells and could inhibit the survival effects of poly I:C and CpG DNA. The NF-κB essential modi fi er-binding domain (NBD)inhibited the survival of activated CD4+T cells induced by poly I:C and CpG DNA, but the AP-1 inhibitor crucumin did not have the same effect. The increased expression of NF-κB induced by poly I:C and CpG DNA in activated CD4+T cells could be inhibited by MP, but the same was not true for the increased expression of AP-1 induced by poly I:C and CpG DNA. Finally, the proliferation of activated CD4+T cells was not affected by poly I:C or MP.
CONCLUSION: The survival of activated CD4+T cells is promoted by TLR ligands, but this effect is inhibited by MP.
Author Af fi liations: Liver Transplantation Center, First Af fi liated Hospital(People's Hospital of Jiangsu Province), Nanjing Medical University, Nanjing 210029, China (Lu YS, Pu LY, Li XC and Wang XH)
Xue-Hao Wang, MD, PhD, Liver Transplantation Center, First Af fi liated Hospital, Nanjing Medical University, Nanjing 210029, China (Email: wang_xuehao@126.com)
© 2010, Hepatobiliary Pancreat Dis Int. All rights reserved.
December 24, 2009
Accepted after revision June 12, 2010
"Question mark" is the key to all sciences.
— Honere de Balzac
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Pancreatic duct stones in patients with chronic pancreatitis: surgical outcomes
- Endoscopic management of postcholecystectomy biliary leakage
- Magnetic resonance imaging-guided percutaneous cryoablation of hepatocellular carcinoma in special regions
- Radiofrequency ablation, heat shock protein 70 and potential anti-tumor immunity in hepatic and pancreatic cancers: a minireview
- Endoscopic retrograde cholangiopancreatography outcome from a single referral center in Iran
- Application of a medical image processing system in liver transplantation
