SWEET蛋白在植物与病原物互作中的功能研究进展
2025-02-06汪洋一舟郭尽新乔凯彬徐珣刘翔宇王凤婷潘洪玉刘金亮






摘要: SWEET蛋白是一类新型糖转运蛋白, 负责介导细胞中糖类的双向跨膜运输, 在植物生长发育过程中具有韧皮部装载, 植物激素转运, 花、 果实和种子的发育, 植物与病原物之间的互作以及植物和微生物之间共生等重要功能, 是植物与病原物互作过程的重要参与者." 总结SWEET蛋白在生物胁迫中的应答机制以及植物与病原物(细菌、 真菌、 线虫和病毒)互作中SWEET基因的代谢特征、 调控途径及特异性防御反应," 并讨论使用基因编辑工具编辑SWEET基因增强植物对病原物的抗性及其在农业领域中的应用. 为深入研究SWEET蛋白参与植物-病原物互作的机制及利用SWEET基因进行抗病育种提供参考.
关键词:" 糖转运蛋白; SWEET蛋白; SWEET基因; 植物-病原物互作; 生物胁迫; 寄主防御; 抗病育种
中图分类号: Q71" 文献标志码: A" 文章编号: 1671-5489(2025)01-0241-12
Research Advances on Function of SWEET Protein in Plant-Pathogen Interactions
WANG Yangyizhou, GUO Jinxin, QIAO Kaibin," XU Xun, LIU Xiangyu, WANG Fengting, PAN Hongyu, LIU Jinliang
(College of Plant Sciences," Jilin University," Changchun" 130062," China)
收稿日期: 2024-11-26.
第一作者简介:"" 汪洋一舟(1999—)," 男, 汉族, 硕士研究生, 从事植物大分子功能结构的研究, E-mail:" wyyz21@mails.jlu.edu.cn.
通信作者简介:" 刘金亮(1978—)," 男," 汉族," 博士, 教授, 博士生导师, 从事植物大分子功能结构的研究, E-mail:" jlliu@jlu.edu.cn.
基金项目:" 国家自然科学基金(批准号: 32172505)和吉林省自然科学基金(批准号: 20230101156JC).
Abstract:"" SWEET (sugars will eventually be exported transporters) proteins are a novel class of sugar transporter proteins that mediate the bidirectional transmembrane transport of sugars in cells and play important functions in plant growth and development," including phloem loading," phytohormone transport," flower," fruit and seed development," interactions between plants and pathogen, and symbiosis between plants and microorganisms." SWEET proteins are important participant in the process of plant-pathogen interactions. We summarize the response mechanisms of SWEET proteins in biotic stresses, as well as the metabolic characteristics," regulatory pathways and specific defense responses of SWEET genes when plants are infected with different pathogens (bacteria," fungi," nematodes and virus). We also discuss" the use of gene editing tools to edit SWEET genes to enhance plant resistance to pathogens and their application in agriculture. The aim is to provide a reference for in-depth research on the mechanism of" SWEET proteins involvement in plant-pathogen interactions and the use of SWEET genes for disease resistance breeding.
Keywords: sugar transporter protein;" SWEET protein; SWEET gene;" plant-pathogen interaction;" biotic stress;" host defense;" disease resistance breeding
在自然条件下, 植物和微生物通过相互接触并识别从而实现复杂的互作过程. 在长久的植物与病原物互作进程中, 寄主植物建立了一套复杂的双层免疫监测系统用于感知并抵抗来自病原物的威胁, 分别为病原相关分子模式触发免疫(pathogen-associated molecular pattern-triggered immunity," PTI)和效应因子触发免疫(effector-triggered immunity," ETI).
1 SWEET蛋白基本特征和功能
1.1 SWEET蛋白的结构特征和分类
糖转运蛋白有3个主要超家族, 包括MFS超家族(major facilitator superfamily)、 钠依赖性葡萄糖转运蛋白和 SWEETs蛋白.
1.2 SWEET蛋白在植物生理中的功能
SWEET基因在蒺藜苜蓿(Medicago truncatula)中被首次发现, 命名为MtN3, SWEET基因广泛存在于原核生物、 植物、 动物和人类中.
2 SWEET蛋白在植物-病原物互作中的功能
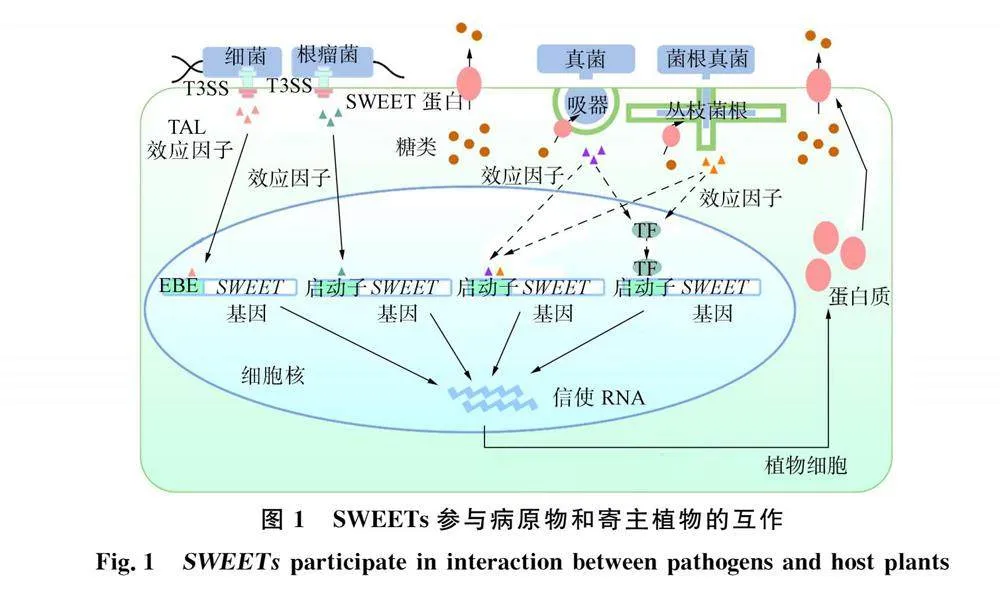
植物编码SWEET蛋白的基因对病原物通常作为感病基因发挥作用, 而大多数病原物在侵入寄主植物时都需从寄主中获取碳源以满足自身生长发育需求. 在病原物和寄主互作过程中, 病原物通过调节寄主植物体内SWEETs的表达水平以影响侵染部位糖外流, 从而帮助自身获取营养, 同时也会影响寄主植物相关防御反应(图1). 为系统了解SWEET蛋白在植物-病原物互作时发挥的功能, 下面主要总结并介绍SWEET蛋白参与植物病原细菌、 真菌、 线虫以及病毒等病原物的相互作用.
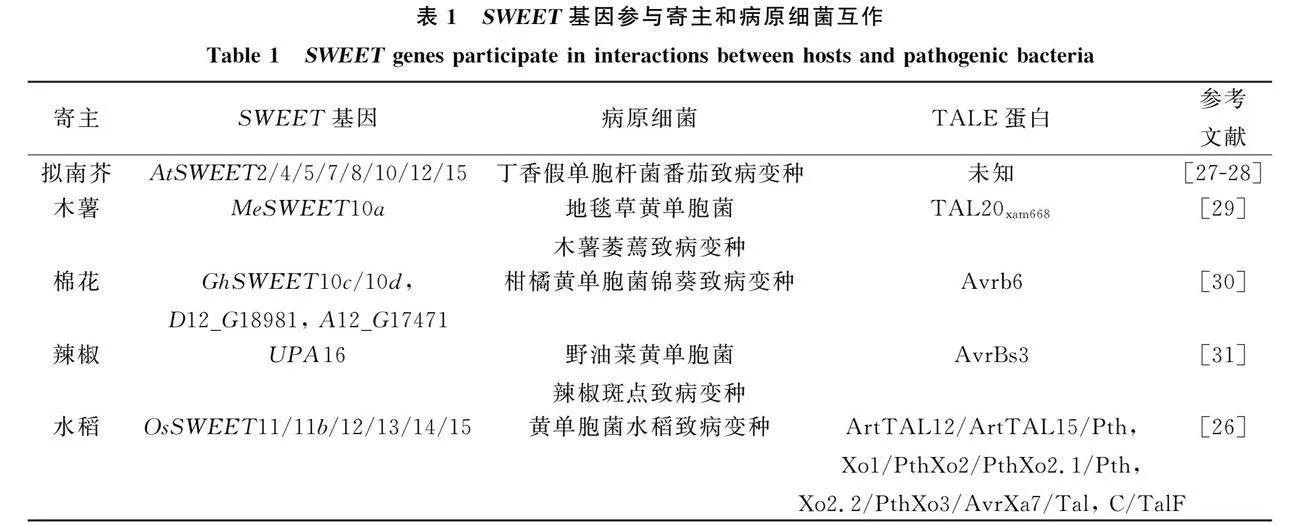
2.1 SWEET蛋白参与植物-病原细菌互作
植物病原细菌侵染寄主可通过Ⅲ型分泌系统(type Ⅲ secretion system, T3SS)将转录激活效应因子(transcription activator-like effectors, TALEs)注入寄主细胞中. TALEs的C端有一个核定位信号域(nuclear localization signal domain," NLS)和激活域(activation domain," AD), 结构中心部位包含1.5~33.5个串联重复序列, 而每个重复序列包含约34个氨基酸. 其中第12位和第13位的重复可变双残基(repeat variant diresidue," RVD)可与多种SWEET基因启动子中效应因子结合元件(effector-binding element," EBE)结合, 诱导SWEET基因表达, 从而转运糖类为病原物提供能量.
2.2 SWEET蛋白参与植物-病原真菌互作
植物病原真菌在侵染寄主时可分泌效应因子直接诱导SWEET基因表达, 也可通过激活转录因子间接诱导SWEET基因表达.
2.3 SWEET蛋白参与植物-线虫互作
根结线虫(Meloidogyne)是一种高度专化的农作物寄生线虫, 主要通过劫持寄主植物的营养物质危害其根部. 在根结线虫侵染植物的过程中, 线虫会诱导植物体内SWEET基因的表达量变化, 尤其在根结部位表达量明显提高, 表明SWEET蛋白参与植物和线虫的互作.
信号途径核心转录因子 ELONGATED HYPOCOTYL5 (HY5)受到南方根结线虫侵染的诱导, 负调控植物对根结线虫的抗性并激活拟南芥AtSWEET11a,AtSWEET12b和AtSWEET15d表达.
2.4 SWEET蛋白参与植物-病毒互作
目前, 仅吉林大学植物与病原物分子互作课题组对SWEET蛋白参与植物-病毒互作进行了研究."" 其中, 马铃薯Y病毒科Y病毒属(Potyvirus)的芜菁花叶病毒(turnip mosaic virus," TuMV)P3蛋白与拟南芥AtSWEET1,AtSWEET4和AtSWEET15蛋白互作.
3 SWEET蛋白参与植物抗病的功能
自开展SWEET蛋白功能研究以来, SWEET基因普遍被认为在植物与病原物互作中作为感病基因(susceptible gene)发挥功能, 其隐形等位基因通常表现为抗病表型. 在病原物侵染植物过程中, 病原能够产生特定效应因子诱导寄主植物中的SWEET基因表达, 从而促进更多的糖类释放到细胞间隙, 为病原物的生长发育提供能量.
随着SWEET基因功能研究的深入开展," 发现在被病原物劫持并为其提供营养外, 部分SWEET蛋白受病原物诱导后会参与增强植物对病原的抗性, 这些SWEET蛋白通过发挥自身的糖转运功能降低质外体中糖类含量, 从而限制病原物的生长. 此外, SWEET蛋白还可通过改变植物体内的糖含量影响防御相关基因的表达, 增强寄主植物的抗性.
3.1 作为感病基因表达产物的SWEET蛋白
水稻中存在大量SWEET基因作为感病基因被病原物利用, 其中OsSWEET11/12 /13/14/15基因在水稻白叶枯病菌侵染时参与病原致病过程, 相关基因表达由黄单胞菌水稻致病变种TAL效应因子(如TalC,AvrXa7,PthXo3和Tal5等)诱导.
综上所述, 大部分植物SWEET蛋白基因能被病菌诱导表达, 并作为感病基因促进病原物侵染寄主, 在表型上表现为植物的易感性增强.
3.2 减少病原可利用糖的SWEET蛋白
腐霉病菌(Pythium irregulare)可引起植物种子、 茎、 根的腐烂和幼苗倒伏, 在拟南芥中, AtSWEET2蛋白主要定位于根表皮液泡膜, 具有转运葡萄糖进入液泡的功能, 在受到腐霉病菌侵染时, AtSWEET2基因会受到显著的诱导表达, 而AtSWEET2突变体植株的根系干质量降低, 且叶片中葡萄糖积累量降低, 植株出现黄化枯萎现象, 此时植株的根系对腐霉的敏感性增强, 说明AtSWEET2可通过限制根系中糖的外排, 从而增强拟南芥对腐霉病菌抗病性.
综上所述, 部分SWEET蛋白可以通过降低质外体中糖含量限制病原物的生长, 从而增强寄主对病原物的抗性.
3.3 诱导植物防御反应的SWEET蛋白
在葡萄(Vitis vinifera)受到腐霉病菌侵染时, 寄主的VvSWEET4基因会受到诱导表达, 而在毛状根中过表达VvSWEET4基因后, 植物对糖的运输能力增强, 同时在高糖水平下, 毛状根中参与类黄酮合成途径的基因表达量上调, 促进了抗真菌特性的黄酮类化合物合成, 从而增强植株对腐霉病菌的抗性.
综上所述, 部分SWEET蛋白导致的糖类物质积累, 不仅可被病原物吸收利用, 同样也可增强植物自身的相关防御反应.
4 SWEET基因在植物抗病育种中的应用
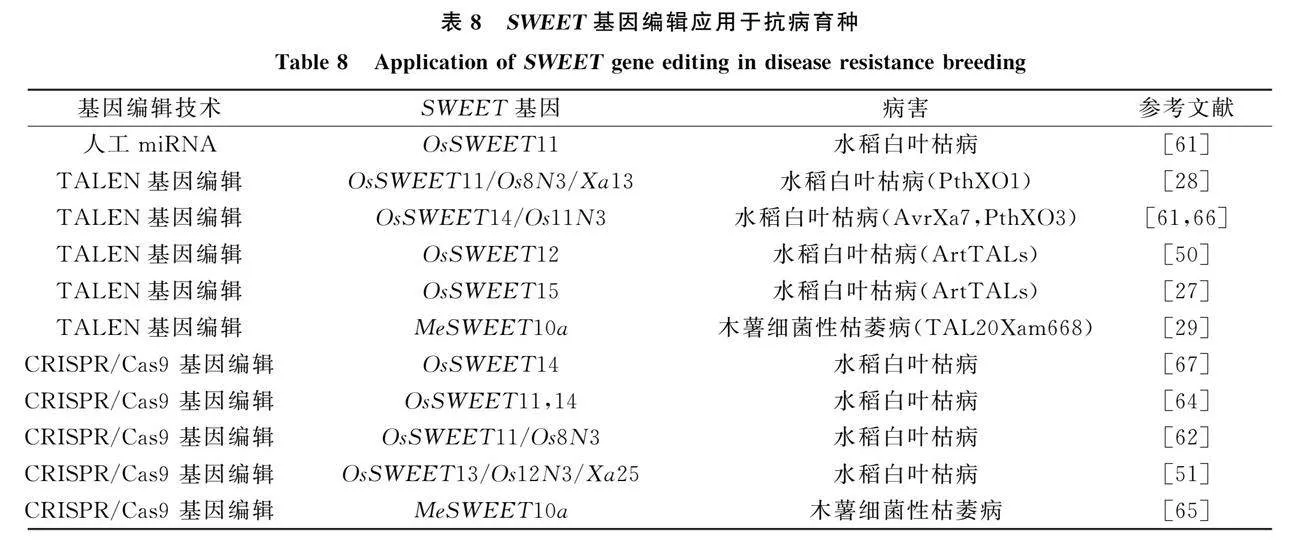
SWEET基因家族广泛参与寄主植物-病原物的相互作用, 但目前仅有少数SWEET基因和病原物的互作机制得到充分解析, 针对这些机制展开的分子生物学改良将有助于植物抗病育种策略的挖掘, 为选育抗病、 高产的优良品系提供理论和技术支持.
目前, 针对SWEET基因的抗病育种方案主要有人工miRNA(artificial microRNA," amiRNA)技术、 转录激活效应因子样核酸酶TALENs(transcription activator-like (TAL) effector nucleases)技术和CRISPR/Cas9技术.
4.1 人工miRNA技术
RNA诱导的基因沉默现象(RNAi)在30多年前首次在植物中被描述, 研究人员通过转录后机制使牵牛花中参与紫色色素合成的基因受到沉默.
高效地使用, 但目前针对SWEET基因使用人工miRNA获得抗病高产理想植株的报道仍较少, 说明相关领域的技术优化仍需深度开发.
4.2 TALENs基因编辑技术
转录激活效应因子样核酸酶(TALENs)蛋白于2009年首次被报道, 来源于植物病原细菌黄单胞菌属(Xanthomonas).
4.3 CRISPR/Cas9基因编辑技术
CRISPR在大肠杆菌(Escherichia coli)基因组中发现并描述了一系列短的重复序列和短序列之间的间隔, 之后在许多细菌和古细菌中也发现了该现象.
目前, CRISPR/Cas9介导的EBEs基因编辑广泛应用于水稻中. 通过CRISPR/Cas9技术同时靶向敲除3个SWEET基因OsSWEET11/13/14的EBE区域, 从而获得对大多数黄单胞菌菌株具有广谱抗性的水稻, 这种编辑方式可在确保水稻获得对黄单胞菌广谱抗性的同时保持产量.
5 总结与展望
近年来, 关于SWEET蛋白在植物和微生物互作尤其是与病原物互作的研究取得了重大进展, 但仍有许多问题需要解决. SWEET蛋白是植物与病原物之间“战斗”的重要参与者, 病原物可诱导SWEET基因的转录控制SWEET蛋白表达, 从而增加寄主植物中碳水化合物含量, 为自身生长发育以及侵染提供能量." SWEET蛋白也会参与调控植物的防御反应, 通过减少侵染部位糖积累阻碍病原物对糖的获取, 同时SWEET基因表达也可使植物体内的糖得到积累, 这些糖类可作为信号分子激活下游信号途径, 从而诱导防御相关基因上调, 抑制病原物对植物侵染. 由于有关SWEET蛋白在植物-病原物互作调控网络以及完整信号通路等方面仍未形成系统性研究, 因此相关领域的深入研究仍需大力开展.
目前, 在已知30多种高等植物的SWEET基因中, 约有10种植物的SWEET基因作为感病基因在植物病原物和寄主互作中发挥作用.
参考文献
[1] ZHANG J, COAKER G, ZHOU J M, et al. Plant Immune Mechanisms: From Reductionistic to Holistic Points of View [J]. Mol Plant, 2020, 13(10): 1358-1378.
[2] NGOU B P M, DING P T, JONES J D G. Thirty Years of Resistance: Zig-Zag through the Plant Immune System [J]. Plant Cell, 2022, 34(5): 1447-1478.
[3] YAO T S, GAI X T, PU Z J, et al. From Functional Characterization to the Application of SWEET Sugar Transporters in Plant Resistance Breeding [J]. J Agric Food Chem, 2022, 70(17): 5273-5283.
[4] LEMOINE R, LA CAMERA S, ATANASSOVA R, et al. Source-to-Sink Transport of Sugar and Regulation by Environmental Factors [J]. Front Plant Sci, 2013, 4: 272-1-272-21.
[5] CHEN H Y, HUH J H, YU Y C, et al. The Arabidopsis Vacuolar Sugar Transporter SWEET2 Limits Carbon Sequestration from Roots and Restricts Pythium infection [J]. Plant J, 2015, 83(6): 1046-1058.
[6] CIERESZKO I. Regulatory Roles of Sugars in Plant Growth and Development [J]. Acta Soc Bot Pol, 2018, 87(2): 3583-1-3583-13.
[7] GUPTA P K. SWEET Genes for Disease Resistance in Plants [J]. Trends Genet, 2020, 36(12): 901-904.
[8] JEENA G S, KUMAR S, SHUKLA R K. Structure, Evolution and Diverse Physiological Roles of SWEET Sugar Transporters in Plants [J]. Plant Mol Biol, 2019, 100(4/5): 351-365.
[9] FORREST L R, KRÄMER R, ZIEGLER C. The Structural Basis of Secondary Active Transport Mechanisms [J]. Biochim Biophys Acta, 2011, 1807(2): 167-188.
[10] JIA B L, ZHU X F, PU Z J, et al. Integrative View of the Diversity and Evolution of SWEET and SemiSWEET Sugar Transporters [J]. Front Plant Sci, 2017, 8: 2178-1-2178-18.
[11] PATIL G, VALLIYODAN B, DESHMUKH R, et al. Soybean (Glycine max) SWEET Gene Family: Insights through Comparative Genomics, Transcriptome Profiling and Whole Genome Re-sequence Analysis [J]. BMC Genomics, 2015, 16: 520-1-520-16.
[12] KRYVORUCHKO I S, SINHAROY S, TORRES-JEREZ I, et al. MtSWEET11, a Nodule-Specific Sucrose Transporter of Medicago truncatula [J]. Plant Physiol, 2016, 171(1): 554-565.
[13] BREIA R, CONDE A, BADIM H, et al. Plant SWEETs: From Sugar Transport to Plant-Pathogen Interaction and More Unexpected Physiological Roles [J]. Plant Physiol, 2021, 186(2): 836-852.
[14] LIN I W, SOSSO D, CHEN L Q, et al. Nectar Secretion Requires Sucrose Phosphate Synthases and the Sugar Transporter SWEET9 [J]. Nature, 2014, 508:" 546-549.
[15] YUAN M, WANG S P. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms [J]. Mol Plant, 2013, 6(3): 665-674.
[16] JONES J D G, VANCE R E, DANGL J L. Intracellular Innate Immune Surveillance Devices in Plants and Animals [J]. Science, 2016, 354: aaf6395-1-aaf6395-8.
[17] JI J L, YANG L M, FANG Z Y, et al. Plant SWEET Family of Sugar Transporters: Structure, Evolution and Biological Functions [J]. Biomolecules, 2022, 12(2): 205-1-205-19.
[18] MIZUNO H, KASUGA S, KAWAHIGASHI H. The Sorghum SWEET Gene Family: Stem Sucrose Accumulation as Revealed through Transcriptome Profiling [J]. Biotechnol Biofuels, 2016, 9: 127-1-127-12.
[19] GAUTAM T, SARIPALLI G, GAHLAUT V, et al. Further Studies on Sugar Transporter (SWEET) Genes in Wheat (Triticum aestivum L.) [J]. Mol Biol Rep, 2019, 46(2): 2327-2353.
[20] MANCK-GÖTZENBERGER J, REQUENA N. Arbuscular mycorrhiza Symbiosis Induces a Major Transcriptional Reprogramming of the Potato SWEET Sugar Transporter Family [J]. Front Plant Sci, 2016, 7: 487-1-487-14.
[21] GUAN Y F, HUANG X Y, ZHU J, et al. RUPTURED POLLEN GRAIN1, a Member of the MtN3/Saliva Gene Family, Is Crucial for Exine Pattern Formation and Cell Integrity of Microspores in Arabidopsis [J]. Plant Physiol, 2008, 147(2): 852-863.
[22] SUN M X, HUANG X Y, YANG J, et al. Arabidopsis RPG1 Is Important for Primexine Deposition and Functions Redundantly with RPG2 for Plant Fertility at the Late Reproductive Stage [J]. Plant Reprod, 2013, 26(2): 83-91.
[23] CHEN L Q, QU X Q, HOU B H, et al. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport [J]. Science, 2012, 335: 207-211.
[24] SEO P J, PARK J M, KANG S K, et al. An Arabidopsis Senescence-Associated Protein SAG29 Regulates Cell Viability under High Salinity [J]. Planta, 2011, 233(1): 189-200.
[25] DURAND M, MAINSON D, PORCHERON B, et al. Carbon Source-Sink Relationship in Arabidopsis thaliana: The Role of Sucrose Transporters [J]. Planta, 2018, 247(3): 587-611.
[26] GUPTA P K, BALYAN H S, GAUTAM T. SWEET Genes and TAL Effectors for Disease Resistance in Plants: Present Status and Future Prospects [J]. Mol Plant Pathol, 2021, 22(8): 1014-1026.
[27] STREUBEL J, PESCE C, HUTIN M, et al. Five Phylogenetically Close Rice SWEET Genes Confer TAL Effector-Mediated Susceptibility to Xanthomonas oryzae pv.oryzae [J]. New Phytol, 2013, 200(3): 808-819.
[28] CHEN L Q, HOU B H, LALONDE S, et al. Sugar Transporters for Intercellular Exchange and Nutrition of Pathogens [J]. Nature, 2010, 468: 527-532.
[29] COHN M, BART R S, SHYBUT M, et al. Xanthomonas axonopodis Virulence Is Promoted by a Transcription Activator-Like Effector Mediated Induction of a SWEET Sugar Transporter in Cassava [J]. Mol Plant-Microbe Interact, 2014, 27(11): 1186-1198.
[30] COX K L, MENG F H, WILKINS K E, et al. TAL Effector Driven Induction of a SWEET Gene Confers Susceptibility to Bacterial Blight of Cotton [J]. Nat Commun, 2017, 8: 15588-1-15588-14.
[31] KAY S, HAHN S, MAROIS E, et al. Detailed Analysis of the DNA Recognition Motifs of the Xanthomonas Type Ⅲ Effectors AvrBs3 and AvrBs3Δrep16 [J]. Plant J, 2009, 59(6): 859-871.
[32] GAO Y, ZHANG C, HAN X, et al. Inhibition of OsSWEET11 Function in Mesophyll Cells Improves Resistance of Rice to Sheath Blight Disease [J]. Mol Plant Pathol, 2018, 19(9): 2149-2161.
[33] GAO Y, XUE C Y, LIU J M, et al. Sheath Blight Resistance in Rice Is Negatively Regulated by WRKY53 via SWEET2a Activation [J]. Biochem Biophys Res Commun, 2021, 585: 117-123.
[34] GAO Y, WANG Z Y, KUMAR V, et al. Genome-Wide Identification of the SWEET Gene Family in Wheat [J]. Gene, 2018, 642: 284-292.
[35] SOSSO D, VAN DER LINDE K, BEZRUTCZYK M, et al. Sugar Partitioning between Ustilago maydis and Its Host Zea mays. L during Infection [J]. Plant Physiol, 2019, 179(4): 1373-1385.
[36] CHONG J L, PIRON M C, MEYER S, et al. The SWEET Family of Sugar Transporters in Grapevine: VvSWEET4 Is Involved in the Interaction with Botrytis cinerea [J]. J Exp Bot, 2014, 65(22): 6589-6601.
[37] ASAI Y, KOBAYASHI Y. Increased Expression of the Tomato SISWEET15 Gene during Grey Mold Infection and the Possible Involvement of the Sugar Efflux to Apoplasm in the Disease Susceptibility [J]. J Plant Pathol Microbiol, 2016, 7(1): 1000329-1-1000329-8.
[38] GEBAUER P, KORN M, ENGELSDORF T, et al. Sugar Accumulation in Leaves of Arabidopsis sweet11/sweet12 Double Mutants Enhances Priming of the Salicylic Acid-Mediated Defense Response [J]. Front Plant Sci, 2017, 8: 1378-1-1378-13.
[39] MIAO H X, SUN P G, LIU Q, et al. Genome-Wide Analyses of SWEET Family Proteins Reveal Involvement in Fruit Development and Abiotic/Biotic Stress Responses in Banana [J]. Sci Rep, 2017, 7(1): 3536-1-3536-15.
[40] WANG L, YAO L N, HAO X Y, et al. Tea Plant SWEET Transporters: Expression Profiling, Sugar Transport, and the Involvement of CsSWEET16 in Modifying Cold Tolerance in Arabidopsis [J]. Plant Mol Biol, 2018, 96(6): 577-592.
[41] SUN M X, ZHANG Z Q, REN Z Y, et al. The GhSWEET42 Glucose Transporter Participates in Verticillium dahliae Infection in Cotton [J]. Front Plant Sci, 2021, 12: 690754-1-690754-13.
[42] LI Y, WANG Y N, ZHANG H, et al. The Plasma Membrane-Localized Sucrose Transporter IbSWEET10 Contributes to the Resistance of Sweet Potato to Fusarium oxysporum [J]. Front Plant Sci, 2017, 8: 197-1-197-15.
[43] WU B H, JIA X Y, ZHU W, et al. Light Signaling Regulates Root-Knot Nematode Infection and Development via HY5-SWEET Signaling [J]. BMC Plant Biol, 2024, 24(1): 664-1-664-12.
[44] 周媛. SWEET糖转运蛋白在南方根结线虫寄生过程中的作用机制研究[D]. 沈阳: 沈阳农业大学," 2020. (ZHOU Y. Studies on the Mechanism of SWEET Sugar Transporers in the Parasitic Process of Meloidogyne incognita[D]. Shenyang: Shenyang Agricultural University, 2020.)
[45] ZHAO D, YOU Y, FAN H Y, et al. The Role of Sugar Transporter Genes during Early Infection by Root-Knot Nematodes [J]. Int J Mol Sci, 2018, 19(1): 302-1-302-15.
[46] 张雅琦. 芜菁花叶病毒p3基因的分子变异及P3蛋白与拟南芥AtSWEET1蛋白的互作研究[D]. 长春: 吉林大学, 2015. (ZHANG Y Q. Molecular Variability of p3 Gene of Turnip mosaic virus and the Interaction between P3 Protein and AtSWEET1 Protein in Arabidopsis thaliana[D]. Changchun: Jilin University, 2015.)
[47] 孙颖, 王艳, 张祥辉, 等. 芜菁花叶病毒编码蛋白与拟南芥AtSWEET1蛋白互作研究 [C]//中国植物病理学会2017年学术年会论文集. 北京:" 中国农业科学技术出版社, 2017: 293. (SUN Y, WANG Y, ZHANG X H, et al. The Interaction between Turnip Mosaic Virus Encoded Proteins and AtSWEET1 Protein in Arabidopsis thaliana [C]//Proceedings of the Annual Meeting of Chinese Society for Plant Pathology. Beijing: China Agricultural Science and Technology Press, 2017: 293.)
[48] 王艳. 芜菁花叶病毒编码蛋白与拟南芥AtSWEET1蛋白互作研究[D]. 长春: 吉林大学, 2017. (WANG Y." The Interaction between Turnip mosaic virus Encoded Proteins and AtSWEET1 Protein in Arabidopsis thaliana[D]. Changchun: Jilin University, 2017.)
[49] 孙玥. 大豆花叶病毒P3和P3N-PIPO蛋白与大豆蛋白的互作研究[D]. 长春: 吉林大学, 2021." (SUN Y." Study on the Soybean Proteins Interacting with P3 and P3N-PIPO Proteins Encoded by Soybean Mosaic Virus[D]. Changchun: Jilin University, 2021.)
[50] LI T, HUANG S, ZHOU J H, et al. Designer TAL Effectors Induce Disease Susceptibility and Resistance to Xanthomonas oryzae pv. oryzae in Rice [J]. Mol Plant, 2013, 6(3): 781-789.
[51] ZHOU J H, PENG Z, LONG J Y, et al. Gene Targeting by the TAL effector PthXo2 Reveals Cryptic Resistance Gene for Bacterial Blight of Rice [J]. Plant J, 2015, 82(4): 632-643.
[52] HU Y, ZHANG J L, JIA H G, et al. Lateral organ boundaries 1 Is a Disease Susceptibility Gene for Citrus Bacterial Canker Disease [J]. Proc Natl Acad Sci USA, 2014, 111(4): E521-E529.
[53] KIM P, XUE C Y, SONG H D, et al. Tissue-Specific Activation of DOF11 Promotes Rice Resistance to Sheath Blight Disease and Increases Grain Weight via Activation of SWEET14 [J]. Plant Biotechnol J, 2021, 19(3): 409-411.
[54] METEIER E, LA CAMERA S, GODDARD M L, et al. Overexpression of the VvSWEET4 Transporter in Grapevine Hairy Roots Increases Sugar Transport and Contents and Enhances Resistance to Pythium irregulare, a Soilborne Pathogen [J]. Front Plant Sci, 2019, 10: 884-1-884-14.
[55] SHAH T, ANDLEEB T, LATEEF S, et al. Genome Editing in Plants: Advancing Crop Transformation and Overview of Tools [J]. Plant Physiol Biochem, 2018, 131: 12-21.
[56] NAPOLI C, LEMIEUX C, JORGENSEN R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-suppression of Homologous Genes in Trans [J]. Plant Cell, 1990, 2(4): 279-289.
[57] HUANG G Z, ALLEN R, DAVIS E L, et al. Engineering Broad Root-Knot Resistance in Transgenic Plants by RNAi Silencing of a Conserved and Essential Root-Knot Nematode Parasitism Gene [J]. Proc Natl Acad Sci USA, 2006, 103(39): 14302-14306.
[58] ROSATTI S, ROJAS A M L, MORO B, et al. Principles of miRNA/miRNA* Function in Plant MIRNA Processing [J]. Nucleic Acids Res, 2024, 52(14): 8356-8369.
[59] LI C Y, WEI J, LIN Y J, et al. Gene Silencing Using the Recessive Rice Bacterial Blight Resistance Gene xa13 as a New Paradigm in Plant Breeding [J]. Plant Cell Rep, 2012, 31(5): 851-862.
[60] BOCH J, SCHOLZE H, SCHORNACK S, et al. Breaking the Code of DNA Binding Specificity of TAL-Type Ⅲ Effectors [J]. Science, 2009, 326: 1509-1512.
[61] LI T, LIU B, SPALDING M H, et al. High-Efficiency TALEN-Based Gene Editing Produces Disease-Resistant Rice [J]. Nat Biotechnol, 2012, 30(5): 390-392.
[62] KIM Y A, MOON H, PARK C J. CRISPR/Cas9-Targeted Mutagenesis of Os8N3 in Rice to Confer Resistance to Xanthomonas oryzae pv. oryzae [J]. Rice, 2019, 12(1): 67-1-67-13.
[63] ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide Sequence of the iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product [J]. J Bacteriol, 1987, 169(12): 5429-5433.
[64] XU Z Y, XU X M, GONG Q, et al. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice [J]. Mol Plant, 2019, 12(11): 1434-1446.
[65] WANG Y J, GENG M T, PAN R R, et al. Editing of the MeSWEET10a Promoter Yields Bacterial Blight Sesistance in Rassava Cultivar SC8 [J]. Mol Plant Pathol, 2024, 25(10): e70010-1-e70010-6.
[66] YU Y H, STREUBEL J, BALZERGUE S, et al. Colonization of Rice Leaf Blades by an African Strain of Xanthomonas oryzae pv. oryzae Depends on a New TAL Effector That Induces the Rice Nodulin-3 Os11N3 Gene [J]. Mol Plant-Microbe Interact, 2011, 24(9): 1102-1113.
[67] ZAFAR K, KHAN M Z, AMIN I, et al. Precise CRISPR-Cas9 Mediated Genome Editing in Super Basmati Rice for Resistance Against Bacterial Blight by Targeting the Major Susceptibility Gene [J]. Front Plant Sci, 2020, 11: 575-1-575-11.
(责任编辑: 单 凝)
