人巨细胞病毒耐药突变及其检测的研究进展
2010-05-30夏长胜
夏长胜,张 正
(北京大学人民医院检验科,北京100044)
HCMV属于疱疹病毒科,β疱疹病毒亚科,巨细胞病毒属,人疱疹病毒5型。HCMV基因组为线性双股DNA分子,长约230 kb,由长单一序列(UL)和短单一序列(US)构成[1,2]。HCMV在自然界广泛存在,正常人群中自然感染普遍。大多数感染者无明显症状,但在婴幼儿和免疫功能受抑制的个体可引起严重疾病。目前常用的抗HCMV药物主要有更昔洛韦(GCV)、磷甲酸钠(FOS)和西多福韦(CDV)等。免疫抑制患者需长期进行抗病毒治疗,使得巨细胞病毒的耐药成为一个严重的问题。临床耐药株大多分离自HCMV感染久治不愈患者的标本[3]。抗病毒药物有毒副作用,如GCV可使中性粒细胞减少,易引发致命性条件感染,而FOS和CDV则有严重的肾毒性。因此,临床在长期使用抗HCMV药物治疗时需监测HCMV的耐药性,以便指导临床制定合理的治疗方案。本文就人巨细胞病毒耐药的分子机制和耐药突变的检测方法进行综述。
1 人巨细胞病毒耐药突变的分子机制
1.1 HCMV UL97基因耐药突变
UL97基因全长2124个碱基,编码707个氨基酸组成的磷酸转移酶[1],其基序具有蛋白激酶特征,并且和其它疱疹病毒的激酶基因具有同源性。GCV是第一个被开发及应用最普遍的治疗HCMV感染的高效药物,它的选择性抗病毒活性依赖于UL97激酶对其进行初始磷酸化。因此UL97基因突变可以引起HCMV对GCV发生耐药。临床绝大多数的GCV耐药株是由于UL97基因突变。UL97基因多个位点突变均可发生GCV的耐药,具体如图1[4](A)及表1所示[5-18]。临床分离的HCMV GCV耐药株是由于UL97基因的460位、520位及590位到607位密码子发生突变引起,绝大多数为发生了M460V、M460I、A594V和 L595S突变所致,其分别占临床GCV耐药分离株的14%、9%、29%和23%[19]。
1.2 HCMV UL54基因耐药突变
UL54基因全长3729个碱基,编码由1242个氨基酸组成的DNA聚合酶[1]。GCV、FOS和CDV均可作为UL54的替代底物发挥抗病毒作用。UL54基因耐药突变可表现为耐GCV、CDV及FOS[20-26],其突变位于高度保守区,如图1(B)及表1所示。UL54耐GCV突变大多同时交叉耐CDV[21,22]。有意思的是这种耐药突变主要是由于长期使用GCV筛选获得,而非使用CDV获得。
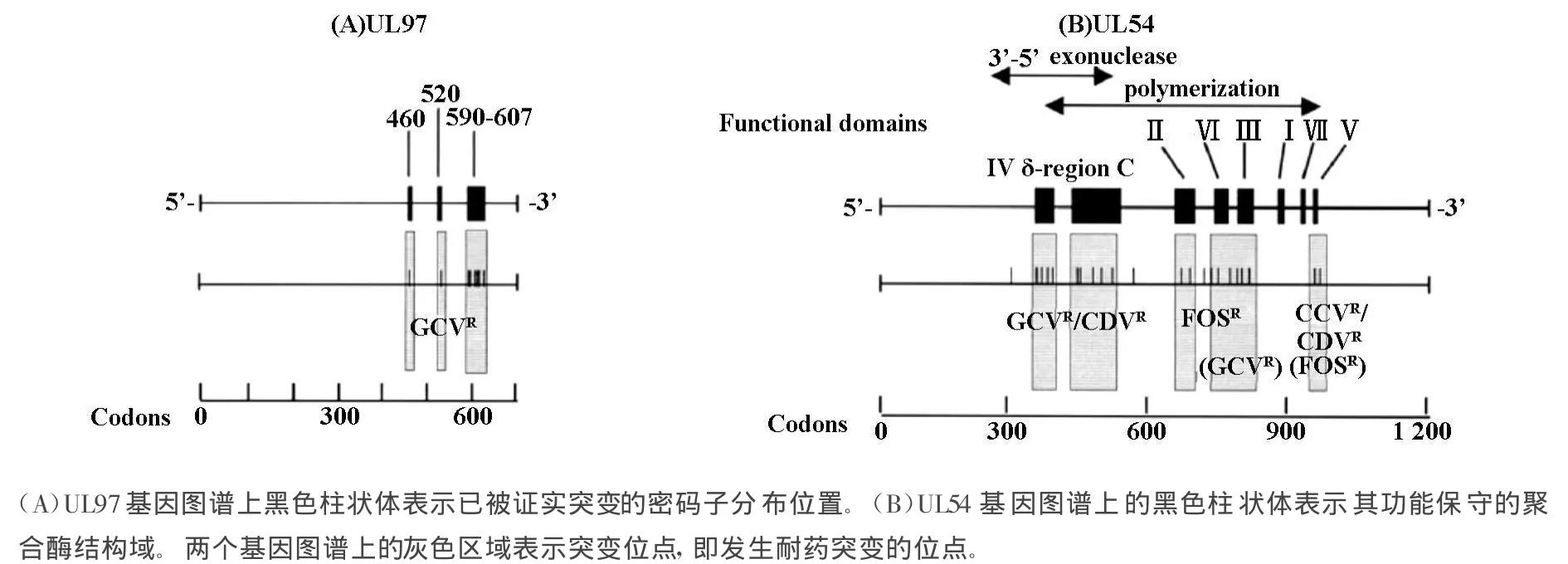
图1 UL97和UL54基因突变图解
2 人巨细胞病毒耐药的检测
2.1 表型检测
表型检测首先需分离培养病毒,然后在系列浓度药物的细胞培养中检测病毒的复制情况。表型检测中的空斑减少试验(PRA)是评价HCMV耐药性的金标法[27]。PRA即病毒在系列浓度药物的培养基中生长7-10天,然后根据各药物浓度下的病毒蚀斑数来确定药物的半数抑制浓度(IC50)。此方法获得实验结果周期较长超过1个月。表型检测除了可检测病毒的蚀斑数,还可通过利用PCR检测病毒DNA、免疫组化检测HCMV pp65抗原表达、流式细胞术检测病毒感染细胞[28]以及报告细胞株检测[29]等方法来确定病毒的耐药性。这些方法可缩短检测周期,但是由于均需要进行病毒培养,从而限制了表型检测在临床上的应用。表现检测中的重组表型检测是确定临床分离株中发现新的基因突变是否引起耐药的参考方法。重组表型检测,即通过同源重组将突变序列转移到参考病毒株上,再进行耐药表型检测。目前技术发展到可以利用独特的限制性内切酶位点进行靶基因位点特异性重组[8],运用报告基因进行快速病毒定量[30]。克隆HCMV全基因组的细菌人工染色体被构建为含UL97的变异体,利用此系统也可以进行重组表型检测[31,32]。表型检测可以确定未知突变能否引起耐药性改变,但表型检测由于需要进行病毒培养,存在如下缺点:①耗时费力;②缺乏统一标准;③室内误差较大。HCMV的耐药表型检测主要运用于研究型实验室,而在临床实验室的应用有限。

表1 HCMV相关的耐药突变
2.2 基因型检测
基因型检测方法主要包括PCR限制性片段长度多态性分析(PCR RFLP)、PCR直接测序法、荧光标记杂交双探针PCR溶解曲线法(FH-PCR-MC)、分子信标PCR溶解曲线法和扩增阻碍突变系统实时PCR法(ARMS RT PCR)等。PCR RFLP方法是根据已知HCMV基因组中与药物抗性有关DNA序列的突变可丢失或创造一个限制性内切酶位点,先进行PCR再利用内切酶酶切分析[5,12,33]。有时基因突变没有改变限制性内切酶的位点时,可通过引物设计引入一个限制性内切酶位点。PCR RFLP检测方法优点是较快速(2天出结果)、敏感(突变株达到10%即可检测到),且不需培养病毒(可直接检测标本);其缺点是只能检测已知的耐药突变,对于未知可能引起的耐药突变无法检测,对于突变株感染和突变株野生株混合感染往往难以区分。此方法依靠凝胶电泳判断结果,敏感性较差,假阴性率较高。由于需要取PCR产物进行酶切及电泳,易造成PCR实验室污染,所以其使用受到一定的限制。直接测序法是指对HCMV的UL97基因或UL54基因进行PCR扩增,再对PCR扩增产物进行测序,通过与参考病毒株DNA序列的比对识别突变位点[28,34-36]。一般认为测序法是较为客观的方法,获得的信息较全面。随着测序服务的专业化和商品化,测序结果变得准确可靠,测序费用变得便宜,因此直接测序法检测HCMV耐药突变具有较高的应用价值。但PCR产物直接测序对感染病毒中优势株的检测效果较好,较难发现非优势株,对混合株的感染检出受限,从而限制了其临床应用。报道显示其仅能检测突变超过25%的突变病毒株[37]。FH-PCR-MC法是基于杂交双探针技术和荧光共振能量转移(FRET)原理,结合融解曲线分析的一种突变检测方法。杂交双探针荧光PCR完成后,以恒定的变温速率缓慢地升高扩增产物的温度,可测定PCR产物的解链温度(Tm值)。由于野生株和变异株的解链温度不同,借此可识别。Göhring等[38-39]利用此种方法成功检测HCMV UL97基因多个密码子的耐药突变。该方法能实时监测PCR后解链过程中荧光信号的变化,操作简便,特异性和准确性较好,能区分野生型、不同变异类型和混合型,但相对敏感性略低,不能对变异株在病毒群体中的比例进行定量,且容易出现假阳性结果。分子信标PCR溶解曲线法和FH-PCR-MC法检测突变的原理相似,不同的是利用分子信标替代了双探针。分子信标不像双探针,由于其茎部不能和模板完全配对结合,因此稳定性较差。Yeo等[40]成功建立了检测UL97基因460密码子突变的分子信标PCR溶解曲线法。除了上述方法,本实验室建立了ARMS PCR结合SYBR GreenⅠ的实时PCR法[41]。实时ARMS PCR方法的关键技术在于引物的设计。一般检测单个位点突变需设计3条引物,1条为公共引物,1条引物为针对突变模板的突变引物,另外一条为针对混合模板的引物。公共引物结合突变引物要求能有效扩增突变模板,对于野生模板不扩增或低效扩增;公共引物结合针对混合模板的引物则可以同时扩增突变和野生模板。ARMS PCR引物设计一般采用引物3'端最后一个碱基错配可引起产物锐减的原理进行设计。但仅一个碱基错配往往不能有效阻止野生株的扩增,此时需要在引物3'端倒数第二个碱基上引入额外的错配碱基才可能达到有效阻止野生株的扩增。ARMS PCR需要试验验证不同突变引物的效果,然后筛选最佳的突变引物[42,43]。ARMS实时PCR则是利用SYBR GreenⅠ、TaqMan探针或分子信标等进行实时监测。SYBR GreenⅠARMS PCR方法简单、费用低廉,但其易受非特异性扩增产物和引物二聚体的干扰,特异性不高。运用TaqMan探针或分子信标的ARMS实时PCR则排除非特异性扩增和引物二聚体的影响,采用突变引物及针对混合模板的引物、公共引物和公共探针或公共分子信标可对病毒突变株占病毒总量的百分比进行检测,并可对突变株进行准确定量[44,45]。总之,基因型检测不需病毒培养,可直接采用临床标本提取DNA检测,比较简便快捷。但基因型检测方法除了直接测序法,其它方法均只能检测已知位点的耐药突变,对于未知的突变信息无法了解。
随着器官、骨髓移植术的开展以及艾滋病的流行等,免疫受抑个体将不断增加,HCMV感染者会越来越多。抗病毒药物的广泛使用将使得HCMV的耐药性变成严重的问题。临床必须不断了解HCMV耐药机制和检测方法,从而监测HCMV的耐药性和制定治疗方案。研究型实验室可以通过表型检测方法确定新的突变是否引起耐药;普通临床实验室则可利用基因型检测方法检测已知的耐药突变,或是直接测序法了解更多的突变信息。
[1]Chee MS,Bankier AT,Beck S,et al.Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169[J].CurrTop Microbiol Immunol,1990,154:125.
[2]Dolan A,Cunningham C,Hector RD,et al.Genetic content of wild-type human cytomegalovirus[J].J Gen Virol,2004,85(Pt 5):1301.
[3]LITTER E,STUART AD,CHEE MS.Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nueleoside analogue ganciclovir[J].Nature,1992,358:160.
[4]Baldanti F,Lurain N,Gerna G.Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs[J].Hum Immunol,2004,65(5):403.
[5]Chou S,Erice A,Jordan MC,et al.Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance[J].J Infect Dis,1995,171(3):576.
[6]Lurain NS,Spafford LE,Thompson KD.Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir[J].J Virol,1994,68:4427.
[7]Hanson MN,Preheim LC,Chou S,et al.Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir[J].Antimicrob Agents Chemother,1995,39:1204.
[8]ChouS,Waldemer RH,Senters AE,et al.Cytomegalovirus UL97 phosphotransferase mutations that affect susceptibility to ganciclovir[J].J Infect Dis,2002,185(2):162.
[9]Sullivan V,Talarico CL,Stanat SC,et al.A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells[J].Nature,1992,358:162.
[10]Chou S,Meichsner CL.A nine-codon deletion mutation in the cytomegalovirus UL97 phosphotransferase gene confers resistance to ganciclovir[J].Antimicrob Agents Chemother,2000,44:183.
[11]Wolf DG,Smith IL,Lee DJ,et al.Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma[J].J Clin Invest,1995,95:257.
[12]Baldanti F,Silini E,Sarasini A,et al.A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate[J].J Virol,1995,69:796.
[13]Baldanti F,Michel D,Simoncini L,et al.Mutations in the UL97 ORF of ganciclovir-resistant clinical cytomegalovirus isolates differentially affect GCV phosphorylation as determined in a recombinant vaccinia virus system[J].Antiviral Res,2002,54(1):59.
[14]Khan FR,Mori S,Eizuru Y,et al.Genetic analysis of a ganciclovir-resistant human cytomegalovirus mutant[J].Antiviral Res,1998,40:95.
[15]Hantz S,Michel D,Fillet AM,et al.Early selection of a new UL97 mutant with a severe defect of ganciclovir phosphorylation after valaciclovir prophylaxis and short-term ganciclovir therapy in a renal transplant recipient[J].Antimicrob Agents Chemother,2005,49(4):1580.
[16]Chou S,Marousek G,Guentzel S,et al.Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease[J].J Infect Dis,1997,176:786.
[17]Marfori JE,Exner MM,Marousek GI,et al.Development of new cytomegalovirus UL97 and DNA polymerase mutations conferring drug resistance after valganciclovir therapy in allogeneic stem cell recipients[J].J Clin Virol,2007,38:120.
[18]Baldanti F,Underwood MR,Talarico CL,et al.The Cys607 >Tyr change in the UL97 phosphotransferase confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised patients[J].Antimicrob Agents Chemother,1998,42:444.
[19]Chou S,Guentzel S,Michels KR,et al.Frequency of UL97 phosphotransferase mutations related to ganciclovirresistance in clinicalcytomegalovirus isolates[J].J Infect Dis,1995,172(1):239.
[20]Baldanti F,Underwood MR,Stanat SC,et al.Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype,while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS[J].J Virol,1996,70(3):1390.
[21]Cihlar T,Fuller MD,Cherrington JM.Characterization of drug resistanceassociated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments[J].J Virol,1998,72(7):5927.
[22]Chou S,Lurain NS,Thompson KD,et al.Viral DNA polymerase mutations associatedwith drug resistance in human cytomegalovirus[J].J Infect Dis,2003,188(1):32.
[23]CihlarT,FullerMD,Mulato AS,et al.A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture[J].Virology,1998,248(2):382.
[24]Weinberg A,Jabs DA,Chou S,et al.Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis[J].J Infect Dis,2003,187(5):777.
[25]Chou S,Marousek G,Parenti DM,et al.Mutation in region III of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy[J].J Infect Dis,1998,178(2):526.
[26]Chou S,Miner RC,Drew WL.A deletionmutation in region V of the cytomegalovirus DNA polymerase sequence confers multidrug resistance[J].J Infect Dis,2000,182:1765.
[27]Landry ML,Stanat S,Biron K,et al.A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates[J].Antimicrob Agents Chemother,2000,44:688.
[28]Scott GM,Isaacs MA,Zeng F,et al.Cytomegalovirus antiviral resistance associatedwith treatment induced UL97(protein kinase)and UL54(DNA polymerase)mutations[J].J Med Virol,2004,74(1):85.
[29]Gilbert C,Boivin G.New reporter cell line to evaluate the sequential emergence of multiple human cytomegalovirus mutations during in vitro drug exposure[J].Antimicrob Agents Chemother,2005,49:4860.
[30]Chou S,Van Wechel LC,Lichy HM,et al.Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene[J].Antimicrob Agents Chemother,2005,49:2710.
[31]Martin M,Gilbert C,Covington E,et al.Characterization of human cytomegalovirus(HCMV)UL97 mutations found in a valganciclovir/oral ganciclovir prophylactic trial by use of a bacterial artificial chromosome containing the HCMV genome[J].J Infect Dis,2006,194(5):579.
[32]Chevillotte M,Schubert A,Mertens T,et al.Fluorescence-based assay for phenotypic characterization of human cytomegalovirus polymerase mutations regarding drug susceptibility and viral replicative fitness[J].Antimicrob Agents Chemother,2009,53(9):3752.
[33]Baldanti F,Simoncini L,Sarasini A,et al.Ganciclovir resistance as a result of oral ganciclovir in a heart transplant recipient with multiple human cytomegalovirus strains in blood[J].Transplantation,1998,66:324.
[34]Sánchez Puch S,Ochoa C,Carballal G,et al.Cytomegalovirus UL97 mutations associated with ganciclovir resistance in immunocompromised patients from Argentina[J].J Clin Virol,2004,30(3):271.
[35]Iwasenko JM,Scott GM,Ziegler JB,et al.Emergence of multiple antiviral resistant CMV strains in a highly immuncompromised patient[J].J Clin Virol,2007,40:152.
[36]Iwasenko JM,Scott GM,Rawlinson WD,et al.Successful valganciclovir treatment of post-transplant cytomegalovirus infection in the presence of UL97 mutation N597D[J].J Med Virol,2009,81(3):507.
[37]Pas SD,de Man RA,Fries E,et al.The dynamics of mutations in the YMDD motif of the hepatitis B virus polymerase gene during and after lamivudine treatment as determined by reverse hybridisation[J].J Clin Virol,2002,25(1):63.
[38]Göhring K,MikelerE,JahnG,et al.Rapid simultaneous detection by real-time PCR of cytomegalovirus UL97 mutations in codons 460 and 520 conferring ganciclovir resistance[J].J Clin Microbiol,2006,44(12):4541.
[39]Göhring K,Mikeler E,Jahn G,et al.Rapid semiquantitative real-time PCR forthe detection of human cytomegalovirus UL97mutations conferring ganciclovir resistance[J].AntivirTher,2008,13(3):461.
[40]Yeo AC,Chan KP,Kumarasinghe G,et al.Rapid detection of codon 460 mutations in the UL97 gene of ganciclovir-resistant cytomegalovirus clinical isolates by real-time PCR using molecular beacons[J].Mol Cell Probes,2005,19(6):389.
[41]Liu JB,Zhang Z.Development of SYBR Green I-based real-time PCR assay for detection of drug resistance mutations in cytomegalovirus[J].J VirolMethods,2008,149(1):129.
[42]Kwok S,Kellogg DE,Mckinney N,et al.Effects of primer-template mismatches on the polymerase chain reaction:human immunodeficiency virus type Imodel studies[J].Nucleic Acids Res,1990,18:999.
[43]Huang MM,ArnheimN,GoodmanMF.Extension of base mispairs by Taq DNA polymerase;implications for single nucleotide discrimination in PCR[J].Nucleic Acids Res,1992,20:4567.
[44]Punia P,Cane P,Teo CG,et al.Quantitation of hepatitis B lamivudine resistant mutants by real-time amplification refractory mutation system PCR[J].J Hepatol,2004,40(6):986.
[45]Ntziora F,Paraskevis D,Haida C,et al.Quantitative detection of the M204V hepatitis B virus minor variants by amplification refractory mutation system real-time PCR combined with molecular beacon technology[J].J Clin Microbiol,2009,47(8):2544.
