酶法脱乙酰化制备壳寡糖的研究进展
2021-07-15周勇
周勇

摘 要:壳寡糖是甲壳素脱除乙酰基和降解后的低分子聚合物,因其具有黏度低、水溶性好、保湿性强、吸收能力强及生物相容性好等特性而具有广泛的应用价值。传统生产壳寡糖工艺存在能耗高、污染重且脱乙酰度不均匀等问题,而酶法制备壳寡糖能有效避免上述问题。当前,酶法制备壳寡糖的关键酶在于甲壳素脱乙酰酶。该文介绍了甲壳素脱乙酰酶的来源与酶学性质,结构与催化机理以及其生产菌株的选育,以期为后续产业化应用提供参考。
关键词:甲壳素;壳寡糖;甲壳素脱乙酰酶;脱乙酰
中图分类号 TS201.2文獻标识码 A文章编号 1007-7731(2021)11-0027-06
Research Progress of Enzymatic Deacetylation to Make Chitosan Oligosaccharides
ZHOU Yong
(COFCO Biochemical Co., Ltd., Bengbu 233010, China)
Abstract: Chitosan oligosaccharides (COS) is a kind of low molecular polymer after deacetylation and degradation of chitin.It has a wide range of application value because of its low viscosity, good water solubility, strong moisture retention, strong absorption capacity and good biocompatibility. The traditional COS preparation process has some problems, such as high energy consumption, heavy pollution and uneven deacetylation degree, while the enzymatic method can effectively avoid the above problems.At present, chitin deacetylase is the key enzyme for enzymatic preparation of COS.This paper mainly introduces the source, enzymatic properties, structure and catalytic mechanism of chitin deacetylase, as well as the screening of its producing strains, in order to provide reference for the subsequent industrial application.
Key words: Chitin; Chitosan oligosaccharides (COS); Chitin deacetylase; Deacetylation
近年来,随着人们生活水平的提高,膳食结构和生活方式发生了一系列改变,导致代谢性疾病的患病率迅猛增长,因此人们更加重视身心健康、自身免疫力提升以及营养健康等高品质生活与环境。从淀粉、肝糖、菊粉、纤维素、几丁质中提取的多种聚合物已经被广泛应用于生物、医学、美容、保健等领域[1,2]。壳寡糖(Chitosan oligosaccharides,COS),又称为低聚葡萄糖胺、低聚氨基葡萄糖、低聚壳聚糖等,因具有黏度低、水溶性好、保湿性强、吸收能力强和生物相容性好等特性而被广泛关注[3]。目前,COS已被广泛应用于保健品、食品添加剂、植物生长调节剂及饲料添加剂等领域。已有研究表明,壳寡糖具有增强免疫力,调节血糖、血压和血脂,排除体内毒素、重金属和多余自由基,抗肿瘤、肥胖、阿尔茨海默病和通风等效果[4-7]。
来自虾蟹等节肢动物、昆虫外壳或真菌细胞壁的甲壳素(又称几丁质),经过脱乙酰处理后得到壳聚糖,经过进一步水解反应破坏壳聚糖分子中的糖苷键可制备获得壳寡糖[8]。其中,脱乙酰化过程主要有化学法、物理法和酶法。化学法是用强酸处理甲壳素上的乙酰基而得到不同聚合度的分子,该方法存在环境污染严重、不同聚合度产物中副产物含量较高且反应条件苛刻等问题;物理法是通过超声波、γ射线或微波辐射使甲壳素分子内的化学键断链从而得到不同聚合度分子,该方法对设备的要求程度高、能耗高且降解效率低。基于甲壳素脱乙酰酶的酶法制备壳寡糖能较好地避免这些问题,是行业发展趋势[8,9]。
甲壳素脱乙酰酶(Chitin deacetylase,简称CDA,E.C.3.5.1.41)是一种催化甲壳素中N-乙酰基-D-葡萄糖胺(GlcNAc)脱除乙酰基转化为壳聚糖的酶[10]。CDA酶解甲壳素作为一种生物转化法,其与传统的化学法相比,不仅可以降低生产能耗,而且在生产过程中不需要强酸强碱处理,可解决生产过程中的环境污染问题。此外,通过生物转化可获得高品质的壳聚糖和壳寡糖产品,表现均匀的乙酰化程度以及具有分布范围窄的分子量[8]。因此,基于过甲壳素脱乙酰酶的酶法转化生产壳寡糖是一种高品质、低成本、节能环保的新型生产方法。本文重点介绍了甲壳素脱乙酰酶的来源与酶学性质、结构与催化机理以及编码基因的克隆表达,以期为后续研究与产业化应用提供参考。
1 CDA的来源及与酶学性质
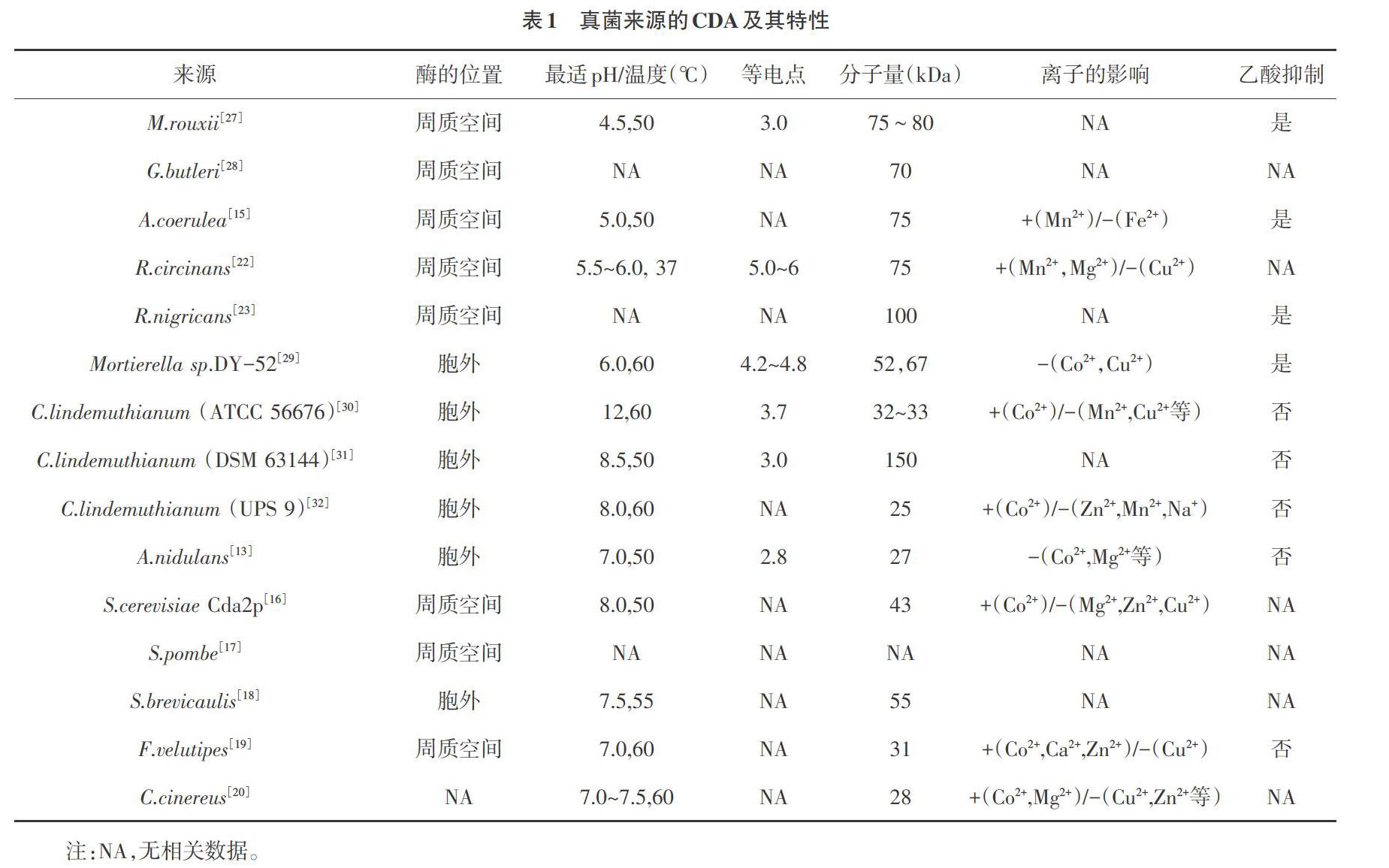
早在1973年,Amki等[11]首次从接合菌纲(Zygonycetes)的鲁氏毛霉(Mucor rouxii)中发现CDA。1982年,Kauss等[12]从植物病原菌菜豆炭疽菌(Colletotrichum lindemuthianum)中分离并得到部分纯化的CDA。随后,越来越多真菌被报道具有CDA活性,如曲霉(Aspergillus)[13, 14]、蓝色犁头霉(Absidia coerulea)[15]、酿酒酵母(Saccharomyces cerevisiae)[16]、粟酒裂殖酵母(Schizosaccharomyces pombe)[17]、短帚霉(Scopulariopsis brevicaulis)[18]、金针菇(Flammulina velutipes)[19]、灰盖鬼伞(Coprinus cinereus)[20]、卵形孢球托霉(Gongronella butleri)[21]、卷柄根霉(Rhizopus circinans)[22]、黑根霉(R.nigricans)[23]、青霉(Penicillium)和尖孢镰刀菌(Fusarium oxysporum)[24,25]等,其酶学特性见表1。目前为止,真菌来源的CDA基本是单肽链糖蛋白,表现出良好的热稳定性,但不同来源的CDA在细胞内的存在位置、分子量、等电点以及发挥作用的最适pH值、最适温度,对金属离子和乙酸的反应都有着较大的差异。例如,M.rouxii的产酶位置在周质空间,而C.lindemuthianum产生的CDA分泌到细胞外,且酶活力不受乙酸的影响;分子量大小由25~150kDa;最适pH分布在4.5~8.0。总体显示,子囊菌门真菌(包括菜豆炭疽菌、曲霉、酿酒酵母、粟酒裂殖酵母和短帚霉)来源的CDA不受底物乙酸抑制,同时在Co2+存在的条件下,可激活该酶的活性,并且均为胞外酶,有利于提取,在工业化生产与应用中具有广泛的前景。此外,不同真菌来源CDA也在其生命活动中发挥了不同的功能。Mouyna等[26]研究显示,并不是所有CDA均参与细胞壁合成或作为毒力因子。
大量菌株筛选研究显示,采用4′-硝基乙酰苯胺显色法发现某些细菌也具有CDA活性,如嗜热脂肪芽孢杆菌(Bacillus stearothermophilus)[33]、解淀粉芽孢杆菌(B. amyloliquefaciens)[34]、地衣芽孢杆菌(B. licheniformis)[35]、蜡样芽孢杆菌(B. cereus M1)[36]、红平红球菌(Rhodococcus erythropolis)[37],以及近年來发现的海水硝酸盐还原菌(Nitratireductor aquimarinus)[38]和马红球菌(Rhodococcus equi)[39]。其中,R. equi来源的CDA大小为36kDa,最适温度和pH分别为30℃、8.0,同时在较宽温度范围(如4~25℃)和较宽pH范围(如6.0~9.0)仍保持稳定。此外,外源添加金属离子,如Sr2+、Mg2+、Na+可提高酶活,而Co2+、Ba2+可抑制酶活[39]。近年来报道的基于醋酸盐受体的荧光筛选法[40],为新环境中高通量筛选CDA产生菌提供了新的途径。
尽管大多数报道的CDA来自真菌或细菌,但昆虫CDA是昆虫几丁质代谢中重要酶系。研究发现,CDA在昆虫在其生长发育过程中,如器官形成、甲壳素修饰、幼虫-蛹和蛹-成虫蜕皮等结构形成中有助于昆虫外骨骼和其他结构的完整性[41, 42]。随着农业领域生物杀虫剂的广泛关注,基于CDA的绿色杀虫剂逐渐成为研究热点。Wu等[41]通过RNA干扰技术使马铃薯甲虫的CDA基因沉默,结果显示幼虫发育期延长,生长发育迟缓,甲壳素含量降低。棉花红蜘蛛(Tetranychus cinnabarinus)来源的不太CDA序列比对显示[43],TecCDA1和TecCDA2均由5个结构域组成相似的催化位点,但通过RT-PCR显示它们在成虫期的表达水平存在差异,表明它们具有不同的功能。
2 CDA的结构与催化机理
甲壳素脱乙酰酶属于糖酯酶家族4(carbohydrate esterase family 4,缩写CE-4)成员之一[44]。该家族的成员具有保守的结构域,命名为“NodB同源结构域”或“多糖脱乙酰酶结构域”,因此,除外CDA,该家族的其他蛋白包括NodB蛋白(EC 3.5.1.-)和肽聚糖脱乙酰酶(EC 3.1.1.-)[45, 46]。CE-4家族成员序列比对显示该家族成员氨基酸序列具有高度一致性[47]。NodB同源域在NodB蛋白(根瘤蛋白)的基因序列中处于功能表达区,推测其与根瘤蛋白的脱乙酰活性密切相关。同时,脱乙酰酶的编码基因内及Bacillus sp.的开放阅读框中均存在相似基因片段,如乙酰木聚糖酯酶基因、木聚糖酶基因、肽聚糖脱乙酰酶以及许多未表征的开放阅读框,且与CDA基因中保守片段相似,说明NodB同源域对CDA的脱乙酰基功能非常重要。
CE-4家族蛋白含有5个保守的催化位点(MT1-5),构成了乙酰脱氢酶结构域的活性位点,包括保守的组氨酸和天冬氨酸残基[47]。其中,C.lindemuthianum来源的CDA活性位点结构图(图1)[47]显示,MT1包括2个天冬氨酸残基(D),1个与锌或者钴离子反应而另一个结合底物释放出来的乙酸;MT2包括2个组氨酸(H)结合金属离子和1个丝氨酸(S)或者苏氨酸(T)与第2个组氨酸形成氢键用于稳定环的结构;MT3形成一侧的活性凹槽,同时具有多种功能,包括结合乙酸、结合锌以及协调天冬氨酸残基的催化;MT4利用色氨酸(W)形成另一侧的活性凹槽;MT5包括亮氨酸(L)和组氨酸(H)构成疏水性口袋结构。CDA常见的脱乙酰基模式为多点进攻模式,其过程如下:CDA先与底物链的任一序列结合,然后从结合位点的非还原端开始,催化脱去乙酰基,本次酶解完毕;然后酶与底物解离,与另一底物链结合进行新一轮的脱乙酰基反应。这一作用方式也与其他CE-4家族成员相近[47, 48]。
3 CDA基因的克隆表达
目前,文献报道并在NCBI公布的CDA基因来自18个属,约29个种。因野生菌难以实现工业化应用,来自C. lindemuthianum[49,32,50],F. velutipes[19],R. circinans[22],A. nidulans[51],V. parahaemolyticus[52]和B. licheniformis[35]等物种的CDA序列已被克隆并在模式菌株中进行异源表达。
以大肠杆菌(Escherichia coli)为表达宿主,早在1999年,Tokuyasu等[49]将C.lindemuthianum 的CDA基因与变铅青链霉菌(Streptomyces lividans)的甲壳素酶信号肽序列结合克隆到大肠杆菌内,得到分泌型CDA,其酶活力与野生型CDA无明显区别。Wang等[51]将A. nidulans来源的CDA基因克隆到pET-28a(+)后转入E.coli BL21中表达,得到纯化的CDA活性为4.17 U/mg,最适温度和pH分别为50℃,pH8.0,但是该酶以包涵体的形式存在,同时其活性受金属离子及乙二胺四乙酸的影响,不利于大规模工业化生产。Sun等[36]利用B. cereus M1的CDA基因序列构建的重组表达载体pCT7-CHISP6H-BcCDA在大肠杆菌E. coli BL21体内高效分泌表达,重组BcCDA的分子量约23kDa,具有脱乙酰基酶活性,上清液中的最大比活力为235.43U/mg,纯化后该酶的比活力高达11608.31U/mg,提高49.31倍。危蓉萍等[53]将蜜蜂螺原体CH-1中的CDA基因克隆至pET-28a(+)表达载体上并在大肠杆菌中克隆表达,测得该酶活力最高可达10.14U/mL,最适温度为50℃左右。Raval等[35]将B. licheniformis来源的CDA基因克隆至pET22b载体后转化至大肠杆菌细胞内表达,酶活为320μmol/min·mL。添加丙三醇可使生物量提高50%,酶活由90μmol/min·mL提高至343.14μmol/min·mL,同时纯化后的CDA比活力为1142±43μmol/min·mL,较粗酶液提高221倍。
以毕赤酵母(Pichia pastoris)为表达宿主,2008年Yamada等[19]将F. velutipes来源的CDA基因利用表达质粒pPICZαA转入P. pastoris X-33后,其重组基因的表达水平较低,约1mg/L培养液,重组蛋白大小为31kDa。Gauthier等[22]将R. circinans来源的含CDA序列的cDNAs克隆至P.pastoris内,在其体内表达得到的蛋白酶具有和野生型相似的活性和大小,其中大小为75kDa,在37℃条件下最适pH为5.5~6,比活力为965.13U/mg。Kang等[50]将C. lindemuthianum来源的CDA基因装入组成型表达载体pHMB905A后转入P. pastoris GS115中,经72h的甲醇诱导表达后,培养物上清液中CDA产量可达110mg/L,酶活为77.27U/mg,大小为33kDa。
以昆虫细胞为表达宿主是一种新兴的表达系统,因具有外源蛋白折叠修饰功能和避免密码子的偏好性使其在表达昆虫来源CDA时更具有天然的优势。赵盼等[54]利用昆虫细胞昆虫Sf9细胞表达飞蝗CDA基因,Western blot结果显示其蛋白分子量61kD左右,酶活检测显示不同基因表达存在差异,最高可达0.354U/μL。因昆虫细胞表达CDA基因成本较高,相较大肠杆菌和酵母表达系统,培养条件苛刻,因此不适用于大规模工业化生产。
4 討论
随着人们对美好生活的向往及高品质生活的追求,以壳寡糖为代表的越来越多的高分子化合物已被广泛应用于医药、生物医学、食品工业、卫生、新型医疗器械和农业等领域。我国在20世纪中叶发起了对壳寡糖的研究,到2000年时已经有10余款保健食品上市,2004年将壳寡糖研究纳入国家“十五”攻关计划项目,2009年将壳寡糖产业列入“十二五”产业计划,2014年正式批准壳寡糖为新食品原料。目前市场上富含壳寡糖的保健品主要用于增强免疫产品约17种、降血糖产品4种、降血脂产品3种、保护肝脏类产品4种、增加骨密度类产品4种、抗氧化产品3种等8大类。这些产品的壳寡糖主要来源于甲壳类动物的外壳,采用传统的化学降解法或物理降解法处理,存在能耗高、污染大、反应条件苛刻、降解效率低下且产品品质不佳的问题。
为了深入贯彻落实绿色发展理念,加快推动绿色生物制造,全面提高资源利用效率,酶法制备壳寡糖已成为行业发展趋势。当前,针对不同制备原料和应用领域,研发人员不断筛选新型CDA,如从海洋环境筛选海洋细菌来源的脱乙酰酶、从昆虫体内筛选相关脱乙酰酶,以满足不同应用领域需求,提高酶的匹配性,同时为后续外源表达提供序列信息。但目前筛选的菌株仍存在菌株退化、CDA性能不稳定及活力低等问题。因此,筛选高活性的CDA仍是工业化应用中需要解决的重要问题。为了提高酶法制备壳寡糖的效率,研发人员不断探索CDA的结构及催化机理,但不同来源CDA其结构类似,但在机体内的基因表达情况及作用机制存在差异,可能存在不同的诱导因素或功能修饰。因此,将产生菌直接应用于催化反应中或利用其编码基因进行异源表达,需要进行进一步基因分析和蛋白结构分析,从而有针对性地进行基因编辑以提高其表达量或者酶活。
参考文献
[1]Baranwal A,Kumar A,Priyadharshini A,et al.Chitosan:An undisputed bio-fabrication material for tissue engineering and bio-sensing applications[J].Int J Biol Macromol,2018,110:110-123.
[2]Wang Wenqian,Meng Qiuyu,Li Qi,et al.Chitosan derivatives and their application in biomedicine[J].International Journal of Molecular Sciences,2020,21(2):487-512.
[3]Kim S K.Chitin,chitosan,oligosaccharides and their derivatives:Biological activities and applications[M].CRC Press,2010.
[4]Parashar P,Mazhar I,Kanoujia J,et al.Appraisal of anti-gout potential of colchicine-loaded chitosan nanoparticle gel in uric acid-induced gout animal model[J].Archives of Physiology and Biochemistry,2019:1-11.
[5]Lodhi G,Kim Y,Hwang J,et al.Chitooligosaccharide and its derivatives:Preparation and biological applications[J].BioMed research international,2014,2014(1):654913.
[6]Imran M,Sajwan M,Alsuwayt B,et al.Synthesis,characterization and anticoagulant activity of chitosan derivatives[J].Saudi Pharmaceutical Journal,2019,28(1).
[7]Naveed M,Phil L,Sohail M,et al.Chitosan oligosaccharide (COS):An overview[J].Int J Biol Macromol,2019,129:827-843.
[8]Gabriel S K,Peters L,Mucalo M.Chitosan:A review of sources and preparation methods[J].Int J Biol Macromol,2020.
[9]楊倩,刘建辉,蒋彤,等.几丁质脱乙酰酶的研究进展[J].食品研究与开发,2017,038(010):200-203.
[10]Ghormade V,Kulkarni S,Doiphode N,et al.Chitin deacetylase:A comprehensive account on its role in nature and its biotechnological applications[J].Current Research,Technology and Education Topics in Applied Microbiology and Microbial Biotechnology,2010,2:1054-1066.
[11]Araki Y,Ito E.A pathway of chitosan formation in Mucor rouxii:Enzymatic deacetylation of chitin[J].Biochem Biophys Res Commun,1974,56(3):669-675.
[12]Kauss H,Jeblick W,Young D H.Chitin deacetylase from the plant pathogen Colletotrichum lindemuthianum[J].Plant Science Letters,1983,28(2):231-236.
[13]Alfonso C,Nuero O M,Santamaria F,et al.Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation[J].Curr Microbiol,1995,30(1):49-54.
[14]Gastebois A,Clavaud C,Aimanianda V,et al.Aspergillus fumigatus:Cell wall polysaccharides,their biosynthesis and organization[J].Future Microbiology,2009,4(5):583-595.
[15]Gao X D,Katsumoto T,Onodera K.Purification and characterization of chitin deacetylase from Absidia coerulea[J].J Biochem,1995,117(2):257-263.
[16]Martinou A,Koutsioulis D,Bouriotis V.Expression,purification,and characterization of a cobalt-activated chitin deacetylase (cda2p) from Saccharomyces cerevisiae[J].Protein Expr Purif,2002,24(1):111-116.
[17]Matsuo Y,Tanaka K,Matsuda H,et al.Cda1+,encoding chitin deacetylase is required for proper spore formation in Schizosaccharomyces pombe[J].FEBS Lett,2005,579(12):2737-2743.
[18]Cai J,Yang J,Du Y,et al.Purification and characterization of chitin deacetylase from Scopulariopsis brevicaulis[J].Carbohydr Polym,2006,65(2):211-217.
[19]Yamada M,Kurano M,Inatomi S,et al.Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris[J].FEMS Microbiol Lett,2008,289(2):130-137.
[20]Bai Y,Wang Y,Liu X,et al.Heterologous expression and characterization of a novel chitin deacetylase,cda3,from the mushroom Coprinopsis cinerea[J].Int J Biol Macromol,2020,150.
[21]Maw T,Tan T K,Khor E,et al.Selection of Gongronella butleri strains for enhanced chitosan yield with uv mutagenesis[J].J Biotechnol,2002,95(2):189-193.
[22]Gauthier C,Clerisse F,Dommes J,et al.Characterization and cloning of chitin deacetylases from Rhizopus circinans[J].Protein Expr Purif,2008,59(1):127-137.
[23]Jeraj N,Kuni[c] B,Lenasi H,et al.Purification and molecular characterization of chitin deacetylase from Rhizopus nigricans[J].Enzyme & Microbial Technology,2006,39(6):1294-1299.
[24]Pareek N,Vivekanand V,Saroj S,et al.Purification and characterization of chitin deacetylase from Penicillium oxalicum saem-51[J].Carbohydr Polym,2012,87(2):1091-1097.
[25]Suresh P V,Sakhare P Z,Sachindra… N M.Extracellular chitin deacetylase production in solid state fermentation by native soil isolates of Penicillium monoverticillium and Fusarium oxysporum[J].Journal of Food Science & Technology,2014,51(8):1594-1599.
[26]Mouyna I,Dellière S,Beauvais A,et al.What are the functions of chitin deacetylases in Aspergillus fumigatus?[J].Frontiers in Cellular and Infection Microbiology,2020,10:28.
[27]Hunt D E,Gevers D,Vahora N M,et al.Conservation of the chitin utilization pathway in the Vibrionaceae[J].Appl Environ Microbiol,2008,74(1):44-51.
[28]Maw T,Tan T K,Khor E,et al.Complete cdna sequence of chitin deacetylase from Gongronella butleri and its phylogenetic analysis revealed clusters corresponding to taxonomic classification of fungi[J].J Biosci Bioeng,2002,93(4):376-381.
[29]Zhao Y,Jo G H,Ju W T,et al.A highly n-glycosylated chitin deacetylase derived from a novel strain of Mortierella sp.Dy-52[J].Biosci Biotechnol Biochem,2011,75(5):960-965.
[30]Tokuyasu K,Ohnishikameyama M,Hayashi K.Purification and characterization of extracellular chin deacetylase from Colletotrichum lindemuthianum[J].Bioscience Biotechnology & Biochemistry,1996,60(10):1598-1603.
[31]Tsigos I,Bouriotis V.Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum[J].J Biol Chem,1995,270(44):26286-26291.
[32]Shrestha B,Blondeau K,Stevens W F,et al.Expression of chitin deacetylase from Colletotrichum lindemuthianum in pichia pastoris:Purification and characterization[J].Protein Expr Purif,2004,38(2):196-204.
[33]Kafetzopoulos D,Thireos G,Vournakis J N,et al.The primary structure of a fungal chitin deacetylase reveals the function for two bacterial gene products[J].Proc Natl Acad Sci U S A,1993,90(17):8005-8008.
[34]He Y,Xu J,Wang S,et al.Optimization of medium components for production of chitin deacetylase by Bacillus amyloliquefaciens z7,using response surface methodology[J].Biotechnol Biotechnol Equip,2014,28(2):242-247.
[35]Raval R,Simsa R,Raval K.Expression studies of Bacillus licheniformis chitin deacetylase in E.Coli rosetta cells[J].Int J Biol Macromol,2017,104(Pt B):1692-1696.
[36]Sun Y Y,Zhang J Q,Wang S J,et al.Cloning and recombinant expression of chitin deacetylase from Bacillus cereus[J].Food Research & Development,2016.
[37]Sun Y,Zhang J,Wu S,et al.Statistical optimization for production of chitin deacetylase from Rhodococcus erythropolis hg05[J].Carbohydr Polym,2014,102:649-652.
[38]Chai J,Hang J,Zhang C,et al.Purification and characterization of chitin deacetylase active on insoluble chitin from Nitratireductor aquimarinus mcda3-3[J].Int J Biol Macromol,2020,152.
[39]Ma Q,Gao X,Tu L,et al.Enhanced chitin deacetylase production ability of Rhodococcus equi cgmcc14861 by co-culture fermentation with Staphylococcus sp.Mc7[J].Frontiers in Microbiology,2020,11.
[40]Pawaskar G M,Pangannaya S,Raval K,et al.Screening of chitin deacetylase producing microbes from marine source using a novel receptor on agar plate[J].Int J Biol Macromol,2019,131:716-720.
[41]Wu J J,Chen Z C,Wang Y W,et al.Silencing chitin deacetylase 2 impairs larval–pupal and pupal–adult molts in Leptinotarsa decemlineata[J].Insect Molecular Biology,2019,28(1):52-64.
[42]Miller S,Shippy T,Tamayo B,et al.Characterization of chitin deacetylase genes in the Diaphorina citri genome[J].2020.
[43]An,Xiangshun,Zhong,et al.Comparative characterization of putative chitin deacetylases from Tetranychus cinnabarinus[J].Bioscience Biotechnology & Biochemistry,2019.
[44]Caufrier F,Martinou A,Dupont C,et al.Carbohydrate esterase family 4 enzymes:Substrate specificity[J].Carbohydr Res,2003,338(7):687-692.
[45]Gilmore M E,Bandyopadhyay D,Dean A M,et al.Production of muramic delta-lactam in Bacillus subtilis spore peptidoglycan[J].J Bacteriol,2004,186(1):80-89.
[46]Vollmer W,Tomasz A.The pgda gene encodes for a peptidoglycan n-acetylglucosamine deacetylase in Streptococcus pneumoniae[J].J Biol Chem,2000,275(27):20496-20501.
[47]Blair D E,Hekmat O,Schuttelkopf A W,et al.Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum[J].Biochemistry,2006,45(31):9416-9426.
[48]Liu Z,Gay L M,Tuveng T R,et al.Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans fgsc a4[J].Sci Rep,2017,7(1):1746.
[49]Tokuyasu K,Kaneko S,Hayashi K,et al.Production of a recombinant chitin deacetylase in the culture medium of Escherichia coli cells[J].FEBS Lett,1999,458(1):23-26.
[50]Kang L,Chen X,Zhai C,et al.Synthesis and high expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris gs115[J].J Microbiol Biotechnol,2012,22(9):1202-1207.
[51]Wang Y,Song J Z,Yang Q,et al.Cloning of a heat-stable chitin deacetylase gene from Aspergillus nidulans and its functional expression in Escherichia coli[J].Appl Biochem Biotechnol,2010,162(3):843-854.
[52]Kadokura K,Sakamoto Y,Saito K,et al.Production of a recombinant chitin oligosaccharide deacetylase from Vibrio parahaemolyticus in the culture medium of Escherichia coli cells[J].Biotechnol Lett,2007,29(8):1209-1215.
[53]危蓉萍,楊东航,卜莹莹,等.蜜蜂螺原体几丁质脱乙酰酶基因的克隆、表达及酶学性质[J].南京农业大学学报,2016,39(03):417-424.
[54]Zhao P,Zhang X,Liu X,et al.Eukaryotic expression,affinity purification and enzyme activity of chitin deacetylase in Locusta migratoria[J].Scientia Agricultura Sinica,2017,50:1057-1066.
(责编:张宏民)
