PBRM1-SWI/SNF complex contributes to cell cycle control via E2F1 in renal cell carcinoma cells
2023-03-02,,,,,,,
, , , , , , ,
(Department of Medical Genetics,Zunyi Medical University,Zunyi Guizhou 563099,China)
[Abstract] Objective The aim of this study is to investigate whether PBRM1 deficiency affects cell proliferation and how this process is regulated at the transcriptional level.Methods We performed RNA interference,RT-PCR,Western blot,MTT,and FACS experiments to analyze cell proliferation and expression of E2F1 family members after PBRM1 knockdown.Meanwhile,the transcription regulation mechanism of E2F1 and PBRM1 was also investigated by the ChIP experiment.Results Our data showed that PBRM1 deficiency in SN12C cells significantly increased the mitotic index (MI),which was accompanied by upregulation of E2F1 transcription factors.PBRM1 was recruited to the E2F1 promoter,co-occurring with active histone modifications.E2F1 was recruited to the PBRM1 promoter and could play a repressive role in PBRM1 expression via binding to its response elements.Conclusion These data suggest that the PBRM1-containing SWI/SNF complex plays an antiproliferative role.Deficiency of PBRM1 along with downstream deregulated genes by PBRM1 and/or E2F members may act synergistically in ccRCC carcinogenesis and development.
[Key words] ccRCC; PBRM1; E2F1 transcription factor; SWI/SNF; cell cycle; histone modification
Using high-throughput screening techniques,researchers have found that genes involved in epigenetic modifications are commonly inactivated in many cancers.This includes a significant number of genes from the switch/sucrose nonfermenting (SWI/SNF) family[1].Besides the DNA-dependent catalytic subunit BRM or BRG1(also known as SMARCA2 and SMARCA4,respectively),the mammalian SWI/SNF contains about 10 other subunits called BRM or BRG1-associated factors (BAFs)[2].One types of SWI/SNF complexes is the PBAF complex that containing only BRG1 as the catalytic subunit and PBRM1 as the specific subunit[3-4].
Kidney cancer accounts for about 2% of all cancer deaths worldwide and clear cell renal cell carcinoma (ccRCC) is the subtype accounting for 75% of clinical cases[5].PBRM1 is highly mutated (~40%) in ccRCC[6].PBRM1 maps to chromosome 3p encoding the polybromo 1 protein or BRG1-associated factor 180 (PBRM1/PB1/BAF180)[7].The PBRM1 protein contains 6 bromodomains that can interact with acetylated proteins.PBRM1 interacts with acetylated histones (i.e.H3K14ac) in vitro and can function as a chromatin targeting subunit[8].
The germlinePBRM1 mutation predisposes to RCC and loss ofPBRM1is associated with worse survival rates of ccRCC patients[9].It is believed that highly mutatedPBRM1 plays a driving role in ccRCC progression[10].It has been speculated that PBRM1 may play a role including genome instability,cell growth advantage,mTOR activation in VHL-deficient mouse models and immune escape[6,11].
Studies on the BAF complex showed that theARID1A-depleted cells slightly but consistently favor the cell cycle,suggesting that ARID1A-containing SWI/SNF complexes have an antiproliferative function,while ARID1B-containing SWI/SNF complexes have an opposite function[12].Since PBRM1 is the specific subunit of the PBAF-SWI/SNF complex while ARID1A and ARID1B are specific subunits of the BAF-SWI/SNF complex,it is highly desirable to know whether PBRM1 functions like either ARID1A or ARID1B.As a tumor suppressor,PBRM1 is assumed to have an antiproliferative function.However,PBRM1 knockdown studies have yielded conflicting results that some ccRCC cells showed no growth advantage afterPBRM1 depletion in some studies[13].Studies also showed that SN12C and UO31 cells have some different features compared to other ccRCC cells[14].To understand the role ofPBRM1 in these specific and seemingly unaffected cells,we investigated howPBRM1 deficiency affects the cell cycle process and the relationship between PBRM1 and E2F transcription factors involved in tight cell cycle control.We report here that for SN12C and UO31 cells,althoughPBRM1 knockdown does not significantly increase cell growth,it does increase the mitotic index (MI).This progression involves the upregulation of E2F transcription factors,and the elevation can be transcriptionally controlled by the PBAF-SWI/SNF complex mediated by acetylated histones.Our data suggest that deficientPBRM1,activated E2F family members,and deregulated downstream genes may act synergistically in the carcinogenesis and development of ccRCC.
1 Materials and methods
1.1 Cell culture and antibodies The kidney cancer cell lines SN12C and UO31,as well as HEK 293T cells were routinely maintained in RPMI medium supplemented with 10% fetal bovine serum,penicillin and streptomycin (100 U/ml)in a 37 ℃ incubator at 5% CO2.The following antibodies were used in this study:PBRM1(A301-591A),PBRM1(A301-590A),E2F1(KH95)(sc-251),H3K9ac (ab10812),H3K27ac (ab4729),H3K4Me3 (ab8580),normal rabbit IgG (sc-2027),and normal mouse IgG (sc-2025).H3K14ac (07-353) was purchased from Millipore (Billerica,MA).The final antibody dilution was as recommended by the manufacturer.
1.2 RNA interference and proliferation assays SiRNAs forPBRM1andE2F1 were used in knockdown experiments.Lipofectamine RNAiMAX Reagent (Invitrogen,13778) was used for RNA interference experiments.KnockdownPBRM1 stable cell lines were established usingPBRM1-shRNA lentiviral particles (Santa Cruz,sc-76075-V) and control shRNA lentiviral particles (Santa Cruz,sc-76075-V).Proliferation assays were performed using MTT reagents (Promega,CellTiter 96® AQueous) according to the manufacturer’s instructions.
1.3 Fluorescence-activated cell sorting (FACS) assay We synchronized cells by growing the cells to full confluence in starvation medium (RPMI medium without addition of fetal bovine serum,FBS).We reseeded the cells on 60 mm dishes and cultured in normal medium.We harvested the cells at the indicated time points and resuspended the cells in 0.5 ml PBS to obtain the single cell suspension,followed by fixing the cells with ice-cold 70% ethanol for 15 min or storing at 4 ℃ for further use.The cells were centrifuged and stained with propidium iodide solution (0.1mg/ml,0.05% Triton X-100,RNase A,5μg/ml).FACS analysis was performed with Cell Quest Pro (BD Biosciences,San Jose,CA).Three biological experiments were carried out to verify the observations.
1.4 RNA extraction,cDNA synthesis and qPCR Gene expression analysis was performed as previously mentioned[15].In brief,total RNA was collected using TRIreagent (Roche Applied Science) and purified with RNeasy kit (QIGEN).Reverse transcription of RNA was performed with iScriptTMcDNA Synthesis Kit (Bio-Rad,170-8891).Real-time PCR was performed with SensiFAST SYBR reagents (Bioline,UK) on the Bio-Rad CFX 96 system.
1.5 Western blotting assay Western blotting assay was performed similarly as previously reported[16].In brief,SN12C cells were lysed in buffer (20 mmol Tris-HCl,pH 7.5,150 mmol NaCl,1.0 mmol EDTA,1.0 mmol EGTA,1% (v/v) Triton X-100,1xProtease Inhibitor Cocktail (Roche Applied Science)) for 30 min on ice with vortexing for 10seconds every 5 min.The lysates were centrifuged,separated by 10% SDS-PAGE,and transferred to a nitrocellulose membrane.The blots were stained with ECL reagents (GE Healthcare,RPN2132) and exposed to X-ray film.
1.6 Chromatin immunoprecipitation (ChIP) and ChIP-seq libraries ChIP assays,PBRM1 and E2F1 ChIP-seq libraries were performed as we previously reported[15-16].For ChIP-seq libraries,adapters were added after CHIP DNA preparation and the DNA was sequenced using the Illumina HiSeq 2 000 platform.The analysis and visualization were the same as previously described[16].
1.7 Dual luciferase reporter assay DNA fragments of E2F1 binding sites were obtained by a synthetic method (GenePharma,Shanghai).It was then cut with XhoI and BglII (New England Biolabs),the product fromE2F1 (760 bp) was cloned into pGL4.11Vector (Luc2P) and that fromPBRM1 (570 bp) was cloned into pGL4.24 Vector (Luc2P/ minP).T4 DNA ligase (New England Biolabs) was used for ligation.HEK 293T cells were plated in 24-well plates at 60% density.The next day,cells were transfected with Lipofectamine 3 000 as recommended by the manufacturer (Thermo Fisher Scientific).Firefly luciferase reporter constructs were co-transfected with the pRenilla luciferase plasmid at a 5∶1 ratio.Assays were performed using the dual luciferase reporter assay system (Promega,Madison,WI) 48 h later.Each experiment was repeated three times.
1.8 Data analysis Relative quantification of gene expression was performed using the 2-ΔΔCtmethod,whereas actin or GAPDH was used as a reference control.Statistical analyzes were performed using GraphPad PRISM® version 6(2-way ANOVA analysis or t-test) and P-values less than 0.05 were considered statistically significant.At least three replicates were used to obtain the data.
2 Results
2.1 Proliferation of SN12C cells was not significantly affected by PBRM1 knockdown After lentivirus infection,we successfully established knockdown cell lines of SN12C and UO31,andPBRM1 expression was significantly reduced at both mRNA (Fig 1A) and protein (Fig 1B) levels.To investigate whether stable knockdown ofPBRM1 affects growth of SN12C cells,we performed MTT assays.The results showed no significant difference after knockdown ofPBRM1 in SN12C cells (Fig 1C).This was also observed in UO31 cells (Fig 1D).
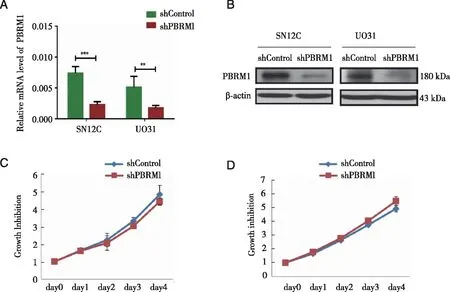
A,B:Established knockdown cell lines of SN12C and UO31,showing a remarkable reduction in PBRM1 expression at mRNA and protein levels;C,D:Stable knockdown of PBRM1 did not significantly affect the growth of SN12C cells and of UO31 cells;t-test; **:P<0.01; ***:P<0.001.
2.2 FACS analysis showing the mitotic index (MI) was increased when PBRM1 was reduced We thought the effect ofPBRM1 knockdown on cell proliferation might be too subtle in a short culture period.To overcome this obstacle,we performed a FACS assay.After synchronization,approximately 80% of the cells were arrested in the G0/G1phase (Fig 2A).Compared to the control,the knockdown cells showed no significant difference up to 18 hours.This difference increased and became most significant at 32 hours,where 36.44% of knockdown SN12C cells were in G2/M phase,while only 29.69% of control cells were in G2/M phase.Consequently,51.26% of the cells in the control sample were in G1phase,while 42.49% of the cells in knockdown cells were in G1phase (Fig 2A).The mitotic index (MI) (S+M/G0+G1) and the graph clearly showed a significant difference between control and knockdown cells.It was most prominent after 28 hours when the knockdown cells showed the maximum MI value (Fig 2B).
2.3 Depletion of PBRM1 up-regulating expression levels of E2F family members To further investigate the molecular mechanism thatPBRM1 deficiency increases MI,we examined the expression ofE2F1 as well as some other members of the E2F family,which play a crucial role in cell cycle control.WhenPBRM1 was deprived,we not only observed that E2F1 was significantly upregulated at the mRNA level,but also included other E2F transcription factors (except E2F7) (Fig 3A).Not surprisingly,some of the E2F transcription factors we examined,such as E2F1 and E2F3,also increased at the protein level (Fig 3B-C).
2.4 Binding of PBRM1 to the E2F1 promoter occurred in conjunction with acetylated histone markers To investigate whetherE2F1 is transcriptionally regulated by PBRM1 and whetherPBRM1 is a target of E2F1,we generated PBRM1 and E2F1 ChIP-seq libraries and performed ChIP qPCR validation.Interestingly,we observed that both PBRM1 and E2F1 bound to theE2F1 promoter region (Fig 4A-B).The PBRM1 binding signal was broadly distributed at the E2F1 promoter near the E2F1 binding site,while the E2F1 binding site was sharp and peaked at TSS.The two loci were close but did not overlap.We performed a PBRM1-ChIP assay and designed primers for two potential PBRM1 binding sites (site 1 and site 2),they showed consistent results as the ChIP-seq result (Fig 4B).
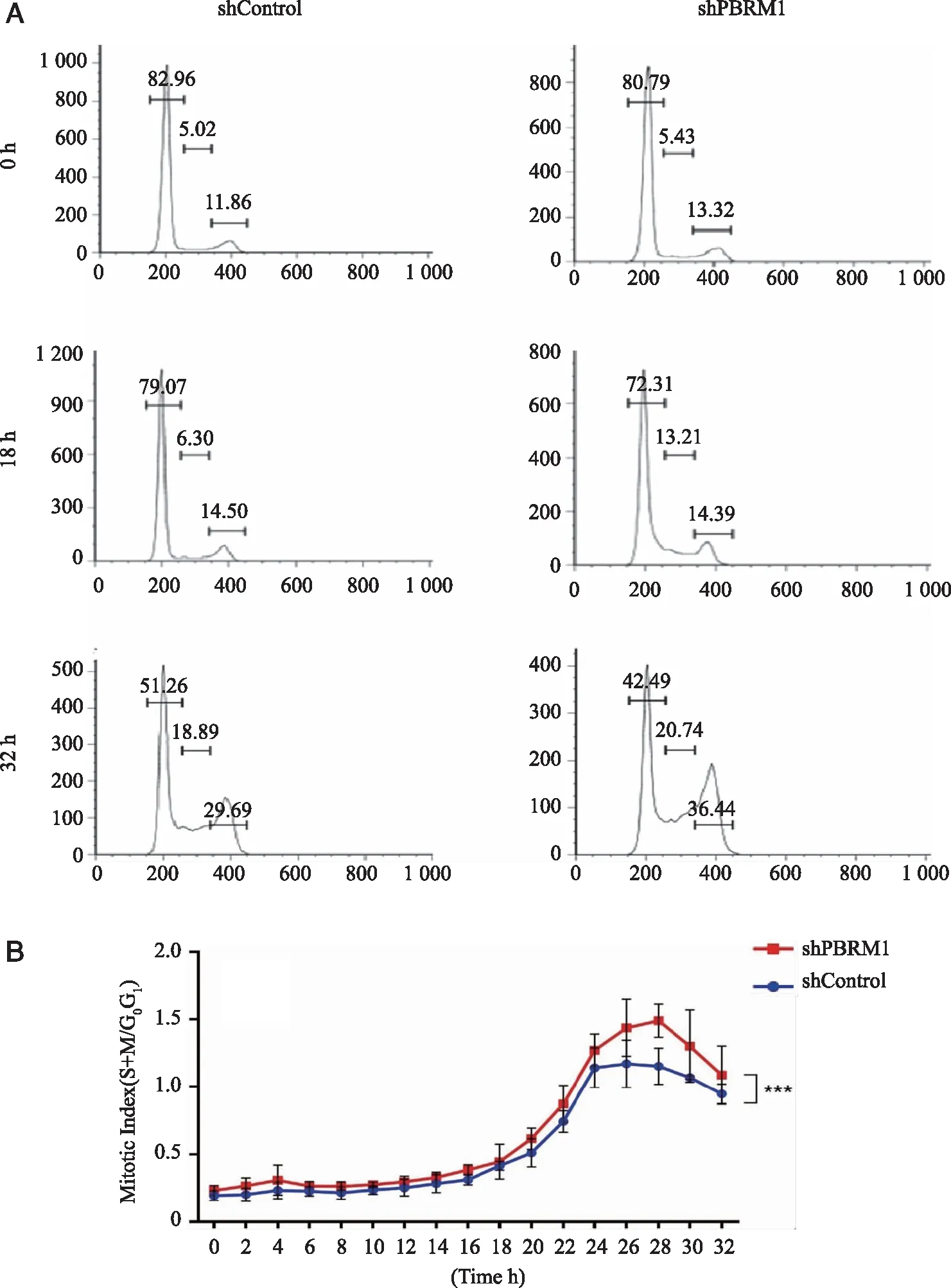
A:The knockdown cells showed significant difference at 32 hours,where 36.44% of the knockdown cells are in the G2/M phase,while only 29.69% for the control.Only one representative result was shown;B:The MI value differed most significantly at 28 hours between control and knockdown cells,indicating that a deficiency of PBAF SWI/SNF complex might favor the cell cycle process of SN12C cells;2-way ANOVA analysis,***:P<0.001.
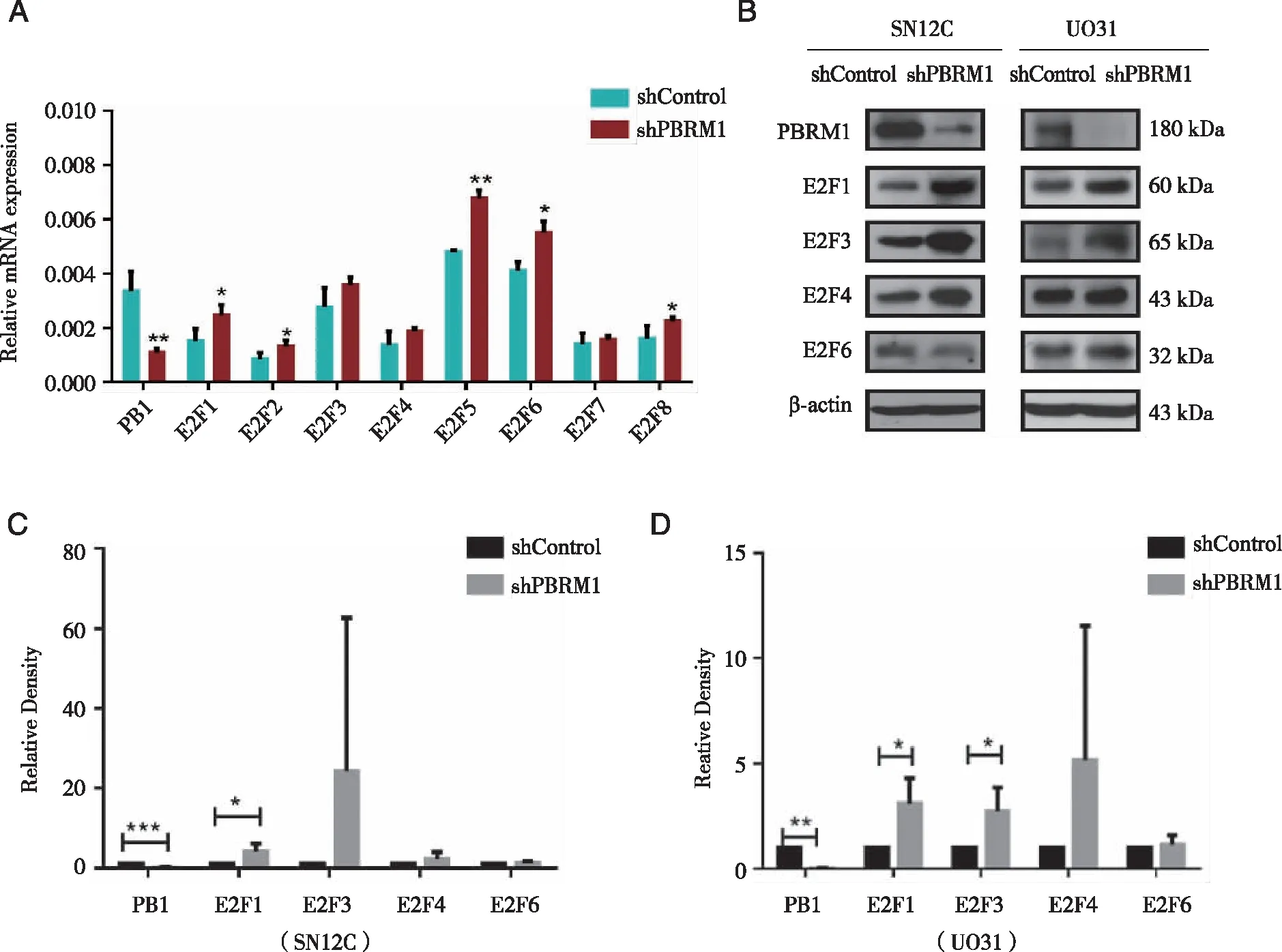
A:QPCR result for E2F1 and other E2F transcription factors.The majority of E2F members were upregulated at the mRNA level after PBRM1 knockdown; B:Western blot analysis for some E2F transcription factors; C:E2F1was consistently up-regulated in both cell lines; *:P<0.05; **:P<0.01; ***:P<0.001.
It is known that PBRM1 could interact with acetylated histones,such as H3K14ace[17].The coexistence of acetylated histone markers would be a prerequisite for binding of PBAF-SWI/SNF to specific regions of DNA.ChIP-qPCR analysis for selected acetylated histone modifications revealed high affinity of H3K9ac,H3K14ac,H3K27ac and H3K4me3 to theE2F1 promoter region,to which PBRM1 can bind (Fig 4C).PBRM1 is a specific subunit of the PBAF-SWI/SNF complex that functions by interacting with acetylated histones.For the reporter gene assay,we did not consider it feasible to study PBRM1 regulatory activity with plasmids lacking histones and related modifications.Thus,we could only examine the functional role of E2F1 binding sites.Our reporter assay showed that the construct containing the E2F1 binding region near E2F1 showed only weak but consistent luciferase expression (P=0.14,Fig 4D-E).
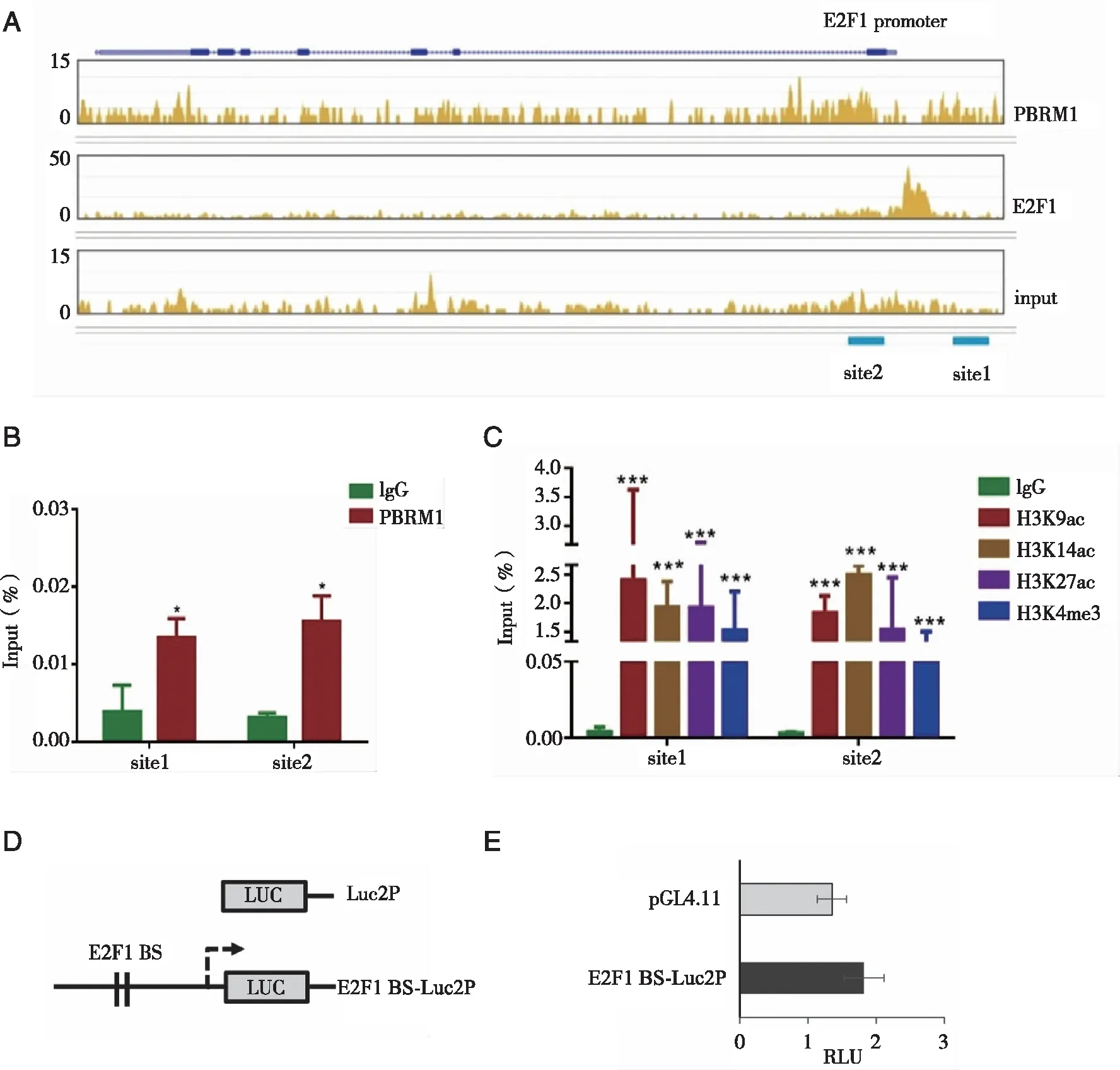
A:Snapshot for PBRM1 and E2F1 ChIP-seq on the E2F1 gene body showing that the PBRM1 binding signal was broadly distributed and the intensity was stronger around the transcription start site (TSS) of E2F1 promoter while E2F1 binding was sharp and peaked at TSS.Schematic for the E2F1 gene body and two PBRM1 binding sites for qPCR amplification;B:ChIP qPCR results showed that PBRM1 was recruited at E2F1 promoter site;C:The sites where PBRM1 can bind around the E2F1 promoter were enriched with histone modifications such as H3K9ac,H3K14ac,H3K27ac and H3K4me3.E2F1 locus:chr20:32262650-32275093 (hg19);D:HEK293T cells were transformed with reporter constructs;E:There is no significant difference between the empty vector and the construct containing a proximal E2F1 binding site.PBRM1 site 1:immediate upstream transcription start site; PBRM1 site2:immediately downstream of TSS; E2F1 BS,E2F1 binding site; RLU:relative luminometer units;*:P<0.05; ***:P<0.001.
2.5 E2F1 was recruited at the PBRM1 promoter site We next examined whetherPBRM1 is also a target of E2F1.Our data clearly showed that E2F1 was remarkably recruited to thePBRM1 promoter (Fig 5A-B),and the reporter assay showed that the luciferase expression of the construct containing theE2F1 binding site was significantly reduced (P=0.000 6)(Fig 5C-D).This suggests a possible repressive effect of this DNA element on the transcriptional control ofPBRM1 exerted by E2F1 and its family members.
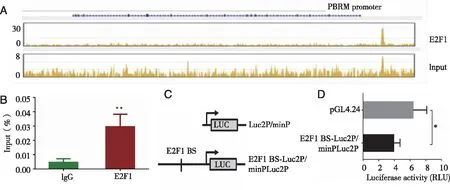
A:Snapshot for E2F1 ChIP-seq on the PBRM1 gene body showing the E2F1 binding signal peaks at the promoter of PBRM1;B:ChIP qPCR showed that E2F1 was remarkably recruited to the PBRM1 promoter site;C:HEK293T cells were transformed with reporter constructs shown;D:The luciferase activity of the construct containing the E2F1 binding site was reduced.PBRM1 locus:chr3:52,579,405-52,719,796 (hg19);*:P<0.05; **:P<0.01.
Furthermore,analysis of the TCGA dataset[18]showed thatE2F1 levels were significantly negatively correlated withPBRM1 mRNA levels in clinical RCC specimens (Spearman’s correlation=-0.216,P=4.85×10-7; TCGA ,Pan Cancer Atlas,Fig 6A).
Based on these data,we hypothesize that the PBRM1-SWI/SNF complex (PBAF,which contains PBRM1) binds to acetylated histones and recruits co-activators or co-repressors and eventually plays a repressive role in the expression of E2F transcription factors in general in ccRCC cells (Fig 6B).Therefore,PBRM1 deletion might increase the levels of E2F transcription factors.PBRM1 is also a target of E2F1,likely including other members of the E2F family.Among these factors,E2F1,E2F2,and E2F3 favor the cell cycle,while other members (such as E2F4 and E2F5) play opposite roles.The cell cycle process and cell proliferation are a balance between these two sets of transcription factors and likely involve downstream target genes regulated by the PBAF-SWI/SNF complex and E2F transcription factors.

A:E2F1 level was significantly negatively correlated with PBRM1 mRNA level in RCC clinical specimens (Spearman’s correlation=-0.216,P=8.07e-7; TCGA,Pan Cancer Atlas);B:The PBRM1-SWI/SNF complex (PBAF,which contains PBRM1) may play a repressive role in the expression of E2F transcription factors in general in ccRCC cells.
3 Discussion
It is expected that PBRM1 could play a crucial role in ccRCC,however,the functional roles and molecular mechanisms are largely unknown prior to the start of this study.Our data clearly showed thatPBRM1 deficiency increased the MI of SN12C cells,consistent with a recent report on ACHN cells thatPBRM1 silencing significantly increased the S-phase population of ACHN cells[19].This indicates that the PBRM1-SWI/SNF complex may play a repressive role in cell cycle control.
It is well known that E2F transcription factors play a crucial role in cell cycle control,so we investigated whether this increase in MI correlates with changes in E2F1 expression levels.Interestingly,we found that mRNA levels of some E2F members increased afterPBRM1 knockdown.Among these,E2F1 is most markedly increased at the protein level.This supports our hypothesis that the increase in MI in ccRCC cells byPBRM1 knockdown is mediated by E2F1 transcription factors.It has been reported that E2F1 is upregulated in a large number of ccRCC cases[20],and this is highly relevant to the high mutant rates ofPBRM1 in ccRCC.Thus,our data provide a possible explanation for the upregulated E2F1 level in ccRCC patients.
Our results showed that the E2F1 promoter is bound by both PBRM1 and E2F1,implying that expression of E2F1 could be co-regulated by both PBAF-SWI/SNF and E2F1.In contrast,thePBRM1 promoter region showed a repressive effect on luciferase expression,which impliesPBRM1 is a target of E2F1 and is suppressed under certain conditions,such as in cells with high proliferation capability.This relationship is consistent with analysis of TCGA clinical data thatE2F1 andPBRM1 mRNA levels were significantly negatively correlated.Our data suggest that the PBRM1-containing PBAF complex plays a repressive role in E2F1 expression and function.Cell cycle control is a delicate balance exerted by different transcription factors,as well as target genes regulated by the PBRM1-containing SWI/SNF complex and E2F transcription factors.
Taken together,our data provide new insights into the tumor suppressor genePBRM1 on cell cycle control and also revealed evidence thatPBRM1 andE2F1 form a regulatory feedback loop at the transcriptional level.This also implies that some E2F family members and their regulated genes could be potential drug targets forPBRM1-deficient ccRCC patients.
