Reporter gene systems for the identification and characterization of cancer stem cells
2021-07-30NohemSalinasJazmArelyRosasCruzMarcoVelascoVelzquez
Nohemí Salinas-Jazmín, Arely Rosas-Cruz, Marco Velasco-Velázquez
Nohemí Salinas-Jazmín, Arely Rosas-Cruz, Marco Velasco-Velázquez, Department of Pharmacology, School of Medicine, Universidad Nacional Autónoma de México, Mexico City 04510, Mexico
Abstract Cancer stem cells (CSCs) are tumor cells that share functional characteristics with normal and embryonic stem cells. CSCs have increased tumor-initiating capacity and metastatic potential and lower sensitivity to chemo- and radiotherapy, with important roles in tumor progression and the response to therapy. Thus, a current goal of cancer research is to eliminate CSCs, necessitating an adequate phenotypic and functional characterization of CSCs. Strategies have been developed to identify, enrich, and track CSCs, many of which distinguish CSCs by evaluating the expression of surface markers, the initiation of specific signaling pathways,and the activation of master transcription factors that control stemness in normal cells. We review and discuss the use of reporter gene systems for identifying CSCs. Reporters that are under the control of aldehyde dehydrogenase 1A1,CD133, Notch, Nanog homeobox, Sex-determining region Y-box 2, and POU class 5 homeobox can be used to identify CSCs in many tumor types, track cells in real time, and screen for drugs. Thus, reporter gene systems, in combination with in vitro and in vivo functional assays, can assess changes in the CSCs pool. We present relevant examples of these systems in the evaluation of experimental CSCs-targeting therapeutics, demonstrating their value in CSCs research.
Key Words: Cancer; Gene reporter systems; Cancer stem cells; Pluripotency transcription factors; Anticancer drugs; Preclinical analysis; Cancer stem cells marker
INTRODUCTION
Cancer stem cells (CSCs) constitute a small population in the heterogeneous tumor mass and have characteristics and functions of cancer cells and stem cells. In addition to the hallmark alterations of cancer cells, CSCs have the capacity to self-renew and generate a pool of transit-amplifying cells that produce tumor-bulk cells. Accordingly,CSCs can seed tumors when transplanted into immunocompromised or syngeneic animals. Also, CSCs mediate metastasis and the resistance to cytotoxic treatments,including radio- and chemotherapy, leading to minimal residual disease and cancer relapse[1-4].
CSCs differ from tumor-bulk cells with regard to phenotype and function. CSCs have a different gene expression profile and, thus, differentially expressed proteins that can be used as markers. CSCs are quiescent, and when they proliferate, they frequently undergo asymmetric cell division. The gene expression and consequent functional characteristics of CSCs are regulated in part by several key transcription factors that control stemness in embryonic and adult stem cells, including POU class 5 homeobox 1 (POU5F1; OCT4), Nanog homeobox (NANOG), Sex-determining region Y-box 2 (SOX2), Kruppel-like factor 4 (KLF4), and MYC proto-oncogene [5,6].
CSCs have been proposed to be the "seeds" for tumor initiation and development,metastasis, and recurrence in many tumors, based on their ability to repopulate tumor heterogeneity[2,7]. Given their critical function in tumor progression and clinical importance, many strategies for identifying CSCs have been described, including the quantification of the fraction of cancer cells that express markers that are associated with the CSCs phenotype; evaluation of the ability of cancer cells to form coloniesin vitro; and assessment of their tumor-initiating potential in xenograft models, the gold standard approach for examining CSCs[8-10].
CSCs are identified by immunophenotyping by analyzing the expression of cellsurface markers. Although this approach is used extensively, it has limited specificity,because dissimilar markers might be expressed in CSCs from disparate tumor subtypes and, in some cases, even between samples of the same subtype[7,11].Conversely, analyzing tumor-initiating capacity by limiting-dilution xenotransplantation (LDX) is expensive and time-consuming and requires many animals, posing an ethical dilemma for researchers. Further, LDX is unsuitable for high-throughput drug screening[9,12,13].
To supplement existing tools for identifying, isolating, and characterizing CSCs,several reporter gene systems have been developed, having proven to be useful in substituting or complementing the identification of CSCs by immunophenotyping[14,15]. Reporter gene systems have become essential tools in analyzing the contribution of CSCs to cancer progression and developing CSCs-selective therapies. In this report,we provide integral information on the advantages and drawback of reporter gene systems for analyzing and studying CSCs. In addition, we review and discuss their use in the development of CSCs-targeting drugs, providing specific examples.
The use of biomarkers in analyzing CSCs
Immunophenotyping is widely used because it is easy and fast, and can be performed without special training. Fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS) with surface markers are the primary strategies for isolating CSCs[21]. FACS can sort by multiple biomarkers simultaneously, has robust specificity, and can be combined with other strategies to analyze the functional characteristics of CSCs, such as fluorescence screening of Hoechst 33342 exclusion[22]. However, FACS requires sterile conditions, and cell sorting is stressful to cells, which can impact their behavior. Further, given that CSCs are a rare population, their sorting requires an excessive number of cells, leading to high experimental costs, and treating cell cultures with trypsin can affect their expression of surface markers[8,23,24].
MACS is a simple antibody-based separation technique that does not requires specialized equipment; however, the number of biomarkers that can be used is limited,and thus, it might be unsuitable for complex CSCs immunophenotypes. The resulting purity is typically higher with FACS, but cell survival rates are better with MACS[14].Both methods are invaluable in CSCs immunophenotyping. For example, leukemia stem cells have been able to be isolated and characterized by FACS[20,25].
However, the expression of CSCs surface markers depends on the type of tumor and the cell of tumor origin, showing heterogeneity between samples[26]. Thus, the immunophenotype of CSCs from a particular tumor can not be applied to all samples.Moreover, the expression of CSC surface markers can change over time or become susceptible to culture conditions[23,24]. For example, enzymatic dissociation of glioblastoma cells modifies the retention of CD133 at their surface[23], and in “stemlike” pancreatic cancer cells, CD133 is upregulated under hypoxic culture conditions[24]. In addition, the use of different commercial monoclonal antibodies (each with a different specificity) complicates the reproducibility of results[15,27]. Given these caveats, surface marker profiles of CSCs are frequently inconsistent between cancer types. Thus, immunophenotyping alone is considered to be insufficient to demonstrate changes in the CSCs pool and has limited use in developing new prognostic and therapeutic options for cancer[11,28,29].
To overcome these issues, non-membrane CSCs biomarkers have been identified,the most prominent of which is aldehyde dehydrogenase (ALDH). ALDH1 catalyzes the oxidation of aldehydes to carboxylic acids and retinol to retinoic acid, allowing detoxification from drugs and reactive oxygen species[30,31]. ALDH is expressed by normal stem cells, and high levels of ALDH1 activity are observed in CSCs, representing a reliable biomarker for identifying this subset of cells in tumors from many tissues, including breast, bladder, embryonal rhabdomyosarcoma, head and neck squamous cell carcinoma, and lung cancer[30]. Higher ALDH1 expression confers resistance to several chemotherapeutic agents, such as cisplatin, etoposide,fluorouracil, and gefitinib[32]. The selection of a population of interest must be based on the expression levels of the enzyme in the tumor cells, given the heterogeneity in CSCs phenotype between tumors[11]. For example, in breast tumors, 2 subpopulations of CSCs have been identified, but only one is ALDH+[33]. Thus, it is possible that different methods enrich distinct subpopulations of CSCs.
ALDH-based staining is also transient and depends on the presence of its substrate,rendering the system suitable only for a limited period[34,35]. To mitigate these disadvantages, Anormaet al[34] developed and tested a turn-on fluorescent probe(AlDeSense)in vitroandex vivo. The methyl acetate (MA) group of AlDeSense MA is hydrolyzed by an intracellular esterase to form AlDeSense, and its aldehyde group is then oxidized to carboxylic acid by ALDH1A1 in CSCs, emitting fluorescence. The authors observed a 3-fold increase in fluorescence in spheres that were formed by purified CSCs. For theex vivoevaluation, they analyzed the lungs of mice that had been injected intravenously with CSCs or non-CSCs through the tail vein to generate metastases. When the lungs were perfused with AlDeSense solution, the signal in the lungs from CSCs-injected mice was higher than in non-CSCs-injected mice. When AlDeSense was injected intratumorally, intratumoral CSCs could be observedin vivousing a whole-body fluorescence imager-but only for 2 weeks postimplantation.
These limitations and the need to track CSCsin vivoduring metastasis,angiogenesis, and CSC-stroma interactions, have prompted the development of new tools, including reporter gene systems.
REPORTER GENE SYSTEMS TO STUDY CSCs
A reporter gene system comprises an easily detectable reporter gene and a regulatory complex of transcriptional control (promoters or enhancers that are constitutive or inducible). The expression of the reporter gene reflects the direct activation of the latter in response to the binding of transcription factors to response elements. Reporters that are under constitutive promoters are used primarily to track cells that have been transduced with the construct[36]. Conversely, inducible reporters are used to monitor biological processes. When a reporter gene construct includes transcriptional control components, it functions as a molecular-genetic sensor that responds to endogenous transcription factors and transcription-regulating complexes that initiate and control reporter gene expression[36,37].
The design and development of reporting systems to analyze such properties as phenotypic plasticity and response to therapy require expertise in genetic engineering[36,38,39]. Moreover, because reporter systems are usually designed to trigger the expression of fluorescent proteins, the incorporation of additional fluorescent dyes into the experiment should be planned carefully to prevent cross-contamination between the signals. Fortunately, there are various fluorescent proteins with a range of excitation and emission spectra (from blue to far red) and distinct structural properties and stability. The selection of the fluorescent protein must also consider its maturation time and half-life in the cell to match the desired application[36,37,40,41]. Alternatively, bioluminescent reporter genes with increased sensitivity can be used forin vivoapplications[36]. The combination of luciferase genes with fluorescent protein-coding genes into a single sequence has provided an additional tool for analyzing cell populationsin vivoandex vivo, because this strategy allows 2 signals to be monitored independently[36,37].
By the time I graduated from college, I was ready to spread my wings. I got a job teaching special education at a school in Coachella, California, a desert town about 170 miles from home. It was no dream job. Low-income housing across the street from the school was a haven3 for drug users. Street gangs hung around the school after dark. Many of my charges, emotionally disturbed 10-to 14-year-old boys, had been arrested for shoplifting, car theft or arson4.
In CSCs research, reporter gene systems have many advantages, because they allow live detection and isolation of CSCs from several tumor types. Further, these systems can be combined (simultaneously or sequentially) with other methods that analyze cell viability or the expression of other biomarkers, strengthening the distinction of CSCsvsnon-CSCs and increasing the reliability of the evaluation of effects of stimuli on either population.
However, the value of a particular reporter gene systems in tracking a particular type of CSCs is directly proportional to its validation usingin vitroandin vivofunctional assays. Several reporting systems have been used to identify CSCs from various tumors (Table 1) and have thus become important tools for the study of CSCs biology[12,42-45].

Table 1 Studies using reporter system based on known cancer stem cells biomarkers

ALDH: Aldehyde dehydrogenase; AFP: Alpha-fetoprotein; TERT: Telomerase reverse transcriptase; s-SHIP: Stem-SH2-domain-containing 5'-inositol phosphatase; LGR5: G-protein-coupled receptor 49, Gpr49; GFP: Green fluorescent protein; RFP: Red fluorescent protein.
Reporter gene systems based on CSCs biomarkers
As discussed, CSCs can be characterized by their expression of various differential markers, including: (1) Cell surface proteins, such as CD133; (2) Enzymes, such as ALDH1; and (3) Transcription factors[7,11]. Many reports support that self-renewal markers, including ALDH1A1, POU5F1, and SOX2, reliably distinguish CSCs several cancers[29,38,46,47]. However, because these molecules are intracellular, antibodybased screens are not compatible with functional assays.
To overcome this limitation, reporter systems that are based on ALDH1A1 expression have been developed by cloning fluorescent proteins under the control of the ALDH1A1 promoter[29,31,35]. These systems have been used to identify CSCs in breast cancer[31,35], colon cancer[35], and oral squamous cell carcinoma[29]. They can live track CSCs, allowing one to study CSCs dynamics in their microenvironment,increasing our understanding of CSCs involvement in the formation of metastases,resistance to therapy, and cancer recurrence. For example, a fluorescent reporter system that is based on control of the fusion protein mNeptune-TK by the ALDH1A1 promoter was used to identify a population (mNeptunehigh) in breast cancer cells that were pluripotent, had high sphere-forming capacity, and were more resistant to chemotherapy and radiotherapy[31]. These mNeptunehighcells were more tumorigenic in immunodeficient mice and generated highly resistant tumors. The reporter system efficiently identified and tracked CSCs in several luminal and mesenchymal breast cancer cell lines[31] and thus might be useful in studying CSCs dynamics in tumors.
A different approach exploits the finding that the glycoprotein CD133 is a CSCs surface marker in various cancers, including breast[11,48], colon[11,49], lung[11], and brain[11,16]. A reporter system that is based on CD133 expression has been developed and used to detect CSCs. Guerra-Rebolloet al[16] transduced human glioblastoma U87 tumor cells with a trifunctional chimeric reporter that expressesRenilla reniformisluciferase, red fluorescent protein, and a truncated version of the herpes simplex virus thymidine kinase sr39tk (tTK), driven by the CD133 promoter. This strategy allowedthem to independently monitor the entire tumor population or tumor cell subpopulations with an active CD133 promoter by bioluminescence imaging or confocal microscopy[16]. When culturing U87 cells that were transduced with this reporter, an increase in the formation of tumorspheres was observed. The expression of the tTK gene from the construct selectively killed replicating cells with an active CD133 promoter on treatment with ganciclovir[16].
Other reporter systems center on the activation of specific signaling pathways in CSCs. The Notch pathway, which maintains pluripotent hematopoietic stem cells by inhibiting their differentiation, is specifically involved in preserving self-renewal and amplification in CSCs, supporting tumor formation and mediating resistance to chemotherapeutic agents and recurrence in various tumor types[50]. However, the function and activity of Notch signaling is context-dependent in many tumors[49] and thus can not be considered a universal marker for CSCs. The use of reporter genes that respond to Notch signaling has facilitated the identification of a subset of cells with stem cell activity in breast[51] and lung cancer[52], in which the function of Notch signaling has been examined extensively. These reporter systems detect and monitor CSCsin vitroandin vivo,allowing the study of drug resistance in diverse experimental models. Hassanet al[52] used a Notch-green fluorescent protein (GFP) reporter construct that is activated when Notch intracellular domain translocate to the nucleus.They identified a subset of lung cancer cells with high Notch activity (GFPbright) with increased ability to form tumorspheres and generate GFPbrightand GFPdimcell populations. Similarly, GFPbrightcells were resistant to chemotherapy and tumorigenic in serial xenotransplantation assays, demonstrating that only cells with active Notch signaling could self-renew[52].
Another reporter gene system has been developed to detect CSCs, in which GFP is driven by the telomerase reverse-transcriptase (TERT) promoter, successfully enriching human osteosarcoma stem cells[53]. These GFP+cells had greater sphereforming ability and enhanced stem cell-like properties, such as invasiveness,metastatic activity, and resistance to chemotherapeutic agentsin vitroandin vivo[53].Further, the subpopulation in which the hTERT promoter was activated had significantly higher tumorigenic activityin vivo. In orthotopic and ectopic transplantations, the GFP+cells consistently formed tumors at a lower number of injected cells;these tumors were phenotypically diverse and could initiate new tumors after serial transplantation[53]. However, certain osteosarcoma cell lines and two-thirds of clinical osteosarcoma samples are telomerase-negative, rendering TERT-dependent labeling unsuitable for some patients.
Reporter gene systems based on CSCs transcription factors
The transcription factors that regulate stemness in normal stem cells are also involved in cancer progression and CSC biology. In mouse embryonic stem cells, these factors form interconnected feed-forward transcriptional loops to establish and reinforce cell type-specific gene expression programs[54,55] (Figure 1A). The ensemble of core transcription factors and their regulatory loops constitutes core transcriptional regulatory circuitry in many signaling pathways that regulate CSCs functions[56](Figure 1B).
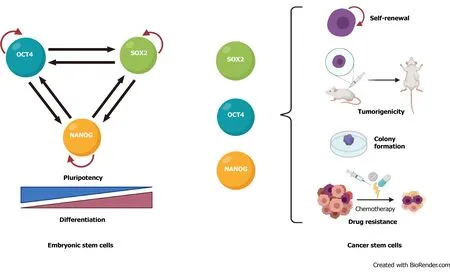
Figure 1 The pluripotency transcription factors POU class 5 homeobox 1, Sex-determining region Y box-2 and Nanog homeobox control stemness. A: In embryonic stem cells, POU class 5 homeobox 1, Sex-determining region Y box-2 and Nanog homeobox form a transcription network that maintain pluripotency and inhibits differentiation. B: In cancer cells, those transcription factors play key roles in controlling the functional characteristics that define cancer stem cells. OCT4: POU class 5 homeobox 1; SOX2: Sex-determining region Y box-2; NANOG: Nanog homeobox.
High expression of OCT4 correlates with self-renewal, chemoresistance, and tumorigenic potential of bladder, breast, and glial cells[57-59] and an unfavorable prognosis in cervical, breast, and esophageal squamous cancers[58,60,61]. SOX2 is important in maintaining self-renewal and tumorigenesis and inhibiting differentiation in CSCs from melanoma, lung adenocarcinoma, and lymphoma tissue[62-64],and its elevated expression correlates positively with drug resistance and poor survival in prostate, breast, and glioma cancer patients[65-69]. Overexpression of NANOG in CSCs promotes tumorigenicity by regulating self-renewal and proliferation in prostate, ovarian, and head and neck squamous cells[4,70-72] and is an unfavorable prognostic marker in colorectal, renal, and rectal cancer patients[73-75].KLF4 is a bifunctional transcription factor that can be an oncogenic or tumor suppressor signal, depending on the type of cancer[76]; lower KLF4 expression contributes to cellular hyperproliferation and malignant transformation in meningioma and prostate cancer[77,78], but upregulation of KLF4 promotes tumor progression in osteosarcoma, breast, and gastrointestinal cancer[79-81]. MYC is usually dysregulated in human cancers, in which it cooperates with other factors during tumorigenesis and promotes invasiveness in CSCs[7,82,83].
Based on their relevance in cancer progression, pluripotent stem cell transcription factors have been used to develop reporter gene systems that are based on their promoters (Table 2). Several promoter-reporter constructs that incorporate portions of theOct4, Sox2,andNanogpromoters have been used widely to monitor the reprogramming of murine somatic cells into an induced pluripotent state[38].However, the expression levels of these transcription factors might be lower in CSCs and vary significantly between samples[84-88]. Moreover, the large promoter regions that are used in such constructs invariably contain response elements for other transcription factors, potentially reducing reporter specificity, limiting their application in identifying CSCs. Further, some of these genes, such asOct4, have alternate transcripts and pseudogenes, complicating their detection[38,39].

Table 2 Studies using reporter system based on CSCs transcription factors.

Thyroid cancer[106]Transactivation assay Transactivation assay Drug sensitivity in vivo Teratomas from neoplastic hPSCs[12]Progenitor assays (clonogenic and multilineage hematopoietic differentiation)Sphere formation Limiting-dilution xenotransplantation Liver (hepatocellular carcinoma)[47]Drug sensitivity in vitro Sphere formation Drug sensitivity in vitro Melanoma[92]Xenotransplantation assay Xenotransplantation assay Drug sensitivity in vitro Sarcoma[18]Drug sensitivity in vivo Sphere formation Drug sensitivity in vitro Breast cancer[43]Transactivation assay Transactivation assay Drug sensitivity in vitro OCT4 GFP; Luminescent protein Teratomas from neoplastic hPSCs[12]Progenitor assays (clonogenic and multilineage hematopoietic differentiation)Sphere formation Limiting-dilution xenotransplantation Breast cancer[38]Drug sensitivity in vitro Sphere formation Limiting-dilution xenotransplantation Sarcoma[42]Drug sensitivity in vitro Sphere formation Limiting-dilution xenotransplantation Drug sensitivity in vivo Prostate cancer[107]Drug sensitivity in vitro Sphere formation Limiting-dilution xenotransplantation Gastric cancer[44]Drug sensitivity in vitro Sphere formation Limiting-dilution xenotransplantation Malignant mesothelioma[108]Drug sensitivity in vitro Sphere formation Limiting-dilution xenotransplantation Head and neck squamous cancer[109]Drug sensitivity in vitro Sphere formation Transactivation assay Limiting-dilution xenotransplantation Drug sensitivity in vitro SOX2-OCT4 GFP; mCherry fluorescent protein;Luminescent protein; Luminescent protein/RFP Glioma[16]

hPSCs: Human pluripotent stem cells; GFP: Green fluorescent protein; RFP: Red fluorescent protein
Thus, systems that use reporter genes under the transcriptional control of promoters that are active specifically in human CSCs have been generated[15,35]. For example, a GFP reporter that is driven by the NANOG promoter was developed to enrich and track ovarian CSCs[72,89]. Wiechertet al[89] introduced this reporter into cisplatinnaïve, high-grade, serous ovarian cancer patient-derived xenografts and ovarian cancer cell lines. GFP+cells expressed higher levels of stem cell transcription factors(NANOG, SOX2, and POU5F1) and CSC surface markers (CD44, CD133, CD117,CD49f, and CD24) and showed increased tumor-initiating potential. GFP+CD49f+patient-derived cells were enriched using the reporter system and CD49f staining.Further, the reporter system allowed the group to visualize dynamic changes in stemness in response to cisplatin treatment and to analyze the self-renewing capacity of cisplatin resistant cells[89].
Another example is a lentiviral reporter system, called SORE6, that was developed by Tanget al[38]. The system comprises 6 concatenated repeats of the SOX2 and OCT4 response elements from the proximal human NANOG promoter, controlling the expression of reporter genes (GFP or mCherry). This tool was validatedin vitroandinvivoin several models of breast cancer, tracking self-renewal, the generation of heterogeneous offspring, tumor- and metastasis-initiating activity, CSCs plasticity, and the response to therapeutics in real time. SORE6+cells underwent asymmetrical cell division, generated SORE6-cells, and initiate tumors in serial transplantation,demonstrating that they have tumor-initiating ability and long-term self-renewal[38].However, no CD44+CD24-cells (the subset commonly reported as breast CSCs) were enriched in the SORE6+fraction, and there was no overlap with the ALDH1-positive population, suggesting that heterogeneity exists even within stem cell populations, as published[90]. Thus, CSCs detection with SORE6 in breast cancer is more robust than with typical biomarkers, rendering the system ideal for the preclinical evaluation of new drugs.
These examples indicate that systems that report the expression and activity of NANOG, SOX2, and OCT4 are valuable tools for studying CSCs, accelerating the development of more efficient and specific reporter systems and transgene delivery strategies. As discussed, the validation of such systems will require extensive and meticulously planned preclinical testing.
USE OF REPORTER GENE SYSTEMS IN THE EVALUATION OF NEW THERAPIES
The number of CSCs affects tumor progression, disease recurrence, promotion of angiogenesis, evasion of the immune system, and resistance to conventional anticancer therapies[1,2,18,91]. Increased CSC content in a tumor has also been associated with a more aggressive form and metastatic type[10,33,92,93]. Although certain therapeutic agents that target CSCs have been described[1,10,11,28], it is evident that new selective treatments should be developed. Several studies have demonstrated the value of reporter gene systems in identifying drugs that target CSCs and determining their mechanisms of action (Table 2). For example, the combination of reporting systems with cell viability-tracking dyes can distinguish agents that induce differentiation and the loss of self-renewing pluripotencyvsthose cause direct cytotoxicity.
In sarcomas, stemness is coordinated by the expression of the pluripotency factor SOX2. Accordingly, the SORE6 reporter system has been used to study the response of CSCs to therapeutics agents in patient-derived cell lines from undifferentiated pleomorphic sarcoma[42]. The simultaneous analysis of SORE6 and caspase-3 activation identified the differential mechanism that was associated with the ability of trabectedin and EC-8042 to reduce the CSC fraction. Trabectedin is an efficient inducer of apoptosis in SORE6+and SORE6-cells, but EC-8042 reduces the percentage of SORE6+cells before the apoptotic effect becomes evident, suggesting that EC-8042 switches off SORE6-related transcriptional activity and CSC-associated properties[42].
Similarly, Páduaet al[44] used the SORE6 reporter system to characterize CSCs from gastric cancer and evaluate small molecules in a high-throughput screen[44]. SORE6+gastric cancer cells from the AGS and Kato III cell lines underwent increased sphere formation and tumorigenicity. Kato III SORE6+cells had higher levels of ALDH1 compared with SORE6−cells, but AGS cells did not express ALDH1. No other CSCmarker was enriched in SORE6+cells from either cell line, consistent with several reports that have demonstrated that stemness transcription factors are better markers of CSCs. In the same work, the authors screened 1200 compounds from the Prestwick chemical library in SORE6+or SORE-cells and observed that monensin induces a reduction in cell number selective towards the SORE6+population[44]. Given that SORE+cells are resistant to 5-FU, the identification of monensin as a gastric CSCtargeting drug might guide the development of future adjuvant therapies.
These examples, with those in Table 2, demonstrate that the transcriptional activity of pluripotency transcription factors can be used as a marker of CSCs in various tumor types. Thus, reporter systems can be implemented as a core component of analyses that identify compounds and molecules that target CSCs.
CONCLUSION
In summary, the appropriate selection of a gene reporter system eliminates common obstacles in the CSCs research, based on their ability to:
Allow direct quantification and isolation of cells with CSCs properties in preclinical tumor models and freshly excised tumors.
Track cells in time and space (in vitroandin vivo) in the analysis of CSCs niches,interactions between CSCs and their microenvironment, and interactions with neighboring cells.
Circumvent direct cell staining procedures and avoid issues with label dilution phenomena.
Track functional properties of CSCs, such as their phenotypic plasticity.
Identify selective agents that target CSCs and could be useful for preclinical testing of anticancer drugs with high sensitivity.
Lastly, it is expected that new reporter gene systems will be generated in the coming years, after the identification of additional CSCs-specific promoters and response elements. Those reporter gene systems could be combined with genetic-editing strategies, such as the CRISPR/Cas9 system, to improve their specificity and reliability by reducing genomic instability due by the integration of indirect genetic markers through viral vectors.
杂志排行
World Journal of Stem Cells的其它文章
- Epigenetic modulators for brain cancer stem cells: Implications for anticancer treatment
- Mechanisms involved in selecting and maintaining neuroblastoma cancer stem cell populations, and perspectives for therapeutic targeting
- Roles of mitochondrial unfolded protein response in mammalian stem cells
- Stem cell therapies in tendon-bone healing
- Exosomal microRNAs from mesenchymal stem/stromal cells:Biology and applications in neuroprotection
- Immunotherapy against programmed death-1/programmed death ligand 1 in hepatocellular carcinoma: Importance of molecular variations, cellular heterogeneity, and cancer stem cells
