QTL Mapping for Plant Height Using Introgression Lines Derived from Zhonghui 8015 and Wild Rice (Oryza rufipogon)
2022-10-25YangQinqinZhangYingxinXuePaoWenXiaoxiaLiuLingXuPengZhanXiaodengCaoLiyongChengShihuaWuWeixun
Yang Qinqin, Zhang Yingxin, Xue Pao, Wen Xiaoxia, Liu Ling, Xu Peng, Zhan Xiaodeng, Cao Liyong, 2, Cheng Shihua, Wu Weixun
Letter
QTL Mapping for Plant Height Using Introgression Lines Derived from Zhonghui 8015 and Wild Rice ()
Yang Qinqin1, #, Zhang Yingxin1, #, Xue Pao1, Wen Xiaoxia1, Liu Ling1, Xu Peng1, Zhan Xiaodeng1, Cao Liyong1, 2, Cheng Shihua1, Wu Weixun1
(China National Center for Rice Improvement / State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 310006, China; Northern Center of China National Rice Research Institute, Shuangyashan 155600, China; These authors contributed equally to this work)
Many excellent genes in wild rice have been lost during the domestication of wild rice to cultivated rice. In this study, introgression lines (ILs) were produced with a wildrice () accession, BJ194, as a donor parent and anrestorer line, Zhonghui 8015 (ZH8015), as a recipient parent to map QTLs for plant height. We identified four QTLs (,,, and) related to plant height distributed on chromosomes 2, 3 and 8. Furthermore, we sequenced and analyzedlocated in the interval of RM15753–RM3525, and found this QTL may be a new locus regulating rice plant height.
Among the agronomic traits of rice, plant height is one of the most important traits because it directly contributes to plant architecture, which affects rice yield (Wang et al, 2018; Wang et al, 2020). In general, higher rice plants are more susceptible to lodging, which reduces yield (Setter et al, 1997; Zhang et al, 2021). Therefore, breeding for dwarf plants based on the dwarf stem genein the 1950s resulted in a green revolution and marked the success of dwarf variety development. Rice yield has been greatly improved, and a higher harvest index has been achieved. Since then,has been called the ‘Green Revolution Gene’ (Peng et al, 1999). However, dwarf and semi-dwarf varieties show reduced photosynthetic efficiency, insufficient growth, and lower crop yield (Peng et al, 1994). Thus, there is no longer a requirement for plants to be short. However, a slight change in this trait drastically influences lodging and crop yields. Therefore, we need to identify more genes that regulate rice plant height and then lay a theoretical foundation for increasing rice yield. Multiple loci with large effects on plant height have been identified and cloned. For example,/encodes GA20 oxidase and participates in the gibberellin synthesis pathway, which in turn regulates plant height (Oikawa et al, 2004; Yano et al, 2012; Wu et al, 2016).is a suitable target gene for high-yield rice breeding. This example demonstrates that discovering genes that regulate rice plant height is significant for breeding.
To identify more genes that regulate plant height and to enrich breeding germplasm resources, three BC4F6ILs named IL66, IL102 and IL145 (Fig. S1), which were higher than ZH8015 under Hangzhou natural long-day conditions (NLDs; Fig. S2), were developed by introgressing chromosomal segments from the wild rice accession BJ194 into ZH8015 through multiple generations of backcrossing and selfing.Crosses between the three selected ILs and ZH8015, followed by selfing, generated three populations (IL66/ZH8015 BC5F2:3, IL102/ZH8015 BC5F2:3, and IL145/ZH8015 BC5F2:3), which were planted in Hangzhou NLDs in 2017 with 150 lines each and used to perform plant height QTL analysis and mapping (Fig. S1). The average value of plant height for each family was used to construct a frequency distribution histogram. Plant heights in the IL66/ZH8015 BC5F2:3population varied from 109.0 to 143.0 cm with an average of 126.2 cm (Fig. 1-A); plant heights in the IL102/ZH8015 BC5F2:3population varied from 89.1 to 162.7 cm, with an average of 122.6 cm (Fig. 1-B); and plant heights in the IL145/ZH8015 BC5F2:3population ranged from 107.4 to 124.6 cm, with an average of 116.3 cm (Fig. 1-C). Plant height for all the three populations was continuously distributed, with skewness values from -0.146 to -0.013. The distributions for IL66/ZH8015 BC5F2:3and IL145/ ZH8015 BC5F2:3were nearly normal, while the allocation for IL102/ZH8015 BC5F2:3had a considerable skewness value (Fig. 1-A to -C). The continuous distributions indicated that plant height is controlled by more than one locus. Thus, it is necessary to conduct QTL analysis of the three populations to identify the causal loci. We also investigated the number of effective tillers for the three groups and found that this trait showed a nearly normal distribution with skewness values ranging from -0.403 to -0.344 (Fig. S3).
To perform QTL analysis and identify the genomic regions associated with the plant-height regulation, we screened for polymorphisms between the three ILs and ZH8015 by 512 simple sequence repeat markers. As a result, 7, 30 and 22 polymorphic markers were identified between IL66/ZH8015, IL102/ZH8015, and IL145/ZH8015, respectively. Using the polymorphic markers, the BC5F2:3families of the three populations were genotyped (Fig. 1-D to -F), and then the combined genotypic and phenotypic data were used to conduct QTL analysis. A total of four plant height QTLs were detected from the three populations (Fig. 1-G).In the IL66/ZH8015 BC5F2:3population,was detected on chromosome 3 between markers RM16152 and RM85. The LOD value forwas 39.8 with an additive effect of 10.8 cm, and it accounted for 67.9% of the phenotypic variance. In the IL102/ZH8015 BC5F2:3population,andwere detected on chromosomes 2 and 8 in the intervals of RM6482–RM12368 and RM6193–RM3480, respectively.had a LOD value of 2.5 with an additive effect of -4.8 cm and explained 7.9% of the phenotypic variance in plant height.had a LOD value of 6.3 with an additive effect of -4.9 cm and explained 77.1% of the phenotypic variance. In the IL145/ZH8015 BC5F2:3population,was mapped on chromosome 3 between markers RM15717 and RM3525, and it had a LOD value of 4.5 with an additive effect of2.4 cm and explained 14.1% of the phenotypic variance (Fig. 1-G). We also performed a QTL analysis of the number of tillers but detected no QTL, which indicates that the QTLs related to plant height were not correlated with the tiller number.
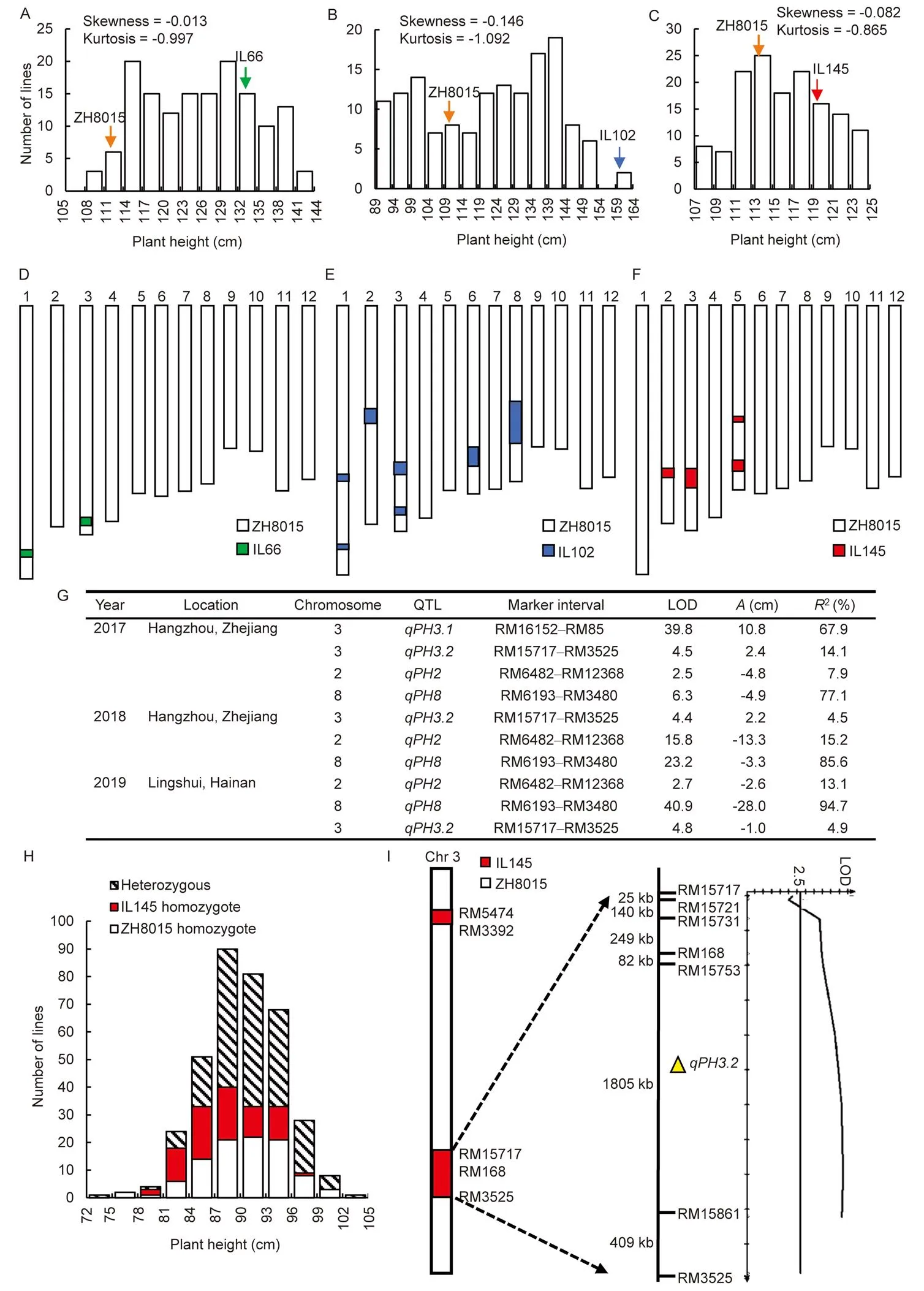
Fig. 1. QTLs for plant height in IL66/ZH8015 BC5F2:3, IL102/ZH8015 BC5F2:3, and IL145/ZH8015 BC5F2:3populations.
A–C, Plant height of IL66/ ZH8015 BC5F2:3(A), IL102/ ZH8015 BC5F2:3(B), IL145/ ZH8015 BC5F2:3(C) in Hangzhou in 2017. D–F, Graphical genotypes of IL66 (D), IL102 (E), and IL145 (F) in the genetic background of ZH8015. G, QTLs detected for plant height under natural long-day conditions in Hangzhou of Zhejiang and natural short-day conditions (NSDs) in Lingshui of Hainan, China. LOD, Log10likelihood ratio;, Additive effect of replacing a ZH8015 allele by anallele;2, Proportion of variance explained. H, Distribution of plant height in near isogenic line-F2populations ofunder NSDs. I, Putative QTL location in IL145/ ZH8015 BC5F5population under NSDs. The validation ofin the IL145/ ZH8015 BC5F5population and the physical distance between two adjacent markers are shown.
ZH8015, Zhonghui 8015. IL66, IL102 and IL145 are three introgression lines.
QTL analysis revealed only one large-effect plant height QTL () in the IL66/ZH8015 BC5F2:3population (Fig. 1-G). IL66 only harbored two wild rice fragments introgressed on chromosomes 1 and 3 in the ZH8015 background (Fig. 1-D). Therefore, we first focused on mapping the plant height QTL in IL66/ZH8015 BC5F2:3population. Among the 150 BC5F2:3families planted in Hangzhou in 2017, 32 heterozygous families with 160 individuals were selected and used for preliminarily mapping of, which was initially located in a 1 327.6-kb interval between markers RM16152 and RM85. Furthermore, 54 heterozygous individuals were chosen to develop small-F2populations. Nine polymorphic markers were developed between RM16152 and RM85, which finally locatedto a 138.8-kb region flanked by RM16217 and RM7389, harbored 24 putative open reading frames (ORFs). We found ORF1 encodes, which contains one nonsynonymous (T122to C122, resulting in Val41to Ala41) and one synonymous single-nucleotide polymorphism (T897to C897) between ZH8015 and IL66 (Fig. S4). The mutation mode was entirely consistent with previous studies in which the near isogenic lines (NILs) with Lemont genotype was shorter than the NILs with Teqing genotype (Wu et al, 2016), supportingas the most likely candidate gene for. Further complementation and CRISPR/Cas9 assays to confirm its function are in progress.
Subsequently, we focused on the IL102/ZH8015 BC5F2:3families because the QTLs in this population have more significant estimated effects on plant height than those in the IL145/ZH8015 BC5F2:3families (Fig. 1-G). We selected two plants from each of the 150 IL102/ZH8015 BC5F2:3families to develop 300 IL102/ZH8015 BC5F3:4families grown under NLDs in Hangzhou of Zhejiang in 2018 (Fig. S1).Consistently,andhave been detected again (Fig. 1-G), indicating that these QTLs are stably expressed in the population and that the alleles derived fromdecreased plant height. We then selected 30 BC5F4plants that were heterozygous forand homozygous for the ZH8015 allele ofin the ZH8015 backgroundto develop 30 NIL-F2populations of. We selected 38 BC5F4plants which were heterozygous forand homozygous for the ZH8015 allele ofin the ZH8015 background to develop 38 NIL-F2populations ofunder natural short-day conditions (NSDs) in Lingshui of Hainan in 2019 (Fig. S5), in whichandwere also detected (Fig. 1-G). QTLs with pleiotropic effects on plant height, heading date, and grain yield have previously been mapped on chromosome 2 () and chromosome 8 () (Yan et al, 2011; Liu et al, 2016), but these genes are not located within the intervals ofand, respectively. Therefore, we speculated thatandrepresent novel loci controlling plant height.
Finally, we carried out plant height QTL mapping of the IL145/ZH8015 BC5F2:3family populations. Two plants from each of the 150 IL145/ZH8015 BC5F2:3families were selected to develop 300 IL145/ZH8015 BC5F3:4families under NLDs in Hangzhou in 2018 (Fig. S1). Consistently,was detected again with a LOD score of 4.4 and an additive effect of 2.2 cm, and it explained 4.5% of the phenotypic variance (Fig. 1-G). The positive additive effect implies that the allele derived fromincreased the plant height. Since plant height is closely related to yield, we also investigated several other agronomic traits. First, we performed phenotypic comparisons of the plant height and internode length of ZH8015 and IL145 (Fig. S6-A). We found that the lengths of all the internodes except for the 5th internode were significantly different between ZH8015 and IL145 (Fig. S6-B). Therefore, IL145 may be taller than ZH8015 because of the more extension in the 1st to 4th internodes. In addition to affecting plant height, we found thatalso affected other agronomic traits such as 1000-grain weight, grain length, grain width, length-width ratio, panicle length, seed-setting rate, and the number of grains in the main panicle (Fig. S7).
Next, 6 BC5F4plants that were heterozygous atwere developed into a BC5F5population of 457 individuals under NSDs in Hainan in 2019 (Fig. 1-H). QTL analysis of this population detectedagain between the markers RM15717 and RM3525 with an additive effect of -1.0 cm and a LOD value of 4.8, explaining 4.9% of the phenotypic variation (Fig. 1-G). Moreover,showed over dominance (|d/a| = 1.42 > 1.20) according to Stuber’s criteria (Stuber et al, 1987), and QTL analysis also revealed thatis a minor-effect locus. Interestingly, the negative additive effect ofin Hainan NSDs showed that the allele derived fromdecreased plant height, indicating that this locus may also be regulated by photoperiod (Fig. S8). Finally, four new polymorphic markers (RM15721, RM15731, RM15753 and RM15861) around RM15717 and RM3525 were used to screen the 457 BC5F5plants to identify recombinants within the target region (Table S1).was mapped to a 2 213-kb region between the markers RM15753 and RM3525 on the long arm of chromosome 3 (Fig. 1-I).
According to the Gramene (www.gramene.org) database, only one gene reported to be related to plant height namedis located in the target interval of(Choi et al, 2012). We first compared the sequences in the coding region between ZH8015 and IL145 and found no differences. Then, we performed sequence comparisons in the promoter region and found 27 single nucleotide polymorphisms and 1 insertion/deletion (Fig. S9-A). To make the results more accurate, we detected the expression levels ofin the two parents, and the results showed no significant difference in their expression levels (Fig. S9-B). Hence,is not the candidate gene for, and we expectedto be a novel gene that regulates plant height with minor effect. In conclusion, asis a minor-effect plant-height QTL, it will be helpful in marker-assisted breeding programs in rice aimed at fine-tuning plant height by developing an ideotype, and further fine-mappingis needed in the future.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (Grant No. 31961143016), the National Rice Industry Technology System of China (Grant No. CARS-01-03), the ‘14th Five-Year Plan’ Major Special Projects for Breeding New Rice Varieties of Zhejiang Province, China (Grant No. 2021C02063-1), the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. CAAS-ASTIP2013-CNRRI), and Hainan Yazhou Bay Seed Laboratory Oratory, China (Grant No. B21HJ0219).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
File S1. Methods.
Fig. S1. Breeding scheme for development of QTL mapping populations.
Fig. S2. Phenotypes of Zhonghui 8015 (ZH8015) and three introgression lines.
Fig. S3. Frequency distribution of tiller number in three populations in Hangzhou in 2017.
Fig. S4. Fine mapping and candidate analysis of.
Fig. S5.Distribution of plant height in near isogenic line-F2populationsofandunder natural short-day conditions in 2019.
Fig. S6. Plant heights of IL145 and Zhonghui 8015 (ZH8015).
Fig. S7. Comparison of different agronomic traits between Zhonghui 8015 (ZH8015) and IL145.
Fig. S8. Comparison of plant height between Zhonghui 8015 (ZH8015) homozygote and IL145 homozygote.
Fig. S9. Promoter sequence analysis ofand transcript levels ofin Zhonghui 8015 (ZH8015) and IL145.
Table S1. Primers used in this study.
Choi M S, Koh E B, Woo M O, Piao R H, Oh C S, Koh H J. 2012. Tiller formation in rice is altered by overexpression ofgene encoding an IAA-conjugating enzyme or exogenous treatment of free IAA., 55(6): 429–435.
Liu J H, Shen J Q, Xu Y, Li X H, Xiao J H, Xiong L Z. 2016., a-like gene, confers drought sensitivity through regulation of senescence in rice., 67(19): 5785–5798.
Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M. 2004. A role of, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice., 55(5): 687–700.
Peng J, Richards D E, Hartley N M, Murphy G P, Devos K M, Flintham J E, Beales J, Fish L J, Worland A J, Pelica F, Sudhakar D, Christou P, Snape J W, Gale M D, Harberd N P. 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators., 400: 256–261.
Peng S, Khush G S, Cassman K G. 1994. Evaluation of a new plant ideotype for increased yield potential.: Cassman K G. Breaking the Yield Barrier: Proceedings of a Workshop on Rice Yield Potential in Favourable Environments. Los Banos, the Philippines: International Rice Research Institute: 5–20.
Setter T L, Laureles E V, Mazaredo A M. 1997. Lodging reduces yield of rice by self-shading and reductions in canopy photosynthesis., 49(2/3): 95–106.
Stuber C W, Edwards M D, Wendel J F. 1987. Molecular marker- facilitated investigations of quantitative trait loci in maize: II. Factors influencing yield and its component traits., 27(4): 639–648.
Wang B, Smith S M, Li J Y. 2018. Genetic regulation of shoot architecture., 69: 437–468.
Wang Y X, Shang L G, Yu H, Zeng L J, Hu J, Ni S, Rao Y C, Li S F, Chu J F, Meng X B, Wang L, Hu P, Yan J J, Kang S J, Qu M H, Lin H, Wang T, Wang Q, Hu X M, Chen H Q, Wang B, Gao Z Y, Guo L B, Zeng D L, Zhu X D, Xiong G S, Li J Y, Qian Q. 2020. A strigolactone biosynthesis gene contributed to the green revolution in rice., 13(6): 923–932.
Wu Y, Wang Y, Mi X F, Shan J X, Li X M, Xu J L, Lin H X. 2016. The QTLencodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems., 12(10): e1006386.
Yan W H, Wang P, Chen H X, Zhou H J, Li Q P, Wang C R, Ding Z H, Zhang Y S, Yu S B, Xing Y Z, Zhang Q F. 2011. A major QTL,, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice., 4(2): 319–330.
Yano K, TakashiT, Nagamatsu S, Kojima M, SakakibaraH, Kitano H,Matsuoka M, Aya K. 2012. Efficacy of microarray profiling data combined with QTL mapping for the identification of a QTL gene controlling the initial growth rate in rice., 53(4): 729–739.
Zhang M H, Mo Z W, Liao J, Pan S G, Chen X F, Zheng L, Luo X W, Wang Z M. 2021. Lodging resistance related to root traits for mechanized wet-seeding of two super rice cultivars., 28(2): 200–208.
31 December 2021;
28 June 2022
Copyright © 2022, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2022.06.001
Wu Weixun (wuweixun@caas.cn); Cheng Shihua (chengshihua@caas.cn)
杂志排行
Rice Science的其它文章
- Transcriptome Analysis Provides Novel Insights into Salt Stress Response in Two Egyptian Rice Varieties with Different Tolerance Levels
- Genetic Dissection of Quantitative Trait Loci for Panicle Traits and Heat Tolerance by High-Density Bin Map in Rice
- Improvement of Rice Production under Drought Conditions in West Africa:Application of QTLs in Breeding for Drought Resistance
- Characterization of a Novel Weak Allele of RGA1/D1 and Its Potential Application in Rice Breeding
- CRISPR/Cas9-Targeted Knockout of Rice Susceptibility Genes OsDjA2 and OsERF104 Reveals Alternative Sources of Resistance to Pyricularia oryzae
- Identification of Potential Zinc Deficiency Responsive Genes and Regulatory Pathways in Rice by Weighted Gene Co-expression Network Analysis
