环境因素所致印迹基因改变与子代器官发育
2022-03-08曲卉柳毅陈雅文汪晖
曲卉,柳毅,陈雅文,汪晖,2
综 述
环境因素所致印迹基因改变与子代器官发育
曲卉1,柳毅1,陈雅文1,汪晖1,2
1. 武汉大学基础医学院药理学系,武汉 430071 2. 发育源性疾病湖北省重点实验室,武汉 430071
印迹基因是由大约100个基因组成的一类特殊子集,主要以亲本单等位基因的方式表达,对胚胎的生长发育具有重要作用。近年来发现,环境因素所引起的印迹基因表观遗传修饰改变可造成胎儿多脏器发育不良甚至成年后多疾病易感,且存在多代遗传效应。本文基于国内外最新研究进展,总结了印迹基因表达改变对个体发育阶段以及生命后期器官功能的影响,提出环境有害因素所致印迹基因表观遗传修饰及表达异常是子代多器官发育不良的重要发生机制,这对于理解个体发育过程中印迹基因表达改变所引起的表型改变及探寻疾病早期防治策略具有重要意义。
印迹基因;环境因素;印迹基因的调控机制;表观遗传修饰;器官发育
印迹基因(imprinted gene)是一类主要以亲本单等位基因方式表达的基因,其表达受到表观遗传的精密调控。由于印迹基因高度参与胎儿的生长发育,且在受精后发生的全基因组去甲基化过程中某些表观遗传修饰会保留并传递给子代,因此印迹基因的异常表观遗传修饰及表达改变可导致子代多疾病易感[1]。研究表明,在配子和胚胎发育过程中,多种环境因素如酒精、香烟、亲代应激等通过改变印迹基因表达来影响胚胎生长发育和能量平衡,导致其成年后代谢性疾病易感及发生[2],并且该过程可能与表观遗传修饰异常有关[3]。本文结合近年来的国内外研究进展,对环境因素所致印迹基因改变与个体多器官发育及其调控机制进行综述,有助于理解表观基因组、基因组和环境之间的相互作用,为发育早期采取新的干预治疗提供思路。
1 印迹基因定义、功能及作用机制
1.1 印迹基因定义、分类及功能
印迹基因是一类以基因簇形式存在的特殊基因,每个簇内通常包含1个印迹控制区域(imprinting control region, ICR)。该ICR的甲基化印迹状态只在胚胎的一条亲源染色体上维持,并且能够调控整个结构域内多个印迹基因的表达。印迹基因分为父源性和母源性印迹基因。父源性印迹基因是指父源性等位基因位点带有印迹,如胰岛素样生长因子2(insulin-like growth factor 2,)和等,其功能为促进胎儿生长,最大化地利用母体资源。母源性印迹基因则相反,如和等,其功能为控制胎儿生长,以节省对母体资源的需求[4]。除直接调节胎儿生长发育外,印迹基因还可以通过调节胎盘的内分泌和运输功能,间接地促进胎儿的生长[5,6]。
1.2 印迹基因发生、维持和消除
来源于不同亲本的印迹基因在个体发育过程中承担着不同的重要功能。印迹基因通过建立、维持和消除基因组印记,参与调控个体的发育过程。印迹基因的表达与亲本等位基因的DNA甲基化印记有关。亲本印记的建立需要生殖细胞首先删除父母表观基因组的DNA甲基化标记,进一步于子代配子形成期间以等位基因特异性方式重建DNA甲基化印记[7,8]。这个DNA甲基化的动态过程包括发生、维持和消除三个部分。在原始生殖细胞(primordial germ cells, PGCs)形成时期,细胞发生DNA去甲基化,以擦除亲本表观遗传“记忆”[9]。进一步,PGCs发育至成熟配子,重新建立起DNA甲基化印记。受精后,DNA甲基化印记会在整个胚胎发育时期和生命后期得以维持[10]。然而,在发育胚胎的PGCs中,先前建立的DNA甲基化印记再次被擦除,以在配子形成的阶段重新建立新的印记,从而完成印迹周期(图1)。这种循环的甲基化和去甲基化过程,将有助于细胞特性的建立、维持和动态变化,以保证所形成的印记在体细胞分裂过程中能稳定遗传,进而使得胚胎可以正常发育。
2 影响印迹基因表达的环境因素
已知印迹的建立、维持和擦除发生在受精和胚胎发育早期[11],在这一过程中表观基因组易受到环境因素的影响,导致印迹基因的表达改变。环境因素主要包括外源性因素(如化学因素、物理因素、生物因素等)和亲代因素(如应激、疾病状况等)。
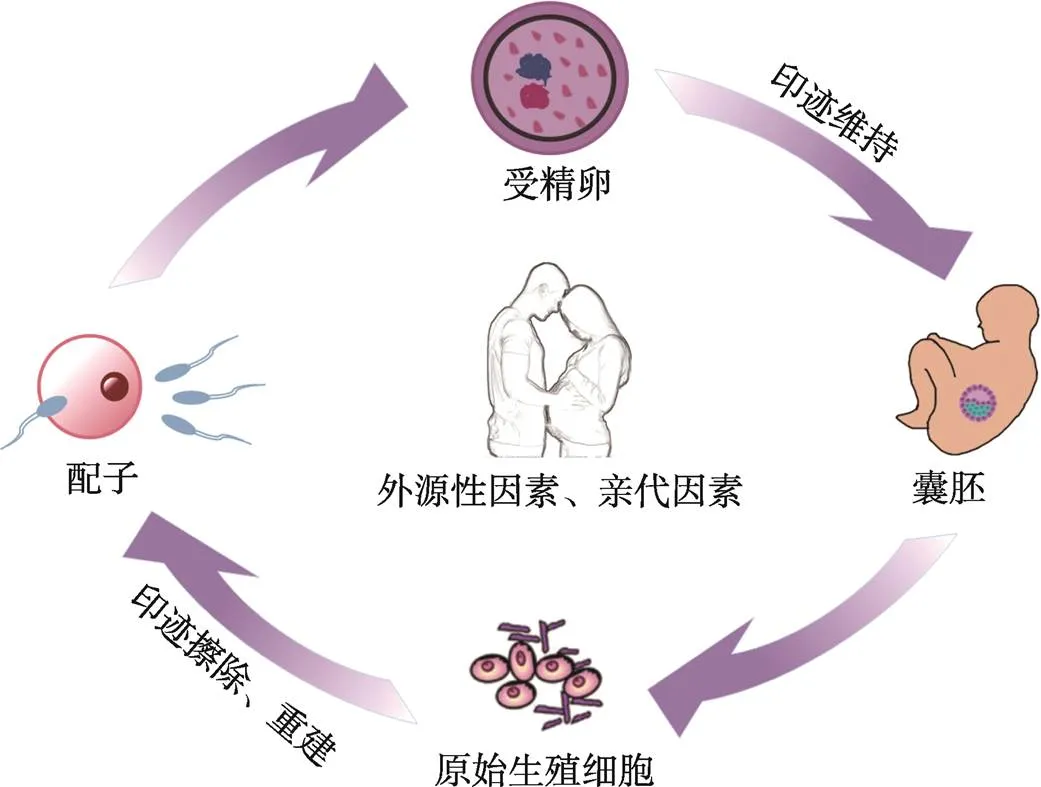
图1 印迹基因发生、维持和消除
随着生活水平的日益提高,酒精、香烟、咖啡因等物质已经成为大多数男性和女性日常生活中不可或缺的一部分。然而,有证据表明,亲代接触酒精、香烟以及咖啡等环境有害因素会导致子代发育等远期危害[12,13]。临床研究和动物模型显示,亲代暴露于多种外源性因素,如化学因素(如酒精、尼古丁等)、物理因素(如电离辐射等)、生物因素(如细菌、真菌等),印迹基因的表观遗传调控方式会发生改变,使其表达持续改变。而这些改变往往可导致子代发育异常,如宫内发育迟缓(intrauterine growth retardation, IUGR)、早期代谢缺陷、大脑神经发育障碍等[14~16]。同时,随着辅助生殖技术(assisted reproductive technology, ART)的快速发展,其安全性也成为社会关注的问题之一。由于ART发生在雌雄配子结合期间和受精后,在这一过程中胚胎冷冻解冻、体外培养、胚胎移植等因素可能会诱导胚胎DNA甲基化印记建立和维持的异常[17],从而增加了不良妊娠结局和出生后成年疾病发生的风险。既往研究表明,ART可导致早产、低出生体重等不良妊娠结局的发生[18,19],并与成年后心血管疾病和代谢性疾病的发生密切相关[20,21]。
亲代的疾病状况也是影响胎儿发育的关键因素。生物体的代谢状态可以直接影响表观遗传修饰[22]。例如,父体体重指数与其精子、胎儿脐带血中DNA甲基化水平之间存在显著的相关性。亲代肥胖可能通过影响生殖细胞中印迹基因的表观遗传修饰,造成胎儿表观基因组改变,从而影响胎儿的发育进程[23]。应激是指各种紧张性刺激物引起个体产生非特异性反应,导致体内生理或心理平衡失调的长期过程。亲代应激是改变胎儿表观遗传标记从而造成胎儿发育不良的另一重要诱因[24]。研究表明,孕妇产前抑郁症会导致胎盘印迹基因的表达改变,从而介导胎盘功能受损,最终导致胎儿生长受限和神经发育不良[25]。在父体中也存在类似的现象[26]。其发生机制可能与亲代下丘脑–垂体–肾上腺轴功能改变及血糖皮质激素水平升高诱导的生殖细胞发生过程中甲基供体或甲基化酶、去甲基化酶表达改变,从而间接影响胎儿表观基因组有关[27]。因此,亲代因素也被认为是调控印迹基因的表观遗传修饰,最终影响子代发育的重要因素之一(表1)。
3 环境因素所致印迹基因改变与器官发育
印迹基因在胎盘和胎儿多器官发育过程中扮演着重要的角色。越来越多的证据表明,在发育早期阶段,环境因素易影响印迹基因的表观遗传修饰及表达,进而编程子代多种器官发育及成年后多疾病易感。这种基因组印记对发育过程具有长期的、决定性的影响,因此探寻环境因素所致印迹基因改变及其调控机制对阐明机体发育编程改变及疾病发生至关重要。
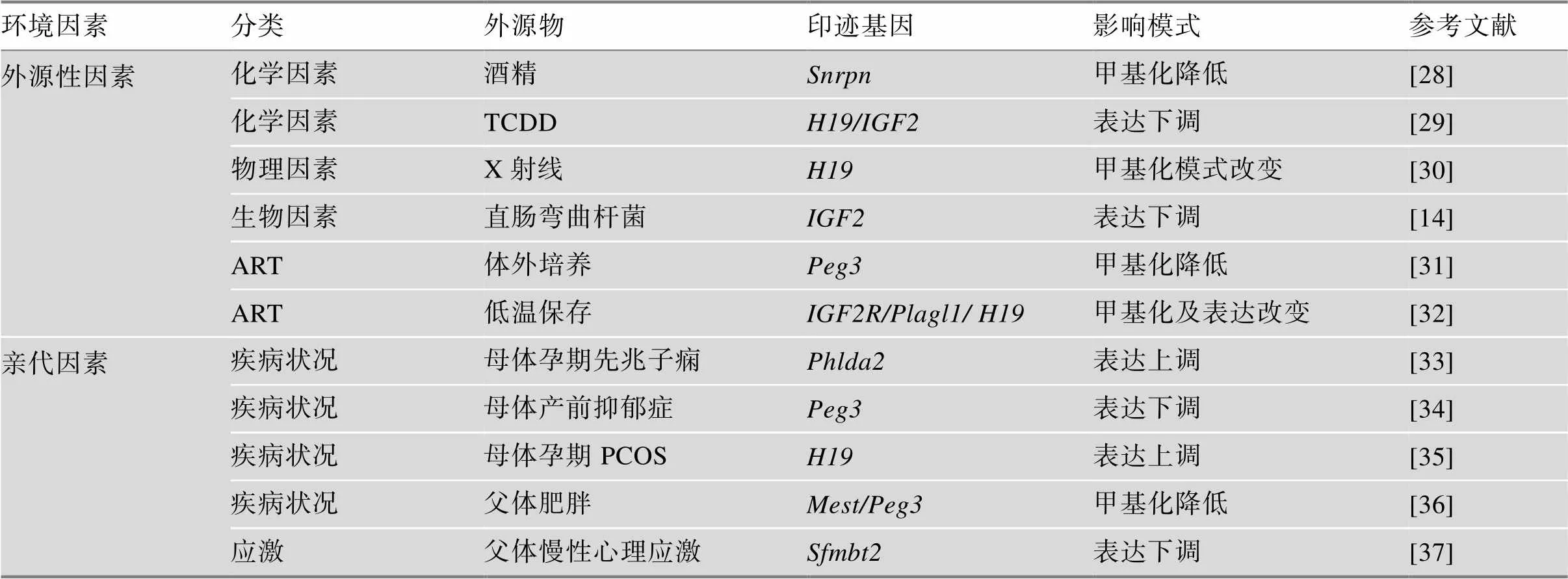
表1 影响印迹基因表达的因素
TCDD:2,3,7,8-四氯苯并-对-二噁英;PCOS:多囊卵巢综合征。
3.1 环境因素所致印迹基因改变与胎盘发育
胎盘是促进母体和胎儿相互作用的器官。在发育早期阶段,环境因素所致印迹基因的表观遗传修饰及表达改变可能通过编程胎盘发育,从而干扰子代的正常发育过程。大量印迹基因在人的胎盘中表达,并在调节胎盘发育和胎儿生长过程中发挥着重要的作用。例如,印迹基因生长因子受体结合蛋白10(growth factor receptor bound protein 10,) 可调控胎盘和胎儿的生长,并参与调控mTOR信号通路,进而参与调节胎盘的转运功能[38],当其在胎盘特异性敲除后可导致子代小鼠过度生长[39]。此外,印迹基因对早期胎盘形成以及对胎儿营养供应方面起着不可或缺的作用[40]。研究显示,主要通过IGFs信号通路促进胎儿的生长发育,其过表达则导致胎盘肥大和胎儿过度生长[41]。Cao等[42]发现,孕期三氯生暴露通过抑制胎盘Akt-mTOR- P70S6K信号通路,影响胎盘发育和营养物质运输,导致胎儿低出生体重。一项队列研究显示,孕期接触三氯生可引起胎盘等多个印迹基因DNA甲基化水平升高[43]。提示,环境因素可能通过诱导胎盘印迹基因DNA甲基化修饰,参与调控mTOR信号通路,引起胎盘中氧气和营养物质运输障碍,导致不良妊娠结局发生。此外,ART可诱导人胎盘印迹基因和表达增加,子代表现为IUGR。其发生机制可能与ART引起的母体高雌激素水平通过降低ICR的DNA甲基化修饰,增加ICR的DNA甲基化水平有关[44]。近几年的研究提示,ART可诱导人胎盘ICR DNA低甲基化,同时伴随ICR组蛋白H3的第9位赖氨酸甲基化(tri-methylation at lysine 9 on histone H3, H3K9me3)、H3K9me2降低和H3K4me2升高,共同作用导致染色质结构疏松从而促进转录[45]。提示,组蛋白修饰和DNA甲基化可能共同作用参与调控ART所致的胎盘中印迹基因表观遗传修饰异常。在出生时胎盘ICR的表观遗传修饰状态可能对胎儿的发育产生显著影响。然而,在这种机制作用下的具体影响尚不清楚。这些研究表明,环境因素可通过影响印迹基因表观遗传修饰及表达,导致胎盘转运功能等异常,最终调控胎盘和胎儿的发育过程。
3.2 环境因素所致印迹基因改变与肝脏发育
肝脏是人体的重要代谢器官, 参与糖脂代谢、生物转化等过程。印迹基因参与调控肝细胞增殖与分化,并影响与胎儿肝脏发育相关基因的表达,从而调控肝脏发育[46,47]。临床研究显示,在人类肝脏相关疾病(如肝纤维化、肝硬化、原发性硬化性胆管炎和原发性胆汁性肝硬化)的患者中,表达显著升高[48]。此外,妊娠期使用二甲双胍可致胎儿肝中表达增加,通过降低肝细胞核因子4α(hepatocyte nuclear factor 4α, Hnf4α)的甲基化水平而增加其表达[49]。而Hnf4α表达增加会阻碍肝脏分化、影响肝结构,并且在上调糖异生相关基因的表达中具有不可或缺的作用[50]。基于此推测,胎儿发育过程中可能通过调控Hnf4α进而影响肝脏的发育及功能。在胎儿肝脏中高度表达的模式提示,孕期二甲双胍暴露引起的表达增加可能是成年代谢性疾病发生的潜在机制之一。Zhu等[51,52]发现,高脂饮食通过抑制组蛋白去乙酰化酶3(histone deacetylase 3, HDAC3)诱导小鼠肝脏表达升高,而作为miR-214竞争性内源RNA,促进叉头状转录因子O1(forkhead box transcription factor, FoxO1)表达,从而抑制糖异生相关基因表达,导致肝脏胰岛素抵抗发生。还有研究显示,在父体慢性应激的小鼠模型中,F1代小鼠肝脏中印迹基因启动子区DNA甲基化水平升高,使内含子miR-466b-3p表达下调,其转录后抑制磷酸烯醇式丙酮酸羧激酶(phosphoenolpyruvate carboxykinase, PEPCK)表达,最终造成F1代小鼠出现高血糖[53]。提示,亲代慢性应激可调控子代肝脏中印迹基因的表观遗传修饰,从而引起子代肝脏发育异常及代谢性疾病易感。以上研究显示,印迹基因主要参与调控胎儿的肝脏发育及出生后的代谢功能,生命早期遭受的环境刺激可对印迹基因产生显著影响,并且可能和肝脏相关疾病的发生密切相关。
3.3 环境因素所致印迹基因改变与脑发育
基因组印记广泛存在于大脑中,并参与调控神经发育、突触功能、社会行为、情绪和认知等。是大脑中研究的较为成熟的母源性印迹基因,其存在于神经突触中,对于调控神经可塑性非常重要。过表达是孤独症谱系障碍(autism spectrum disorder, ASD)发生的重要因素之一[54]。研究显示,可通过影响结节性硬化症复合物2(tuberous sclerosis complex 2, TSC2)表达,参与调控mTOR信号通路[55]。在ASD人脑中,mTOR信号传导的中断,导致正常发育树突棘修剪所需的自噬水平下调,表现为突触功能缺陷[56]。提示,发育过程中大脑的突触功能障碍可能与介导神经元mTOR信号传导异常有关。近几年,研究人员开始探索在人类大脑神经元中表达异常与染色质相关的全基因组效应。研究发现,患有15q重复综合征的人脑中表达升高,通过调节组蛋白H2A.Z单泛素化,引起大脑发育相关基因差异甲基化区域的DNA甲基化水平改变[57]。提示,疾病因素所致大脑发育相关基因的表达异常可能起始于脑神经元中表达改变,其潜在机制可能与组蛋白泛素化引起的DNA甲基化修饰异常有关,但其具体的信号通路还需进一步探究。此外,印迹基因表达改变也会影响海马的发育过程。例如,母体摄入甜菜碱可诱导F1代海马中启动子区出现高甲基化,进而抑制表达及其下游AKT信号通路,并伴随雌激素受体相关基因表达降低,最终导致子代海马发育异常[58]。已知雌性大鼠海马中雌激素参与调控和IGFBP2表达,因此甜菜碱所致胎儿海马发育异常相关的编程机制可能与雌激素结合其核受体后,引起下游表观遗传修饰改变有关。以上研究表明,印迹基因通过参与调控神经发育相关信号通路,影响全基因组DNA甲基化以及大脑发育相关基因的表达,在大脑发育多个过程中发挥着重要的作用,其异常表达可能引起神经发育障碍,最终导致神经相关疾病的发生。
3.4 环境因素所致印迹基因改变与性腺发育
性腺是下丘脑–垂体–性腺轴的重要组成部分。印迹基因是性腺发育中重要的调控因子。与女性颗粒细胞中促性腺激素受体的表达有关,能够促进卵泡的增殖、成熟以及雌二醇、孕酮的产生[59]。研究发现,宫内暴露于TCDD的雌性大鼠卵巢中和表达降低,表现为血雌二醇水平升高、卵泡刺激素生成减少及原始卵泡数减少。然而,卵巢中ICR的甲基化水平并未发生改变,只有少数CpG位点甲基化水平升高[60]。这表明TCDD所致雌性子代卵巢发育毒性可能与和低表达有关,并且还存在其他调控机制。此外,印迹基因簇是调控促性腺激素重要的印迹区域。有文献报道,妊娠期全氟辛酸暴露可通过降低睾丸中印迹基因簇的靶基因(等)表达水平,导致雄性子代血清睾酮水平降低及睾丸发育异常[61]。已知,印迹基因簇存在DMRDMR等多个差异甲基化区域(differentially methylated region, DMR)。这提示,全氟辛酸可能诱导印迹基因簇中DMR甲基化发生改变,进而对睾酮的生成及生殖功能产生影响。总之,上述一系列证据提示,环境因素对印迹基因的调控具有长期的影响,不仅可以直接影响子代性腺的发育,还可以通过调节性腺激素的生成继而影响其生殖功能。在这一过程中,表观遗传遗传修饰扮演着重要的角色,可能是不良环境因素介导不育症等疾病发生的关键机制。
3.5 环境因素所致印迹基因改变与其他器官发育
印迹基因还参与了其他器官的发育,如肾上腺、骨和垂体腺等。本实验室前期研究发现,孕期咖啡因暴露可致子代大鼠肾上腺甾体合成功能抑制,其发生机制与宫内母源性高浓度糖皮质激素激活肾上腺受体(glucocorticoid receptor, GR),降低CTCF表达并抑制其与结合,同时增加DNA甲基转移酶3a(DNA methyltransferase 3a, Dnmt3a)表达并促进其与结合,进而导致高甲基化及低表达有关[62]。已知宫内机体糖皮质激素水平是决定胎组织成熟及其出生后命运的关键[63]。糖皮质激素可能通过调控印迹基因的表达,参与调控环境因素所致的子代肾上腺发育不良。还有研究证实,环境因素所致印迹基因的表达改变会影响骨发育过程。例如,父体摄入低蛋白饮食,胎盘等印迹基因表达升高,并伴随Dnmt1、Dnmt3a等表达升高,胚胎表现为骨矿物质沉积[64]。此外,印迹基因可以调控垂体腺的发育及相关激素的分泌。敲除小鼠表现出生长发育迟缓、出生后追赶性生长和早期肥胖等表型[65,66],而过表达小鼠在禁食期间垂体生长激素产生增加、脂质代谢增强[67]。已有文献指出,在个体早期发育过程中,环境因素可诱导表达发生改变[61]。提示,环境因素可能通过调控垂体生长激素的产生,进而对机体的代谢功能产生影响。然而,关于环境因素相关的印迹基因改变对其它器官发育的影响报道较少,仍有待进一步研究。
4 环境因素所致印迹基因改变的多代遗传效应及其机制
众所周知,亲代在备孕期间接触不良环境因素可影响子代生长发育,导致其出生后成年疾病的发生,并可能持续多代。在过去几年中,越来越多的证据显示,不良环境暴露相关的印迹基因表观遗传修饰改变在经历两次重编程事件后仍得以保留[68]。因此推测,父体和母体配子中印迹基因的表观遗传修饰改变,可能是解释子代表型改变及多代遗传效应发生的重要依据。例如,暴露于苯并吡喃的雄性F0代大鼠精子中和等印迹基因甲基化水平发生改变,且F1、F2代精子中这两个印迹基因的甲基化水平与F0代具有良好的一致性[69]。提示,环境因素引起生殖细胞中的印迹基因DNA甲基化改变并可能传递多代。还有研究显示,在父体慢性应激的小鼠模型中,F0代精子和F1代肝脏中印迹基因启动子区DNA甲基化水平升高,F1代小鼠出现高血糖[53]。此外,当给予亲代慢性应激和GR拮抗剂RU486后,可发现相关表观遗传修饰及子代高血糖现象被逆转。提示,糖皮质激素参与调控亲代慢性应激所致生殖细胞中印迹基因的表观遗传修饰异常及子代多疾病易感。此外,ZFP57是印迹基因重要的调控因子,环境因素还可能通过ZFP57调控印迹基因的表达,从而介导多代遗传效应的发生。Legoff等[70]发现,雄性F1代宫内杀虫剂暴露可引起其生殖细胞中ZFP57表达降低,并且ZFP57的靶基因在H3K27me3区域富集,这种甲基化改变在F2代和F3代中依旧存在。已知组蛋白修饰是配子形成和胚胎植入前的关键调控步骤[71]。因此推测,环境因素可能通过ZFP57影响印迹基因的H3K27me3水平,该修饰可通过生殖细胞传递给子代,从而干扰子代的发育过程。除了DNA甲基化和组蛋白修饰两种表观遗传调控方式以外,非编码RNA对印迹基因的调控也越来越受到学者们的广泛关注。非编码RNA是一类不编码蛋白质的RNA,它们可在RNA水平上行使各自的功能,并广泛参与生命活动中重要的生物功能,如生物个体的发育、生殖、细胞重编程等,与人类疾病的发生密切相关。Short等[72]发现,接受皮质酮治疗的雄性小鼠精子中多种miRNA含量改变,并预测到其可与结合,导致子代海马中表达增加。这一现象表明,环境因素可能通过影响精子中miRNA的含量,诱导精子中印迹基因的表达改变,从而实现印迹基因从亲代到子代的传递。由于miRNA还可以调控其他表观遗传修饰[73],因此另一种可能机制为:在环境因素的作用下,精子中发生变化的miRNA通过进一步调控DNA甲基化或组蛋白修饰,该修饰如果在多代遗传中得以保留,则可能对连续几代胎儿的正常发育产生影响。综上所述,环境因素可诱导生殖细胞中印迹基因的表观遗传修饰改变,这种表观遗传标记可能逃逸生殖细胞重编程,通过减数分裂稳定地遗传至子代的生殖细胞和其他脏器,为发育源性疾病早期编程并实现多代遗传提供了可行的道路。
5 结语与展望
在个体的生长发育过程中,印迹基因对出生前、后各器官的正常发育及功能调节具有重要作用。印迹基因的改变可引起多器官发育不良,并诱发多种疾病的发生。基因组印记是驱动胎儿生长的表观遗传决定因素,其可对环境中各种来源的暴露作出反应,进而通过表观遗传修饰影响基因的表达和子代发育(图2)。然而,环境因素所致子代发育异常的多代遗传效应及其性别差异的分子机制仍有待进一步完善。未来有望将印迹基因作为疾病的潜在生物标志物,采用高灵敏度的甲基化检测技术进行疾病的早期诊断。由于表观遗传学的可逆性,有可能通过开发新的分子靶向药物逆转表观遗传修饰改变来治疗相关疾病。此外,由于基因组印迹是在发育早期出现的,脐带血和胎盘中的印迹基因可用作胚胎发育障碍预警和干预的靶标,并辅以有效的干预方式,进而为这些发育障碍相关疾病提供防治策略。
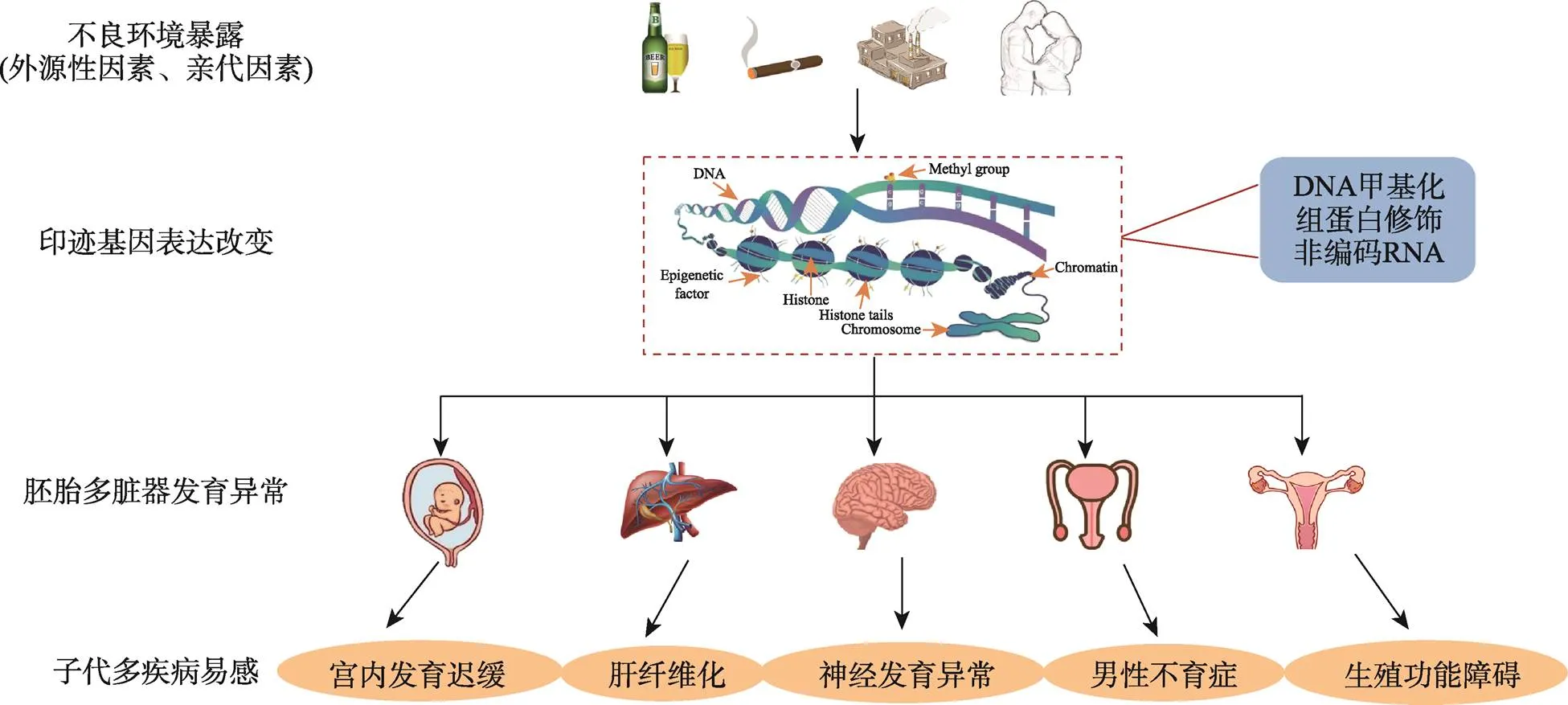
图2 印迹基因在胚胎多脏器发育和疾病易感中的作用
[1] Zoghbi HY, Beaudet AL. Epigenetics and human disease., 2016, 8(2): a019497.
[2] Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity., 2002, 22(15): 5585–5592.
[3] Azzi S, Brioude F, Le Bouc Y, Netchine I. Human imprinting anomalies in fetal and childhood growth disorders: clinical implications and molecular mechanisms., 2014, 20(11): 1751–1763.
[4] Perez JD, Rubinstein ND, Dulac C. New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain., 2016, 39: 347–384.
[5] Charalambous M, Da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life., 2007, 14(1): 3–12.
[6] Wu Y, Feng X, Gao L, Jiao BW. Imprinted genes: important regulators in development., 2016, 38(6): 508–522.
吴瑜, 冯旭, 高岚, 焦保卫. 印记基因: 发育中的重要调节因子. 遗传, 2016, 38(6): 508–522.
[7] Wang L, Zhang J, Duan JL, Gao XX, Zhu W, Lu XY, Yang L, Zhang J, Li GQ, Ci WM, Li W, Zhou Q, Aluru N, Tang FC, He C, Huang XX, Liu J. Programming and inheritance of parental DNA methylomes in mammals., 2014, 157(7): 979–991.
[8] Bartolomei MS, Tilghman SM. Genomic imprinting in mammals., 1997, 31: 493–525.
[9] Leseva M, Knowles BB, Messerschmidt DM, Solter D. Erase-maintain-establish: natural reprogramming of the mammalian epigenome., 2015, 80: 155–163.
[10] Van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence?, 2016, 30(7): 2457–2465.
[11] Weaver JR, Susiarjo M, Bartolomei MS. Imprinting and epigenetic changes in the early embryo., 2009, 20(9–10): 532–543.
[12] Pintican D, Strilciuc Ş, Armean SM, Mihu D. Effects of ethanol, nicotine and caffeine gestational exposure of female rats on lung and brain tissues in fetuses: morphological and biological study., 2019, 60(2): 643–651.
[13] He B, Wen YX, Hu SW, Wang GH, Hu W, Magdalou J, Chen LB, Wang H. Prenatal caffeine exposure induces liver developmental dysfunction in offspring rats., 2019, 242(3): 211–226.
[14] Niller HH, Minarovits J. Patho-epigenetics of infectious diseases caused by intracellular bacteria., 2016, 879: 107–130.
[15] Costa LG, Cole TB, Dao K, Chang YC, Coburn J, Garrick JM. Effects of air pollution on the nervous system and its possible role in neurodevelopmental and neurodegenerative disorders., 2020, 210: 107523.
[16] Kitsiou-Tzeli S, Tzetis M. Maternal epigenetics and fetal and neonatal growth., 2017, 24(1): 43–46.
[17] Mani S, Ghosh J, Coutifaris C, Sapienza C, Mainigi M. Epigenetic changes and assisted reproductive technologies., 2020, 15(1–2): 12–25.
[18] Qin JB, Sheng XQ, Wu D, Gao SY, You YP, Yang TB, Wang H. Adverse obstetric outcomes associated with in vitro fertilization in singleton pregnancies., 2017, 24(4): 595–608.
[19] Chen M, Heilbronn LK. The health outcomes of human offspring conceived by assisted reproductive technologies (ART). J, 2017, 8(4): 388–402.
[20] Pothineni NV, Kovelamudi S, Kantipudi S. Assisted reproductive techniques and cardiovascular risk., 2019, 73(1): 117–118.
[21] Heber MF, Ptak GE. The effects of assisted reproduction technologies on metabolic health and disease., 2021, 104(4): 734–744.
[22] Portha B, Grandjean V, Movassat J. Mother or father: who is in the front line? Mechanisms underlying the non-genomic transmission of obesity/diabetes via the maternal or the paternal line., 2019, 11(2): 233.
[23] Potabattula R, Dittrich M, Schorsch M, Hahn T, Haaf T, El Hajj N. Male obesity effects on sperm and next- generation cord blood DNA methylation., 2019, 14(6): e0218615.
[24] Szyf M. DNA methylation, behavior and early life adversity., 2013, 40(7): 331–338.
[25] Kim J, Frey WD, He HZ, Kim H, Ekram MB, Bakshi A, Faisal M, Perera BPU, Ye A, Teruyama R. Peg3 mutational effects on reproduction and placenta-specific gene families., 2013, 8(12): e83359.
[26] Lei JZ, Nie Q, Chen DB. A single-cell epigenetic model for paternal psychological stress-induced transgenerational reprogramming in offspring., 2018, 98(6): 846–855.
[27] O'neill RJ, Vrana PB, Rosenfeld CS. Maternal methyl supplemented diets and effects on offspring health., 2014, 5: 289.
[28] Liang F, Diao L, Liu J, Jiang N, Zhang J, Wang HJ, Zhou WH, Huang GY, Ma D. Paternal ethanol exposure and behavioral abnormities in offspring: associated alterations in imprinted gene methylation., 2014, 81: 126–133.
[29] Zhang XL, Ji MM, Tan XM, Yu KL, Xu LJ, Chen GY, Yu ZL. Role of epigenetic regulation of Igf2 and H19 in 2,3,7,8-tetrachlorobenzo-p-dioxin (TCDD)-induced ovarian toxicity in offspring rats., 2019, 311: 98–104.
[30] Zhu B, Huang XH, Chen DJ, Lu YC, Chen Y, Zhao JY. Methylation changes of H19 gene in sperms of X-irradiated mouse and maintenance in offspring., 2006, 340(1): 83–89.
[31] De WE, Mak W, Calhoun S, Stein P, Ord T, Krapp C, Coutifaris C, Schultz RM, Bartolomei MS. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies., 2014, 90(2): 22.
[32] Yan Z, Li Q, Zhang L, Kang BJ, Fan W, Deng T, Zhu J, Wang Y. The growth and development conditions in mouse offspring derived from ovarian tissue cryopreservation and orthotopic transplantation., 2020, 37(4): 923–932.
[33] Nomura Y, John RM, Janssen AB, Davey C, Finik J, Buthmann J, Glover V, Lambertini L. Neurodevelopmental consequences in offspring of mothers with preeclampsia during pregnancy: underlying biological mechanism via imprinting genes., 2017, 295(6): 1319–1329.
[34] Janssen AB, Capron LE, O'donnell K, Tunster SJ, Ramchandani PG, Heazell AEP, Glover V, John RM. Maternal prenatal depression is associated with decreased placental expression of the imprinted gene peg3., 2016, 46(14): 2999–3011.
[35] Ghasemi M, Heidari Nia M, Hashemi M, Keikha N, Fazeli K, Taji O, Naghavi A. An association study of polymorphisms in the H19 imprinted gene in an Iranian population with the risk of polycystic ovary syndrome., 2020, 103(5): 978–985.
[36] Soubry A, Murphy SK, Wang F, Huang Z, Vidal AC, Fuemmeler BF, Kurtzberg J, Murtha A, Jirtle RL, Schildkraut JM, Hoyo C. Newborns of obese parents have altered DNA methylation patterns at imprinted genes., 2015, 39(4): 650–657.
[37] Wu L, Lu Y, Jiao Y, Liu B, Li SG, Li Y, Xing FY, Chen DB, Liu X, Zhao JJ, Xiong XL, Gu YY, Lu JL, Chen XJ, Li XY. Paternal psychological stress reprograms hepatic gluconeogenesis in offspring., 2016, 23(4): 735–743.
[38] Charalambous M, Cowley M, Geoghegan F, Smith FM, Radford EJ, Marlow BP, Graham CF, Hurst LD, Ward A. Maternally-inherited Grb10 reduces placental size and efficiency., 2010, 337(1): 1–8.
[39] Wang LX, Balas B, Christ-Roberts CY, Kim RY, Ramos FJ, Kikani CK, Li CL, Deng CX, Reyna S, Musi N, Dong LQ, Defronzo RA, Liu F. Peripheral disruption of the Grb10 gene enhances insulin signaling and sensitivity in vivo., 2007, 27(18): 6497–6505.
[40] Kent LN, Ohboshi S, Soares MJ. Akt1 and insulin-like growth factor 2 (Igf2) regulate placentation and fetal/ postnatal development., 2012, 56(4): 255–261.
[41] Forbes BE, Blyth AJ, Wit JM. Disorders of IGFs and IGF-1R signaling pathways., 2020, 518: 111035.
[42] Cao XY, Hua X, Wang XL, Chen L. Exposure of pregnant mice to triclosan impairs placental development and nutrient transport., 2017, 7: 44803.
[43] Jedynak P, Tost J, Calafat AM, Bourova-Flin E, Busato F, Forhan A, Heude B, Jakobi M, Rousseaux S, Schwartz J, Slama R, Vaiman D, Philippat C, Lepeule J. Pregnancy exposure to synthetic phenols and placental DNA methylation - an epigenome-wide association study in male infants from the EDEN cohort., 2021, 290: 118024.
[44] Chen XJ, Chen F, Lv PP, Zhang D, Ding GL, Hu XL, Feng C, Sheng JZ, Huang HF. Maternal high estradiol exposure alters CDKN1C and IGF2 expression in human placenta., 2018, 61: 72–79.
[45] Choux C, Petazzi P, Sanchez-Delgado M, Hernandez Mora JR, Monteagudo A, Sagot P, Monk D, Fauque P. The hypomethylation of imprinted genes in IVF/ICSI placenta samples is associated with concomitant changes in histone modifications., 2020, 15(12): 1386–1395.
[46] Yamamoto Y, Nishikawa Y, Tokairin T, Omori Y, Enomoto K. Increased expression of H19 non-coding mRNA follows hepatocyte proliferation in the rat and mouse., 2004, 40(5): 808–814.
[47] Chang S, Hur SK, Naveh NSS, Thorvaldsen JL, French DL, Gagne AL, Jobaliya CD, Anguera MC, Bartolomei MS, Kalish JM. Derivation and investigation of the first human cell-based model of beckwith-wiedemann syndrome., 2020: 1–11.
[48] Yoshimura H, Matsuda Y, Yamamoto M, Kamiya S, Ishiwata T. Expression and role of long non-coding RNA H19 in carcinogenesis., 2018, 23: 614–625.
[49] Deng J, Mueller M, Geng TT, Shen YY, Liu Y, Hou P, Ramillapalli R, Taylor HS, Paidas M, Huang YQ. H19 lncRNA alters methylation and expression of Hnf4α in the liver of metformin-exposed fetuses., 2017, 8(12): e3175.
[50] Nyirenda MJ, Dean S, Lyons V, Chapman KE, Seckl JR. Prenatal programming of hepatocyte nuclear factor 4alpha in the rat: A key mechanism in the 'foetal origins of hyperglycaemia'?., 2006, 49(6): 1412–1420.
[51] Zhu X, Wu YB, Zhou J, Kang DM. Upregulation of lncRNA Meg3 promotes hepatic insulin resistance via increasing FoxO1 expression., 2016, 469(2): 319–325.
[52] Zhu X, Li HQ, Wu YB, Zhou J, Yang GW, Wang WD. lncRNA MEG3 promotes hepatic insulin resistance by serving as a competing endogenous RNA of miR-214 to regulate ATF4 expression., 2019, 43(1): 345–357.
[53] Wu L, Lu Y, Jiao Y, Liu B, Li SG, Li Y, Xing FY, Chen DB, Liu X, Zhao JJ, Xiong XL, Gu YY, Lu JL, Chen XJ, Li XY. Paternal psychological stress reprograms hepatic gluconeogenesis in offspring., 2016, 23(4): 735–743.
[54] Copping NA, Christian SGB, Ritter DJ, Islam MS, Buscher N, Zolkowska D, Pride MC, Berg EL, Lasalle JM, Ellegood J, Lerch JP, Reiter LT, Silverman JL, Dindot SV. Neuronal overexpression of Ube3a isoform 2 causes behavioral impairments and neuroanatomical pathology relevant to 15q11.2-q13.3 duplication syndrome., 2017, 26(20): 3995–4010.
[55] Sun JD, Liu Y, Moreno S, Baudry M, Bi XN. Imbalanced mechanistic target of rapamycin C1 and C2 activity in the cerebellum of angelman syndrome mice impairs motor function., 2015, 35(11): 4706–4718.
[56] Tang GM, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue ZY, Arancio O, Peterson BS, Champagne F, Dwork AJ. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits., 2014, 83(5): 1131–1143.
[57] Dunaway KW, Islam MS, Coulson RL, Lopez SJ, Vogel Ciernia A, Chu RG, Yasui DH, Pessah IN, Lott P, Mordaunt C, Meguro-Horike M, Horike SI, Korf I, Lasalle JM. Cumulative impact of polychlorinated biphenyl and large chromosomal duplications on DNA methylation, chromatin, and expression of autism candidate genes., 2016, 17(11): 3035–3048.
[58] Yang Y, Jiang WD, Yang S, Qi FL, Zhao RQ. Transgenerational inheritance of betaine-induced epigenetic alterations in estrogen-responsive IGF-2/IGFBP2 genes in rat hippocampus., 2020, 64(8): e1900823.
[59] Baumgarten SC, Convissar SM, Zamah AM, Fierro MA, Winston NJ, Scoccia B, Stocco C. FSH regulates IGF-2 expression in human granulosa cells in an AKT-dependent manner., 2015, 100(8): E1046– E1055.
[60] Zhang XL, Ji MM, Tan XM, Yu KL, Xu LJ, Chen GY, Yu ZL. Role of epigenetic regulation of Igf2 and H19 in 2,3,7,8-tetrachlorobenzo-p-dioxin (TCDD)-induced ovarian toxicity in offspring rats., 2019, 311: 98–104.
[61] Song PY, Li DY, Wang XD, Zhong XH. Effects of perfluorooctanoic acid exposure during pregnancy on the reproduction and development of male offspring mice., 2018, 50(8): e13059.
[62] He Z, Zhang JZ, Chen GH, Cao JG, Chen YW, Ai C, Wang H. H19/let-7 axis mediates caffeine exposure during pregnancy induced adrenal dysfunction and its multi- generation inheritance., 2021, 792: 148440.
[63] Busada JT, Cidlowski JA. Mechanisms of glucocorticoid action during development., 2017, 125: 147–170.
[64] Watkins AJ, Sirovica S, Stokes B, Isaacs M, Addison O, Martin RA. Paternal low protein diet programs preimplantation embryo gene expression, fetal growth and skeletal development in mice., 2017, 1863(6): 1371–1381.
[65] Cleaton MA, Dent CL, Howard M, Corish JA, Gutteridge I, Sovio U, Gaccioli F, Takahashi N, Bauer SR, Charnock- Jones DS, Powell TL, Smith GCS, Ferguson-Smith AC, Charalambous M. Fetus-derived DLK1 is required for maternal metabolic adaptations to pregnancy and is associated with fetal growth restriction., 2016, 48(12): 1473–1480.
[66] Traustadottir GÁ, Lagoni LV, Ankerstjerne LBS, Bisgaard HC, Jensen CH, Andersen DC. The imprinted gene delta like non-canonical notch ligand 1 (Dlk1) is conserved in mammals, and serves a growth modulatory role during tissue development and regeneration through notch dependent and independent mechanisms., 2019, 46: 17–27.
[67] Charalambous M, Da Rocha ST, Radford EJ, Medina- Gomez G, Curran S, Pinnock SB, Ferron SR, Vidal-Puig A, Ferguson-Smith AC. DLK1/PREF1 regulates nutrient metabolism and protects from steatosis., 2014, 111(45): 16088–16093.
[68] Matsuzaki H, Kuramochi D, Okamura E, Hirakawa K, Ushiki A, Tanimoto K. Recapitulation of gametic DNA methylation and its post-fertilization maintenance with reassembled DNA elements at the mouse Igf2/H19 locus., 2020, 13(1): 2.
[69] Zhang WP, Yang J, Lv Y, Li SL, Qiang M. Paternal benzo[a]pyrene exposure alters the sperm DNA methylation levels of imprinting genes in F0 generation mice and their unexposed F1-2 male offspring., 2019, 228: 586–594.
[70] Legoff L, Dali O, D'cruz SC, Suglia A, Gely-Pernot A, Hémery C, Kernanec PY, Demmouche A, Kervarrec C, Tevosian S, Multigner L, Smagulova F. Ovarian dysfunction following prenatal exposure to an insecticide, chlordecone, associates with altered epigenetic features., 2019, 12(1): 29.
[71] Xu RM, Li C, Liu XY, Gao SR. Insights into epigenetic patterns in mammalian early embryos., 2021, 12(1): 7–28.
[72] Short AK, Fennell KA, Perreau VM, Fox A, O'bryan MK, Kim JH, Bredy TW, Pang TY, Hannan AJ. Elevated paternal glucocorticoid exposure alters the small noncoding RNA profile in sperm and modifies anxiety and depressive phenotypes in the offspring., 2016, 6(6): e837.
[73] Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture., 2013, 34(4): 753–764.
Alteration of imprinted genes and offspring organ development caused by environmental factors
Hui Qu1, Yi Liu1, Yawen Chen1, Hui Wang1,2
Imprinted genes are a special subset of about 100 genes, which are mainly expressed in the form of parental monoallelic genes, and play important roles in the growth and development of embryos. In recent years, it has been found that epigenetic modification of imprinted genes induced by environmental factors can cause fetal multi-organ dysplasia and even susceptibility to multiple diseases in adulthood, which also exhibit multi-generational inheritance. In this review, we summarize the effects of expression changes of imprinted genes on ontogenetic development and organ functions in late stage of life, and propose that abnormal epigenetic modification and expression of imprinted genes caused by environmental deleterious factors are important mechanisms for explaining the multi-organ dysplasia in offspring. Such mechanisms are greatly significant for understanding the phenotypic changes caused by alteration of imprinted gene expression during ontogeny and exploring early prevention and treatment strategies of diseases.
imprinted genes; environmental factors; regulatory mechanisms of imprinted genes; epigenetic modification; organ development
2021-10-01;
2021-12-21;
2021-12-22
国家重点研发计划重点专项(编号: 2020YFA0803900)资助[Supported by the Key Project of National Key R&D Program of China (No. 2020YFA0803900)]
曲卉,在读硕士研究生,专业方向:肾上腺发育毒理。E-mail: 1937211743@qq.com
汪晖,教授,博士生导师,研究方向:亲代应激与子代多代发育编程及疾病易感,药物靶标与新药研究。E-mail: wanghui19@whu.edu.cn
10.16288/j.yczz.21-302
(责任编委: 杜茁)
