Effect of simulated gastrointestinal digestion on antioxidant, and anti-inflammatory activities of bioactive peptides generated in sausages fermented with Staphylococcus simulans QB7
2024-01-24HongyingLiHongingFanZihanWangQiujinZhuJianpingWu
Hongying Li, Honging Fan, Zihan Wang, Qiujin Zhu, Jianping Wu,
a Laboratory of Animal Genetics, Breeding and Reproduction in the Plateau Mountainous Region, Ministry of Education, College of Life Sciences, Guizhou University, Guiyang 550025, China
b Department of Agricultural, Food and Nutritional Science, University of Alberta, Alberta T6G 2P5, Canada
c Department of Animal and Food Sciences, University of Kentucky, Lexington 40546, United States
d School of Liquor and Food Engineering, Guizhou University, Guiyang 550025, China
Keywords: Gastrointestinal digestion Sausages Bioaccessibility Anti-inf lammatory activities
ABSTRACT Dry-fermented sausages are a good source of bioactive peptides, whose stability against gastrointestinal (GI)digestion determines their bioaccessibility. This study focused on evaluating the effect of peptide extracts from sausages fermented with Staphylococcus simulans QB7 during in vitro simulated GI digestion, including peptide prof iles and antioxidant and anti-inf lammatory activities. Peptides present in sausages were degraded during digestion, with molecular weight reduced from > 12 kDa to < 1.5 kDa. Besides, the content of amino acids increased from 381.15 to 527.07 mg/g, especially tyrosine being found only after GI digestion. The anti-inf lammatory activities were increased after GI digestion, however, the changes in antioxidant activities were the opposite. A total number of 255, 252 and 386 peptide sequences were identified in undigested,peptic-digested and GI-digested samples, respectively. PeptideRanker, BIOPEP-UWM and admetSAR were used to further predict the functional properties and intestinal absorption of the identified peptide sequences from GI digestion. Finally, 18 peptides were discovered to possess either antioxidant or anti-inf lammatory capacities.
1. Introduction
Dry-fermented sausages are one of the most popular meat products, where proteolysis and lipid hydrolysis are the most important biochemical reactions during fermentation and the ripening processes of sausage manufacturing[1-2]. Among these, large meat proteins are degraded into peptides and free amino acids by endogenous enzymes present in meat and microbial enzymes from added starter cultures[3-4]. Therefore, dry-fermented sausages are good sources of bioactive peptides that exert biological benef its for human health.
Previous studies demonstrated that Spanish and Belgian dry-fermented sausages showed high angiotensin converting enzyme(ACE) inhibitory activities, whereas the Belgian sample presented high 1,1-diphenyl-2- picrylhydrazyl (DPPH) radical-scavenging activity and reducing power[5]. Kong et al.[6]reported that pork sausages fermented usingLactobacillusplantarumCD101 andStaphylococcus
simulansNJ201 generated antioxidant and ACE inhibitory peptides;similar f indings were also observed in other studies[7]. However, some peptides possessing goodinvitrobioactivities may fail to exert theirinvivoeff icacies due to their gastrointestinal (GI) instability[8-9]. Thus,to better predict a peptide’sinvivoeff icacy, simulated GI digestion systems are commonly applied to study their GI stability and also monitor the release of newly-generated peptides, considering that monitoring these changesinvivois challenging and invasive[10-11].
Our previous studies showed that usingS.simulansQB7 as a starter culture could promote the formation of antioxidant and anti-inflammatory peptides during sausage ripening, and the ripening time significantly affected peptide content, antioxidant,and anti-inflammatory activities. Specifically, the antioxidant and anti-inflammatory activities reached the peak value on the 16thday(unpublished data). However, the GI stability of peptides on the 16thday of sausages fermented withS.simulansQB7 remains unknown yet. Therefore, the objective of the study was to determine the GI stability of antioxidant and anti-inflammatory peptides derived fromS.simulansQB7-fermented sausages. Pepsin and pancreatin were used to simulate GI digestion. The antioxidant and anti-inflammatory activities were assessed in a human endothelial cell line (EA.hy926).Peptides before and after GI digestion were characterized by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
2. Materials and methods
2.1 Chemicals and reagents
Pepsin (from porcine gastric mucosa, 1 064 U/mg) and pancreatin(porcine pancreas, 1 983 U/mg) were obtained from Sigma (Oakville,ON, Canada). Chromatographic grade water, acetonitrile (ACN),and trifluoroacetic acid (TFA) were purchased from Fisher Scientific(Ottawa, ON, Canada). Dulbecco’s modified Eagle’s medium(DMEM), and fetal bovine serum (FBS) were purchased from Gibco Invitrogen (Burlington, ON, Canada). EA.hy926 (CRL-2922) was provided by the American Type Culture Collection (Manassas, VA,USA). Nonessential amino acids (NEAA), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and penicillin-streptomycin were obtained from Gibco Invitrogen (Burlington, ON, Canada).Tumor necrosis factor α (TNF-α) was obtained from R&D Systems(Minneapolis, MN, USA). Dihydroethidium (DHE) was purchased from Biotium (Fremont, CA, United States).
2.2 Preparation of fermented sausages
S.simulansQB7 (CICC No. 11117 s), which was isolated from Chinese fermented sausages and stored in the China Center of Industrial Culture Collection (CICC), was used as the starter and cultivated in the mannitol salt (MSA) broth at 37 °C for 24 h[12]. Cell pellets were collected by centrifugation (10 000 ×g) and washed three times with sterile water before being resuspended in sterile water.
Three independent batches of sausages fermented withS.simulansQB7 were prepared. Fresh lean pork and back fat were purchased from a local supermarket (Walmart) in Guiyang, Guizhou,China. Sausages were prepared using 80% lean pork, 20% back fat, 3%sugar, 2% salt, 2% Chinese liquor, 0.05% sodium erythorbate (food grade), and 0.01% sodium nitrite (food grade). After homogenizing the mixture, the meat batter was stuffed into natural porcine casings.Prepared sausages were fermented at 30 °C for 2 days under (90 ± 5)%relative humidity and then ripened at 15 °C for 4 days under (80 ± 5)%relative humidity[7].
2.3 Preparation of peptide extract from sausages
Peptides were extracted according to a previous method[5]. Briefly,50 g of sausage sample was minced and homogenized in 200 mL of 10 mmol/L HCl by a high-speed disperser (XHF-DY, Ningbo Co.,China), which was repeated three times (10 s, 22 000 r/min) on the ice. The homogenate was centrifuged (20 min, 10 000 ×g) at 4 °C, the supernatant was removed, and the pellet was deproteinized by adding 3 times the volume of ethanol (100%) and the resultant solution was maintained at 4 °C for 20 h. Then, the sample was centrifuged again(20 min, 10 000 ×g) at 4 °C. The supernatant was filtered through a 0.45-μm nylon membrane filter (Millipore, Bedford, MA) and dried in a rotatory evaporator, before being freeze-dried and stored at –20 °C until further analysis.
2.4 Simulated GI digestion
Simulated GI digestion followed procedures described by Fan et al.[13].Peptides extracted from sausage fermented withS.simulansQB7 were dissolved in ddH2O at 5% protein (m/V). Then, samples were digested by pepsin (1% E/S,m/mprotein) for 1.5 h (pH 2.0, 37 °C).Subsequently, the digest was re-adjusted to pH 7.5; the digested sample was halved with one half as peptic digestion group (pepsin)and the other half further digested by pancreatin (1% E/S) for 1.5 h at 37 °C as GI digestion group (pepsin + pancreatin). The reaction was then terminated by maintaining the mixture at 95 °C for 10 min. Digestion was performed in a 15 mL polypropylene tube and the temperature was controlled by a reciprocal shaking water bath(Fisher Scientific). Each digest was prepared individually in triplicate.Control group samples were prepared in the same steps except for the digested enzyme solution replaced by ddH2O used as undigested sample (S). The samples were dried in lyophilized and stored at–20 °C for subsequent assays.
2.5 Protein content, hydrolysis yield, and degree of hydrolysis (DH)
Protein content was measured by a Dumas method, which was determined by a TruSpec CN carbon/nitrogen determinator (Leco Corp., St. Joseph, MI, USA), using a conversion factor of 6.25 as described by Adler-Nissen et al.[14]. Dry matter was analyzed by drying the samples at 110 °C overnight. Ash content was determined by placing samples at 550 °C for 24 h. The degree of hydrolysis (DH)was measured using the 2,4,6-trinitro-benzenesulfonicacid (TNBS)method, where the DH was defined as the percentage (%) of cleaved peptide bonds compared to the total peptide bonds of the original protein extract[14]. Hydrolysis yield was calculated based on the following equation:
2.6 Size exclusion chromatography
Molecular weight distribution was analyzed by size exclusion chromatography using a Superdex peptide 10/300 GL column (at room temperature), connected with an AKTA explorer 10XT system(GE Healthcare, Uppsala, Sweden). Samples were dissolved in 30%ACN (in ddH2O,V/V) containing 0.1% TFA. Samples (100 μL,1 mg/mL) were injected into the column and eluted at an isocratic gradient at a flow rate of 0.5 mL/min. Peaks were detected at 220 nm. Cytochrome C, aprotinin, vitamin B12, and (Gly)3were used as molecular weight markers.
2.7 Amino acids analysis
Peptides extracted (25 mg) from sausage fermented withS.simulansQB7 were dissolved in 3.0 mL of 6 mol/L HCl, and then mixed thoroughly to ensure the entire samples were wetted with HCl. After that, samples were hydrolyzed at 110 °C for 24 h. Subsequently, the samples were analyzed by high-performance liquid chromatography (HPLC) with a Supelcosil LC-18 (150 mm ×4.6 mm, 3 μm). HPLC parameters were as follows: gradient elution with mobile phase A (100%–0% for 40 min, 0.1 mol/L sodium acetate buffer) and then mobile phase B (0%–100% for 40 min,100% methanol); the volume of injected sample, 3 μL; flow rate,1.1 mL/min; column temperature, 35 °C.
2.8 Desalting protocol, cytotoxicity, and bioactivity detection
2.8.1 Desalting protocol
Prior to treatment with cells, samples were desalted according to the protocol described by Fan et al.[15]. Briefly, samples were dissolved in ddH2O (2 mg/mL) and loaded onto the Sep-Pak 35 cctC18cartridge(WAT043350, Waters, Milford, MA, USA). The cartridge was first washed with double column volumes (CV) of ddH2O for removing salt. Then 50% ACN (2 × CV) and 100% ACN (1 × CV) were added sequentially to wash the cartridge and collect the eluent, which was vacuum-evaporated, freeze-dried, and stored at –20 °C for subsequent assays.
2.8.2 Cell culture of EA.hy926 cells
EA.hy926 cells (from 6−10 passages) were cultured based on the procedures described elsewhere[16]. Firstly, EA.hy926 cells (passages 6–10) were grown in the growth DMEM medium (containing 10%FBS, 2.5% HEPES, 1% penicillin-streptomycin, and 1% NEAA) in a 37 °C incubator at 5% CO2and 100% humidity. The growth medium was changed every 2 days. After reaching 80% of confluence, the cells were placed in a quiescing medium (the same recipe as that of the growth medium but containing 1% FBS) for all the experiments in this study.
2.8.3 Cytotoxicity of EA.hy926 cells
Cytotoxicity was evaluated using the alamarBlue™ fluorescence assay (Thermo Fisher Scientific, Burlington, ON, Canada). Cells were seeded on a 96-well plate at 1.0 × 104cells/well. After reaching 80% of confluence, cells were treated with 5 mg/mL of samples(dissolved in the growth medium) for 24 h. Then, culture media were replaced with 200 μL of 10% alamarBlue solution (dissolved in culture medium) for 4 h of incubation (protected from direct light).The absorbance was measured at 560 nm using a SpectraMax M3 microplate reader (Molecular Devices, CA, USA). The control group samples were prepared without peptide samples.
2.8.4 Antioxidant assay
Antioxidant activity was evaluated in EA.hy926 cells by detecting superoxide generation and lipid peroxidation by referring to the method described by Fan et al.[17].
2.8.5 Anti-inflammatory assay
The inflammatory activity was evaluated in EA.hy926 cells upon 6 h of TNF-α induction. Cells were treated with 2.5 mg/mL of samples for 24 h, and then 10 ng/mL of TNF-α was added to the solution for co-treatment for 6 h. Then, the production of cyclooxygenase 2(COX2) and vascular cell adhesion molecule 1 (VCAM-1) was determined. The dose of the samples was selected based on previous studies[12,16]and the results of cytotoxicity. Cells were lysed using boiling Laemmli buffer containing 50 mmol/L dithiothreitol(DTT) and 0.2% Triton-X-100. Cell lysates were loaded onto a 9%separating gel and transferred to a nitrocellulose membrane (diameter 0.45 μm, 1620115, Bio-Rad, Montreal, QC, Canada) for incubation with antibodies. Protein bands of COX2 (ab15191, Abcam, ON, Canada) and VCAM-1 (ab134047, Abcam, ON, Canada) were normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH, ab8245, Abcam).Donkey-anti-rabbit 800 CW or donkey-anti-mouse IRDye 680 RD secondary antibodies (Licor Biosciences, Lincoln, NE, USA) were used to visualize the fluorescent protein bands in a Licor Odyssey BioImager,and quantification of protein bands was performed using Image Studio Lite 5.2 (Licor Biosciences, Lincoln, NE, USA).
2.9 Peptide identification
Peptide sequences were identified using an Atlantis dC18UPLC column (75 μm × 150 mm, 3 μm, Waters, Milford, MA, USA) on nano LC-ESI-MS/MS as previously described in Liang et al.[16].
2.10 In silico prediction of peptides identified from GI digestion: biological activity and intestinal absorption
2.10.1 In silico prediction of potential biological activity of the peptides
The potential biological activity of the peptides identified from GI-digested samples was predicted using PeptideRanker (http://bioware.ucd.ie/compass/biowareweb/) tool. It is a web-based server to predict the probability of biological activity of peptide sequences[18].PeptideRanker tool gives the peptide score in the range of 0–1. A score > 0.5 represents a good potential in biological activity[19]. The selected potential biological peptide sequences were subjected to further analysis.
2.10.2 In silico prediction of peptide potential function
BIOPEP-UWM online tool (https://biochemia.uwm.edu.pl/)was used to predict the functional properties of peptides with potent potential biological activity[20-21]. The procedure was as follows: First,“Bioactive peptides” tool option was adopted in the BIOPEP database;Then “ANALYSIS” was selected, followed by selecting the “FOR YOUR SEQUENCE” option in the “PROFILES OF POTENTIAL BIOLOGICAL ACTIVITY” tool, in which peptide sequences with PeptideRanker score > 0.5 were uploaded; Finally, the “Report”button was clicked to search and analyze the potential antioxidant and anti-inflammatory activity.
2.10.3 In silico prediction of peptide potential intestinal absorption
Amino acid sequences of peptides screened above were converted into the Simplified Molecular Input Line Entry Specification(SMILES) strings using “SMILES” application at the BIOPEP-UWM website[20]. The procedure was as follows: First, “Bioactive peptides”tool option was adopted in the BIOPEP database, and “ANALYSIS”was selected, then the “SMILES” tool was selected to upload the peptide sequences. Finally, the “SHOW SMILES” button was clicked to search the SMILES strings of peptides. DmetSAR online tool(http://lmmd.ecust.edu.cn/admetsar2) was used to predict intestinal absorption according to Wang et al.[22].2.11 Statistical analysis
Data of hydrolysis yield, protein content, and DH were performed in triplicate and were analyzed using one-way analysis of variance(ANOVA) by IBM SPSS Statistics Version 23 (Chicago, IL, USA)followed by Duncan’s multiple range tests. Data collected from the cellular study were expressed as means ± standard errors (SEMs) of six independent experiments and were analyzed by one-way ANOVA followed by Dunnett’s multiple tests using GraphPad Prism 9.
3. Results and discussion
3.1 Effect of in vitro GI digestion on hydrolysis yield, protein content, and DH
Table 1 showed the changes in protein content, hydrolysis yield,and DH during GI digestion. The protein content was significantly decreased to 38.50% after peptic digestion (P< 0.05) and further reduced to 34.30% after pancreatic digestion (P< 0.05). DH was 10.89% after peptic digestion and was further increased to 17.83%after pancreatic digestion (P< 0.05). Meanwhile, the hydrolysis yield was reduced from 43.32% to 36.60% after pancreatic digestion(P< 0.05). Pepsin belongs to endopeptidase, which can cleave Phe,Tyr or Trp residue at the C-terminus, while pancreatin consists of multiple enzymes including trypsin, elastase, chymotrypsin, and carboxypeptidases[23]. These enzymes work jointly and increase the efficiency in cleaving proteins into peptides and free amino acids[24].Similar studies have reported that DH was increased after GI digestion while protein content was decreased, which indicated that more peptide bonds were broken down into amino acids during pancreatic digestion than peptic digestion[25-26].
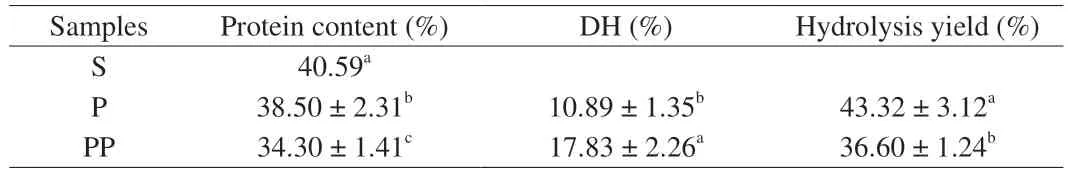
Table 1 Protein content, DH and hydrolysis yield of sausages and its GI-digested samples.
3.2 Effect of in vitro GI digestion on the peptide profile
Size exclusion chromatography was used to analyze the peptide profile (Fig. 1). GI digestion decreased the proportion of fractions with molecular weight (MW) of > 12 kDa but increased those with that of < 1.5 kDa. It was seen that the fraction withMWof > ~ 6 kDa was significantly degraded after peptic digestion and was almost completely degraded after pancreatic digestion. These results indicated that GI digestion further produced low-molecular weigh peptides from sausage. A similar study was reported by Paolella et al.[27]that high amounts of small peptides were identified in the peptide fraction withMWof < 3 kDa, which was extracted from 18 and 24 months aged Parma hams afterinvitroGI digestion. Previous studies demonstrated that small peptides possessed better biological activities including antioxidant and anti-inflammatory properties[28].
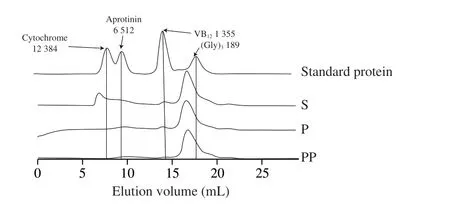
Fig. 1 Molecular weight distribution of peptides from sausages prepare with S. simulans QB7 before and after digestion by size exclusion chromatography.S, P, and PP indicates non-, pepsin-, and pepsin + pancreatin digestion,respectively. Values (189-12 384) in Fig. 1 indicate the molecular weight (Da)of (Gly)3, VB12, aprotinin, and cytochrome C.
3.3 Amino acids analysis
Table 2 showed the changes in amino acid composition during GI digestion. Compared to undigested samples, the content of total amino acids was increased to 433.47 mg/g after peptic digestion (P<0.05),and its content was further increased to 527.07 mg/g after pancreatic digestion (P<0.05). In this study, GI digestion significantly increased the content of essential amino acids (EAA) such as His, Ile, Leu,Lys, Met, Phe, Thr, and Val (P<0.05). A similar trend was observed in the content of hydrophobic amino acids (HAA) including Ala,Val, Ile, Leu, Phe, and Met. Interestingly, Tyr was found only after pancreatic digestion. It has been reported that pepsin is effective in cleaving peptide bonds between hydrophobic and preferably aromatic amino acids such as Phe, and Tyr, while trypsin in pancreatin cleaves Arg or Lys. In addition, chymotrypsin cleaves Phe, Tyr, and Leu, while elastase cleaves Ala and other aliphatic amino acids[29].Therefore, it can be inferred that Tyr generated from sausages peptide was the result of chymotrypsin hydrolysis. Meanwhile, GI digestion significantly increased the content of positively-charged amino acids(PCAA, Arg, His, and Lys) and negatively-charged amino acids(NCAA, Asp, Glu, and Ser) (P< 0.05). The content of PCAA was higher than that of NCAA in all three samples (P< 0.05). Previous reports have demonstrated that PCAA could mitigate inflammation[30],while NCAA could attenuate oxidative stress[31-32]. However, the content of Glu, Ser, and Arg showed no significant change among the three group samples (P> 0.05). These results indicated that peptides containing these amino acids were stable and resistant to GI digestion.

Table 2 Effect of simulated GI digestion on the composition of amino acids in sausages peptide (mg/g).
3.4 Effect of in vitro GI digestion on antioxidant and antiinflammatory activities
3.4.1 Cytotoxicity in EA.hy926 cells
Prior to assessing the antioxidant and anti-inflammatory activities of the samples in EA.hy926 cells, their effects on cell viability were assessed (Fig. 2). Incubation of these samples for 24 h at the concentration of 5 mg/mL, which was higher than the tested concentration (2.5 mg/mL) in the subsequent experiments,demonstrated no negative impact on cell viability in EA.hy926 cells(Fig. 2) (P> 0.05). Therefore, these samples were considered not toxic against EA.hy926 cells at the doses used in the present study(2.5 mg/mL). Similarly, Fan et al.[13]reported that treatment with chicken muscle protein hydrolysate at the same concentration exerted no toxicity to EA.hy926 cells. Other researchers also reported no effect on the viability of various cells after treatment with other sources of meat-derived peptides, such as peptides derived from pork myofibrillar protein (4 mg/mL, in HEK-293 cells)[33]and those derived from connective tissue (4 mg/mL, in Vero cells)[34].
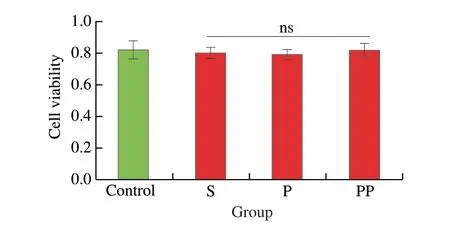
Fig. 2 Effect of treatment with fermented sausage-derived peptides on the viability of EA.hy926 cells. Cells were treated with samples at the concentrations of 5 mg/mL over 24 h, before being analyzed by the alamarBlue cell viability assay. Data were expressed as means ± SEMs. ns, not significant(P > 0.05) vs. TNF-α group. S, P, and PP indicates non-, pepsin-, and pepsin +pancreatin digestion, respectively.
3.4.2 Antioxidant activity
The ability in inhibiting lipid peroxidation and superoxide generation was one of the indicators of the antioxidant activity of peptides. Fig. 3A displayed the changes in MDA levels in TNF-αstimulated EA.hy926 cells after treatment with three group samples.TNF-α significantly increased MDA levels (P< 0.000 1), which could be significantly inhibited by the undigested group (S) and pepsindigested group (pepsin) (P< 0.000 1); however, the GI digestion group (pepsin + pancreatin) was only able to slightly reduce TNFα-induced MDA accumulation (P> 0.05). The inhibitory effect on superoxide generation in TNF-α-stimulated EA.hy926 cells was presented in Figs. 3B and 3C. Both undigested and pepsin digested samples inhibited superoxide generation, which was similar to that of MDA (P< 0.000 1). These results demonstrated that antioxidant peptides in sausages fermented withS.simualansQB7 were susceptible to GI digestion, and thus were degraded into smaller peptides or amino acids with low or without antioxidant capacity. Intestinal enzymes possess more cleavage sites, resulting in more extensive hydrolysis of sausage peptides, thus reducing the antioxidant activity[35]. Indeed, the changes in amino acid composition aligned with the content of free amino acids, which were increased after GI digestion.

Fig. 3 Effect of treatment with undigested and digested sausage-derived peptides on TNF-α-induced oxidative stress in EA.hy926 cells. Cells were treated with undigested and digested samples (2.5 mg/mL) for 1 h before TNF-α (10 ng/mL) stimulation for another 6 h, followed by the detection of (A) malondialdehyde(MDA), (B) superoxide level and (C) superoxide fluorescence intensity in EA.hy926 cells. Data were expressed as means ± SEMs and normalized to the untreated group (Untr). ****P < 0.000 1 vs. TNF-α group. S, P, and PP indicates non-, pepsin-, and pepsin + pancreatin digestion, respectively.
Previous studies demonstrated that different samples have different susceptibility during GI digestion. For example, Fan et al.[13]prepared three chicken muscle protein hydrolysates using thermoase,protex 26L and pepsin, respectively, whose antioxidant activity experienced very different changes during GI digestion. The either enhanced or reduced antioxidant activity after GI digestion suggested that the fate of a peptide’s antioxidant activity largely depended on its constituting amino acids and their respective positions. Megías et al.[36]reported that an ACE-inhibitory peptide, LVYPFPGPIPNSLPQNIPP,significantly lost its activity after GI digestion. The antioxidant peptide SNAAC was degraded into SNAA after GI digestion,which led to a significant loss in antioxidant capacity[37]. However,Wang et al.[38]demonstrated that GI digestion of Xuanwei and Jinhua hams significantly scavenged free radicals such as DPPH.Gallego et al.[10]reported that GI digestion decreased peptides identified from dry-cured ham to scavenge DPPH free radicals and iron reduction, while increased their ability to scavenge ABTS cation free radical. Some peptides are more resistant to GI digestion due to their special structures. Their activity remained stable after GI digestion, such as the collagen derived tripeptide[39].
3.4.3 Anti-inflammatory activity
COX2 and VCAM-1 are two inflammatory mediators in endothelial cells[40-41]. COX2 induces inflammation by catalyzing the conversion of arachidonic acid into prostanoids[42]. VCAM-1 recruits monocytes and macrophages to inflammation sites in the vascular wall[43]. The effect of sausage peptides on COX2 and VCAM-1 expressions was evaluated in TNF-α-induced EA.hy926 cells.As seen in Fig. 4, TNF-α stimulation significantly increased the expression of COX2 and VCAM-1 in EA.hy926 cells (P< 0.000 1).However, both of their expressions were significantly inhibited by the treatment with peptide samples (S, pepsin and pepsin + pancreatin)(P< 0.000 1), while there was no marked difference among S, pepsin and pepsin + pancreatin groups (P> 0.05). These results indicated that peptides inhibiting COX2 protein expression remained stable during GI digestion. Interestingly, inhibiting VCAM-1 expression was first strengthened after peptic digestion and then attenuated after intestinal digestion (P< 0.000 1). Consequently, pepsin-digested peptides possessed a higher inhibition in VCAM-1 expression than that of the undigested sample (P< 0.05). It could be inferred that peptides present in sausages inhibiting COX2 or VCAM-1 expression were different peptide sequences. Therefore, peptides that inhibited COX2 expression were likely more resistant to GI digestion, while those inhibiting VCAM-1 expression were less GI-resistant and were degraded into smaller peptides, which maintained excellent VCAM-1-inhibiting capacities. Indeed, as Fan et al.[17]reported, peptides regulate COX2 or VCAM-1 expression via different signaling pathways in TNF-α-simulated vascular endothelial cells. In addition,peptide stability is important in dictating its anti-inflammatory activity[44]. These phenomena further indicated the complexity of peptides in the modulation of cellular events.
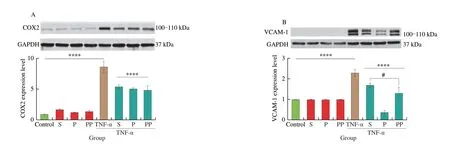
Fig. 4 Effect of treatment with undigested and digested sausage-derived peptides on TNF-α-induced inflammation in EA.hy926 cells. Cells were treated with peptides (2.5 mg/mL) for 24 h before TNF-α (10 ng/mL) stimulation for another 6 h, followed by the detection of (A) COX2 and (B) VCAM-1 expression. Protein bands were normalized to GAPDH. Data were expressed as means ± SEMs and normalized to the untreated group (Untr). ****P < 0.000 1 vs. TNF-α group; #P < 0.05 vs. TNF-α + P group. S, P, and PP indicates non-, pepsin-, and pepsin + pancreatin digestion, respectively.
The inhibitory effects on the expression of VCAM-1 have been reported by many hydrolysates from other sources such as chicken,egg white, and soybean[45]. Earlier studies demonstrated that peptides consisting of PCAA and hydrophobic amino acids, especially at the terminal position, usually possess high anti-inflammatory activity[46-47].
3.5 Peptide profile analysis
The samples during GI digestion were analyzed using LC-MS/MS for peptide sequences. As can be seen in Fig. 5, there were certain differences in peptide amount at different digestion phases. A total number of 256 and 252 peptides were detected in undigested and peptic digestion samples, while a total number of 386 peptides were detected after intestinal digestion. The number of peptides decreased after digestion with pepsin compared to that before digestion. One possible reason was a lack of pepsin cleavage site in the peptides or the formation of hydrophilic peptides that could be washed out from the column during the desalting process. Antioxidant and anti-inflammatory peptides are generally short sequences(2–20 amino acids in length), with molecular weights of approximately 400–3 000 Da[47]. The results of the present study showed that peptide sequences contained about 3–10 amino acids and their molecular weights were approximately 300–2 000 Da. Most peptides identified were derived from major meat proteins, actin, myosin, and troponin T.There were 34 common peptides identified in three samples, a total of 24 common peptides were detected in S and peptic group, and 45 common peptides in S and pepsin + pancreatin groups. There were 24 common peptides were identified in pepsin and pepsin + pancreatin groups. More peptides were released after pancreatin digestion. After assessment by principal component analysis (PCA) (Fig. 6), the peptides in the pepsin + pancreatin group exhibited more difference than S and pepsin group samples. The most abundant peptides were observed in pepsin + pancreatin group samples. The potential biologically of peptides identified from pepsin + pancreatin group samples were predicted using PeptideRanker tool. PeptideRanker is an online server that can rank the peptide sets according to structure-function patterns[18]. The potential biological active peptide sequences were showed in Table 3. A total of 130 peptides were ranked higher than 0.5 in PeptideRanker scores. Among them, peptides WPWMK, GPAPWGFR, FVF, GPGPWG, LGFP,MALF, FIFNL, FLFNL, IGFP, APPAPPPFE, APPHIF, ADAPMF,SGIPAGWMG, FRYLM, and GPYGPPG had the maximum PeptideRanker scores (0.91–0.99). The functions of the potential biologically active peptides were further analyzed based on the BIOPEP-UWM database. As seen in Table 4, there were 10 potential antioxidant peptides and 8 potential anti-inflammatory peptides.Peptide sequences, including LGFP, LPML, LPMI, FRYIM, LPMLQ,FRYLM, LPMIQ, FRYLMG, WLPVVR, PHQYPALTP, with PeptideRanker score of > 0.5, indicated their antioxidant capacity.The biological activity of a peptide depends on its amino acid position in its peptide sequence as well as the amino acid composition[48]. It is well documented that short peptides containing phenylalanine and glycine have the greatest correlation with their PeptideRanker score and are more likely to be predicted as bioactive[18]. In the present study, LGFP, the highest peptide rank score in antioxidant capacity,contained Phe (F) and Gly (G) in the sequences. Moreover, the amino acid composition of the identified potential antioxidant peptides had a high content of hydrophobic amino acid residues Leu (L), Pro (P), Val(V), Ala (A), Ile (I) and Tyr (Y), which accounts for a large proportion in the identified antioxidant peptides[49]. Previous studies have reported that peptides with a higher proportion of hydrophobic amino acids play an important role in antioxidant capacity[50-51]. Liu et al.[52]reported that hydrophobic amino acids located at the N-terminus had an important effect on antioxidant capacity of peptides.The sequences, including IPPLP, ELPFY, IPPIP, IPPEIWEL,FDIPPPPMD, SFNRTAMPYG, ANIFSIPYFQ, and VPPLIPPKIP had the potential for anti-inflammatory capacity. Interestingly, these sequences were only identified in pepsin + pancreatin group samples.This indicated that pancreatic enzymes promoted the generation of anti-inflammatory peptides. Moreover, these peptide sequences were rich in hydrophobic amino acids and PCAA, which explained the higher anti-inflammatory activity in pepsin + pancreatin group samples. Most currently-reported anti-inflammatory peptides contained PCAA and hydrophobic amino acids such as Arg (R), Lys(K), Trp (W), Phe(F), Val (V), Leu (L), Ile (I), Ala (A), Pro (P), and Met (M); these amino acids can be found in the N-terminal, C-terminal,or in the middle of the peptide sequence[48].
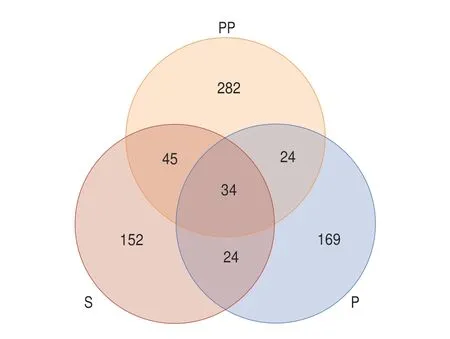
Fig. 5 The Venn analysis of peptides during GI digestion.
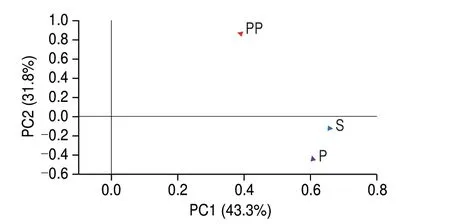
Fig. 6 The PCA analysis of peptides during GI digestion.
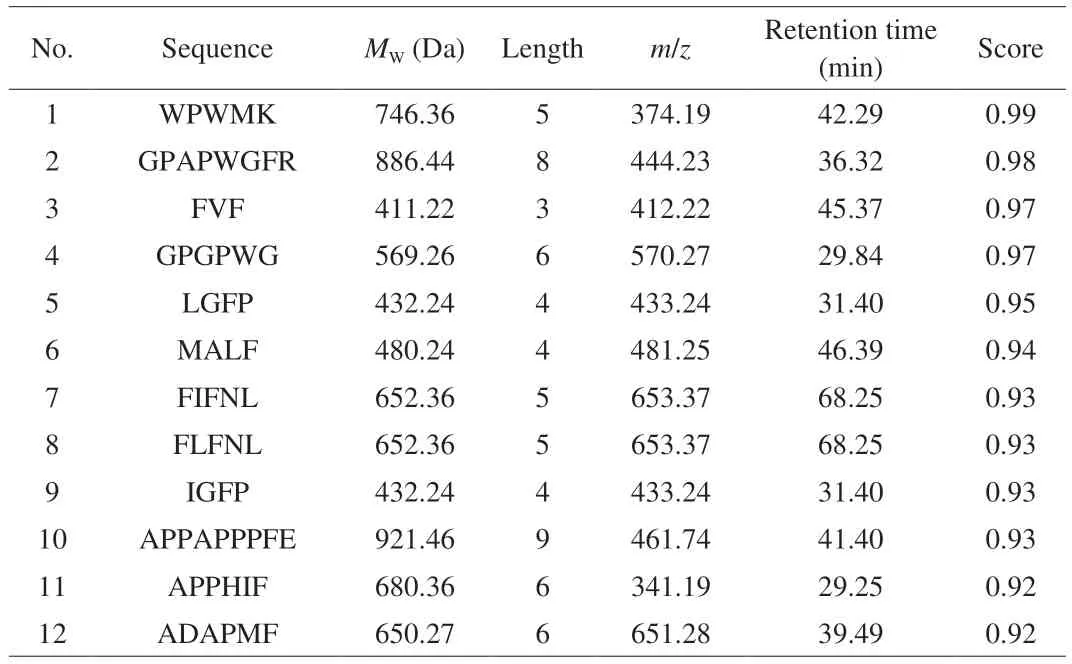
Table 3 The peptide sequences with potential biologically activities identified from GI digestion group.
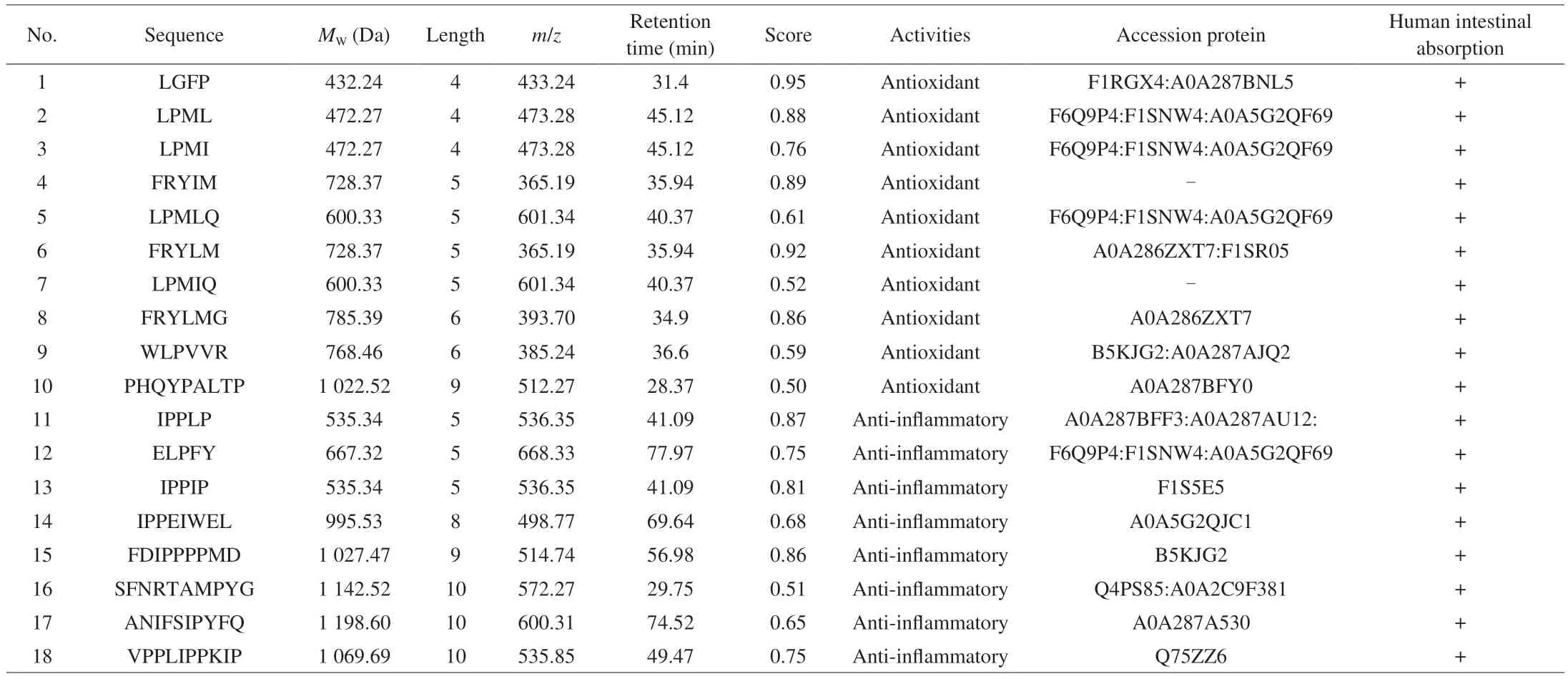
Table 4 The peptide sequences with potential antioxidant and anti-inflammatory capacities predicted according to the BIOPEP-UWM database.
Although a peptide is GI stable, it still may not be absorbed by the GI tract. So, using the admetSAR (http://lmmd.ecust.edu.cn/admetsar1/predict/, accessed on 28 March 2023) to further predict the intestinal absorption of the 18 potential antioxidant and antiinflammatory peptides. As seen in Table 4, these 18 potential antioxidant and anti-inflammatory peptides could be absorbed by human intestinal monolayer predicted usingin silicotool.Bioactivities of the peptides predicted usingin silicotools need to be confirmed experimentally in the near future.
4. Conclusion
Simulated GI digestion increased the anti-inflammatory activity ofS.simulansQB7-fermented sausage peptides, but decreased their antioxidant capacities. Moreover, the content of total amino acids was increased after GI digestion. A total number of 255, 252, and 386 peptide sequences were identified in undigested, pepsin-digested, and GI-digested samples, respectively. There are 130 peptide sequences that possessed potential biological activities after GI digestion.Among them, 18 peptides were predicted to possess antioxidant and anti-inflammatory properties. These peptides were also predicted to be able to cross the intestinal monolayer and thus be absorbed. Our study tends to open a new revenue for the utilization of sausages for the development of value-added functional food ingredients/products.Our next phase is to characterize the responsible peptides fromS.simulansQB7-fermented sausages, and further elucidate the cellular mechanisms underlying these two biological activities in vascular endothelial cells.
Conflict of interest
Jianping Wu is an editorial board member forFood Science and Human Wellnessand was not involved in the editorial review or the decision to publish this article. No conflict of interest exists in the submission of this manuscript.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (32272359), and Natural Science and Engineering Research Council of Canada (NSERC,RGPIN-2018-04680). Hongying Li is the recipient of the scholarship from the China Scholarship Council (202106670005).
杂志排行
食品科学与人类健康(英文)的其它文章
- GUIDE FOR AUTHORS
- Call for Papers from Food Science of Animal Products
- Ascophyllum nodosum and Fucus vesiculosus ameliorate restenosis via improving inf lammation and regulating the PTEN/PI3K/AKT signaling pathway
- Alleviatory effect of isoquercetin on benign prostatic hyperplasia via IGF-1/PI3K/Akt/mTOR pathway
- Adsorption, in vitro digestion and human gut microbiota regulation characteristics of three Poria cocos polysaccharides
- Voluntary wheel running ameliorated the deleterious effects of high-fat diet on glucose metabolism, gut microbiota and microbial-associated metabolites
