Genome-wide identification and expression analysis of Argonaute gene family from longan embryogenic callus
2021-06-24CHENRongzhuSHENXuZHANGShutingZHAOHuaCHENXiaohuiXUXiaopingHUOWenZHANGZihaoLINYulingLAIZhongxiong
CHEN Rong-zhu,SHEN XuZHANG Shu-tingZHAO HuaCHEN Xiao-huiXU Xiao-pingHUO WenZHANG Zi-haoLIN Yu-lingLAI Zhong-xiong
1 Institute of Horticultural Biotechnology,Fujian Agriculture and Forestry University,Fuzhou 350002,P.R.China 2 Department of Pharmacy,Zhangzhou Health Vocational College,Zhangzhou 363000,P.R.China
Abstract Argonaute (AGO) proteins are the core of the RNA-induced gene silencing complex which regulate a wide variety of processes in plants,from organ development to abiotic stress responses. They have been identified in many plants,but little is known in longan (Dimocarpus longan Lour.),and how AGO functions in the signaling pathways in plant embryos in response to changing environmental stimuli remains unclear. In the present research,a genome-wide analysis of the AGO gene family members and their roles in somatic embryogenesis (SE),zygotic embryogenesis (ZE),tissue developmental processes,and responses to hormones,light and abiotic stress in longan were conducted. Ten longan AGO genes were identified genome-wide and divided into four clades. They were distributed on chromosomes 1,4,8,10,12,13,14,and 15,and had 2-23 introns. The expression profiling implied that DlAGOs regulated early and middle embryogenesis,as well as developmental processes of seed,flower,and stem in longan. In addition,the transcript levels of DlAGOs in response to exogenous hormones,light and abiotic stress showed differences in expression patterns. These results provide the useful information for further elucidation of RNAi-mediated gene silencing in longan embryogenic callus (EC).Keywords:Argonaute family,longan embryogenesis,expression profile,hormones,stress response
1.Introduction
RNA silencing is a highly conserved mechanism in most eukaryotes,which regulates specific gene expression.Argonaute (AGO) proteins are catalytic components of the RNA-induced gene silencing complex (RISC) that guides sRNAs to their targetsviasequence complement,resulting in target mRNA cleavage,translation repression,or chromatin modification (Baulcombe 2004;Vaucheret 2006;Chapman and Carrington 2007;Moazed 2009). Therefore,the functional diversification of RNA silencing is directly related toAGOs. AGO proteins are strongly conserved,are composed of 700-1 200 amino acid residues and generally contain four domains,namely the N-terminal,PAZ,Mid and Piwi domains (Vaucheret 2008). Previous studies indicated that the PAZ domain can bend small RNA into a specific binding pocket,whereas the Piwi domain has a cutting function,similar to ribonuclease H (RNase H) cleaving target RNAs,which has four important amino acid residues (Asp-Asp-His-His),but only AtAGO1,AtAGO4 and AtAGO7 inArabidopsis thalianahave catalytic activity (Baumberger and Baulcombe 2005;Parker and Barford 2006;Qiet al.2006;Tolia and Joshua-Tor 2007;Montgomeryet al.2008;Karlowskiet al.2010).
There are 10 AGO proteins inArabidopsis,which can be divided into four major groups phylogenetically:AGO1/AGO10,AGO4/AGO6,MEL1/AGO5,and AGO2/AGO7 (Morelet al.2002;Zhenget al.2007). Two of them are associated with different forms of RNA silencing. One is AtAGO1,which is associated with the action of miRNAs,trans-acting siRNAs (ta-siRNAs) and transgene-derived siRNAs,participating in leaf polarity,lateral organ development,cleaning up viral RNA,and also binding to chromatin to boost gene expression in response to plant hormones and stresses (Baumberger and Baulcombe 2005),while another is AtAGO4,which recruits endogenous 24-nt siRNAs affecting epigenetic silencing (Zilbermanet al.2003,2004;Rowleyet al.2011;Yeet al.2012). In addition,AtAGO7 collaborates with miR390,is involved in the generation of ta-siRNAs from TAS3,and has a function in the plant from the juvenile to the adult stage (Hunteret al.2003),while AtAGO10 has the function of meristem maintenance (Jiet al.2011;Manavellaet al.2011).
Plant RNA silencing participates in a wide variety of biological processes including plant development,stress responses,metabolism and the maintenance of genome integrity. In recent years,research on theAGOgene family has given us a better understanding of RNA silencing. TheAGOgene family was identified in more and more flowering plants. InArabidopsis thaliana,10AtAGOswere identified (Kapooret al.2008). There are 19AGOgenes in rice,and onlyOsAGO2showed specific up-regulation in response to cold,salt and dehydration stress (Qianet al.2011). In maize,theAGOfamily comprises 18 members,and 15SlAGOs were identified in tomato (Baiet al.2012). Additionally,in an emerging model medicinal plant,Salvia miltiorrhiza,there are 10AGOgenes,and the expression levels ofSmAGOsin various tissues were investigated (Shao and Lu 2013). ThirteenAGO(VvAGOs) genes are in grapevine,which show different expression patterns during berry development and abiotic stress responses (Zhaoet al.2015). Similarly,cucumber possesses sevenAGOgenes which play key roles in the stress response (Ganet al.2017),and a total of 27BnAGOs have been identified in the allopolyploid speciesBrassica napusbased on the genome database (Zhaoet al.2016). Moreover,12CaAGOshave been identified in pepper (Qinet al.2018). Finally,13AGOgenes were found inCitrus sinensis,andCsAGO4ashowed the highest expression levels in embryos and it has an important role during fruit abscission (Agustínet al.2019).
Longan (Dimocarpus longanLour.) is an important evergreen fruit tree which is planted in tropical and subtropical areas of China. The fruit is rich in vitamins and minerals,so it is used as a source of nutrients and supplements (Yanget al.2011;Sudjaroenet al.2012).The longan embryogenesis directly determines the fruit size,quality,setting rate and yield. Therefore,the regulation of embryo development is an urgent problem in longan production. However,longan zygote embryos (especially early stage) are extremely difficult to obtain and highly heterozygous,which seriously inhibits research on the longan embryo molecular mechanisms. Somatic embryogenesis (SE) is similar to zygotic embryogenesis (ZE),including morphology,transcription level and protein level (Viola and Ben 2004),therefore,the acquisition of somatic embryos of longan lays a foundation for the study of longan molecular mechanisms. The embryo development,fruit quality,and genetic regulation of longan are the focus of current research (Linet al.2017).
At present,the transcriptional regulation ofAGOgenes during SE and ZE,and the pathways involved in the responses to hormones and stress in somatic embryos remain unclear. In addition,the function ofAGOgene family in longan has not been studied. Therefore,theAGOgene family was identified and analyzed in this study using the longan genome database. We also investigated the expression patterns ofAGOgenes using quantitative real-time PCR (qRT-PCR) during early stage of SE,ZE,in various tissues of Honghezi (HHZ) longan,under different hormone,light,and abiotic treatments. The results provide useful information for illuminating the biological functions of theDlAGOs.
2.Materials and methods
2.1.Plant materials and treatments
The synchronized embryogenic cultures at early stages,including the embryogenic callus (EC),incomplete compact pro-embryogenic cultures (ICpEC),globular embryos (GE),Honghezi (HHZ) longan tissues or organs,and different development stages of zygotic embryos for qRT-PCR were obtained following previously published methods (Lai and Chen 1997;Chen and Lai 2002;Chen Y Ket al.2018). The samples were frozen in liquid nitrogen immediately,and then stored at-80°C prior to RNA isolation.
Samples of 0.3 g 18-day subculture longan EC were transferred to MS liquid medium (2% sucrose) supplemented with 2,4-dichlorophenoxyacetic acid (2,4-D,0.1,0.3,0.5,and 1.0 mg L-1),kinetin (KT,0.25,0.5,0.75,1.0,and 2.0 mg L-1),salicylic acid (SA,25,50,75,and 100 mg L-1),and methyl jasmonate (MeJA,25,50,75,and 100 mg L-1) with shaking culture at 25°C,110 r min-1,in dark conditions for 24 h. The 19-day subculture longan EC were treated with 150 mmol L-1sodium chloride (NaCl),150 mmol L-1mannitol,10% (w/v) polyethylene glycol 4000 (PEG-4000),and 10 μmol L-1abscisic acid (ABA) for different times (1,2,4,8,and 12 h) in liquid MS medium with agitation at 25°C,110 r min-1under dark conditions. Meanwhile,EC were cultured in MS liquid medium as control. EC treated with different light qualities (dark,red,yellow,blue,green and white) was carried out at 25°C for 24 h. All the above treatments were repeated three times. All samples were collected respectively and quickly frozen in liquid nitrogen,then stored at-80°C prior to isolating the total RNA.
2.2.Genome-wide identification of AGO genes in longan
The third generation of longan genome data was used (unpublished data). We obtained the amino acid sequences of AGO inArabidopsis thalianafrom the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/) and searched for the longan orthologous genes with TBtools (Chen Y Ket al.2018) and NCBI’s online BLASTP tool. Conserved domains of AGO proteins in longan were analyzed and annotated using the Simple Modular Architecture Research Tool (SMART,http://smart.emblheidelberg.de/) and the Pfam database (https://pfam.xfam.org/). The online software ExPASy ProtParam (https://web.expasy.org/protparam/) was used to predict and analyze the physico-chemical properties of DlAGO proteins,such as isoelectric point (PI) and molecular weight (MW).
2.3.Chromosomal localization and phylogenetic analysis
TheAGOgene chromosomal location images were made according to genome annotations from the third generation of longan genome data,using the TBtools (Show Gene on Chromosome) (Chen C Jet al.2018). The phylogenetic tree was built for the AGO protein family using MEGA v5.0 and the neighbor-joining (NJ) method with the bootstrap test replicated 1 000 times.
2.4.Gene structure,conserved motif and protein domain analysis
TheDlAGOgene structures were determined by comparing their full-length coding sequences (CDS) with the general feature format (gff) file using TBtools. The conserved motifs of the DlAGO proteins were determined by MEME (http://meme-suite.org/tools/meme) and were re-edited using the TBtools Software,while the domains of longanAGO proteins were analyzed through SMART,Pfam database and TBtools.
2.5.RNA extraction and qRT-PCR analysis
Total RNA was isolated from longan embryogenesis cultures and EC under various treatments using TransZol Up Plus RNA Kit (TransGen,Beijing,China),while total RNA of longan tissues and different stages of ZE were extracted by the Universal Plant Total RNA Extraction Maxi Kit (Spin-Column) (Bioteke Corp.,Beijing,China). We used 1.0% agarose gel electrophoresis and a Nano Drop Lite spectrophotometer (Thermo Electron Corp.,Shanghai,China) to check the integrity and concentration,and then the cDNA was synthesized using a PrimeScript®RT reagent Kit (TaKaRa,Beijing,China). The specific primers for qRTPCR were performed by DNAMAN6.0 (Appendix A). The qPR-PCR was processed on the Light cycler 480 System (Roche Applied Science,Basel,Switzerland),and carried out in a 20-μL volume containing 10 μL of 2×SYBR PremixExTaqTM(TaKaRa),1 μL diluted cDNA (10×),0.8 μL forward and reverse primers (100 nmol),and 7.4 μL ddH2O.The qRTPCR reaction profile was as follows:94°C for 30 s,40 cycles of 94°C for 10 s and 60°C for 30 s.EF-1α,eIF4α,andFe-SODwere used as reference genes to calculate the relative expression at different stages of SE,KT,MeJA,SA,light and abiotic treatments,whileFe-SODwas used as reference gene,and the 2-ΔΔCtmethod was applied to calculate the relative expression levels ofDlAGOsafter longan EC were treated with 2,4-D (Schmittgen and Livak 2008). The relative expressions ofDlAGOsat different stages of longan ZE and in different tissues of“HHZ”longan were calculated usingUBQandEF-1αas internal controls. The averageFPKM values of transcriptome data of embryogenic cultures (EC,ICpEC and GE) ofDlAGOsfamily genes were calculated to predict the expression profile (Chenet al.2020) (Appendix B). Heatmaps were constructed using the transformed log2(FPKM+1) values.
2.6.Statistical analysis
Each experimental data point represents three independent biological replicates,and all data are expressed as mean±standard deviation (SD). To assess significant differences between control and treatment conditions atP<0.05,experimental data were tested using SPSS oneway ANOVA.
3.Results
3.1.Identification and structural organizational analysis of AGO genes in longan
In order to identify theAGOfamily genes,the amino acid sequences ofArabidopsiswere usedas queries in BLAST analyses,and 10 genes encoding AGO proteins were identified in the third generation of the longan genome database. Based on the sequence similarity with the correspondingArabidopsisor rice AGOs,the identifiedAGOgenes are named fromDlAGO1toDlAGOMEL1(Table 1).
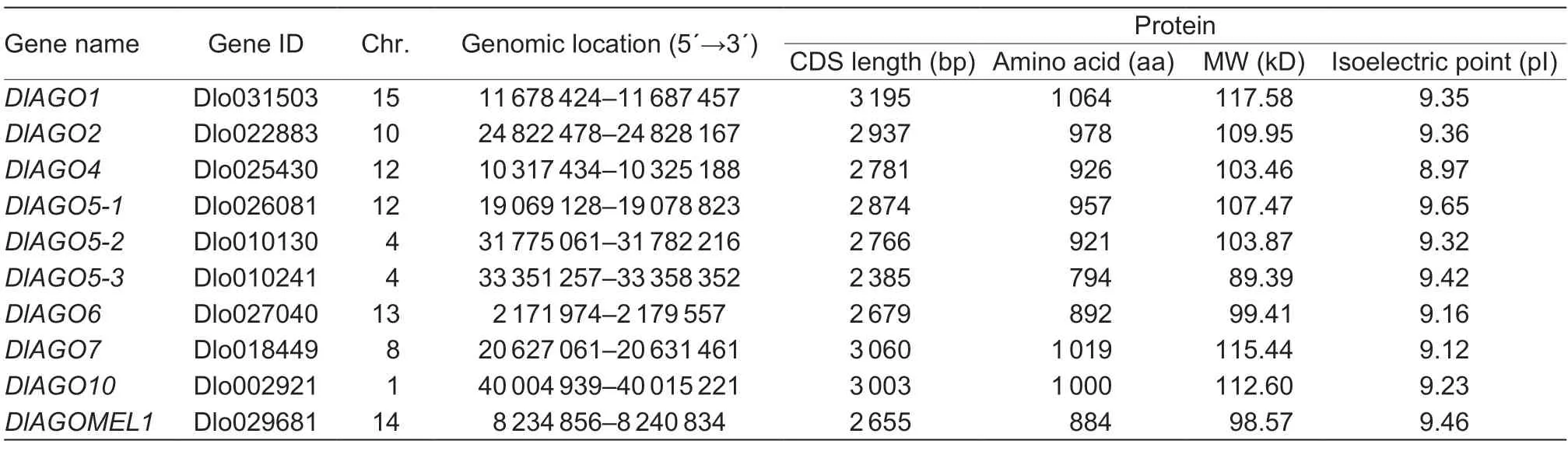
Table 1 Characterization of AGO genes in longan
In order to further understand theAGOgene family,the gene characteristics,including lengths of the coding sequences (CDS),protein sequence,protein isoelectric point (PI) and molecular weight (MW),were analyzed.These genes coded for basic proteins of about 100 kDa,with PI ranging from 8.97 to 9.65. The CDS lengths of theDlAGOsranged from 2 385 (DlAGO5-3) to 3 195 bp (DlAGO1),and the sizes of encoded potential proteins varied between 794 and 1 064 aa (Table 1). Searching longan AGO proteins for conserved domains using SMART and Pfram database showed that all DlAGOs have N-terminus,PAZ domain and Piwi domain,whereas the AgoL2 domain was present in eight DlAGOs but missing in DlAGO2 and DlAGO7. Also,the Mid domain is found in six of the DlAGO proteins,except for DlAGO2,DlAGO5-3,DlAGO6 and DlAGO7,but only DlAGO1 has the Gly-rich-Ago1 domain in the starting end,which was consistent with theArabidopsis thalianaAGO protein. The characteristics of conserved domains of the AGO proteins in longan andArabidopsiswere further analyzed and the results showed that some AGO proteins contained the same conserved domains,such as DlAGO5-1/DlAGO5-2/AtAGO5,DlAGO1/AtAGO1,DlAGO10/AtAGO10,DlAGO4/AtAGO4,DlAGO6/AtAGO6;but some AGO proteins did not have the same conserved domains,such as DlAGO2/AtAGO2,DlAGO7/AtAGO7 (Fig.1). The information and sequences for each conserved domain of DlAGOs are shown in Appendices C and D.
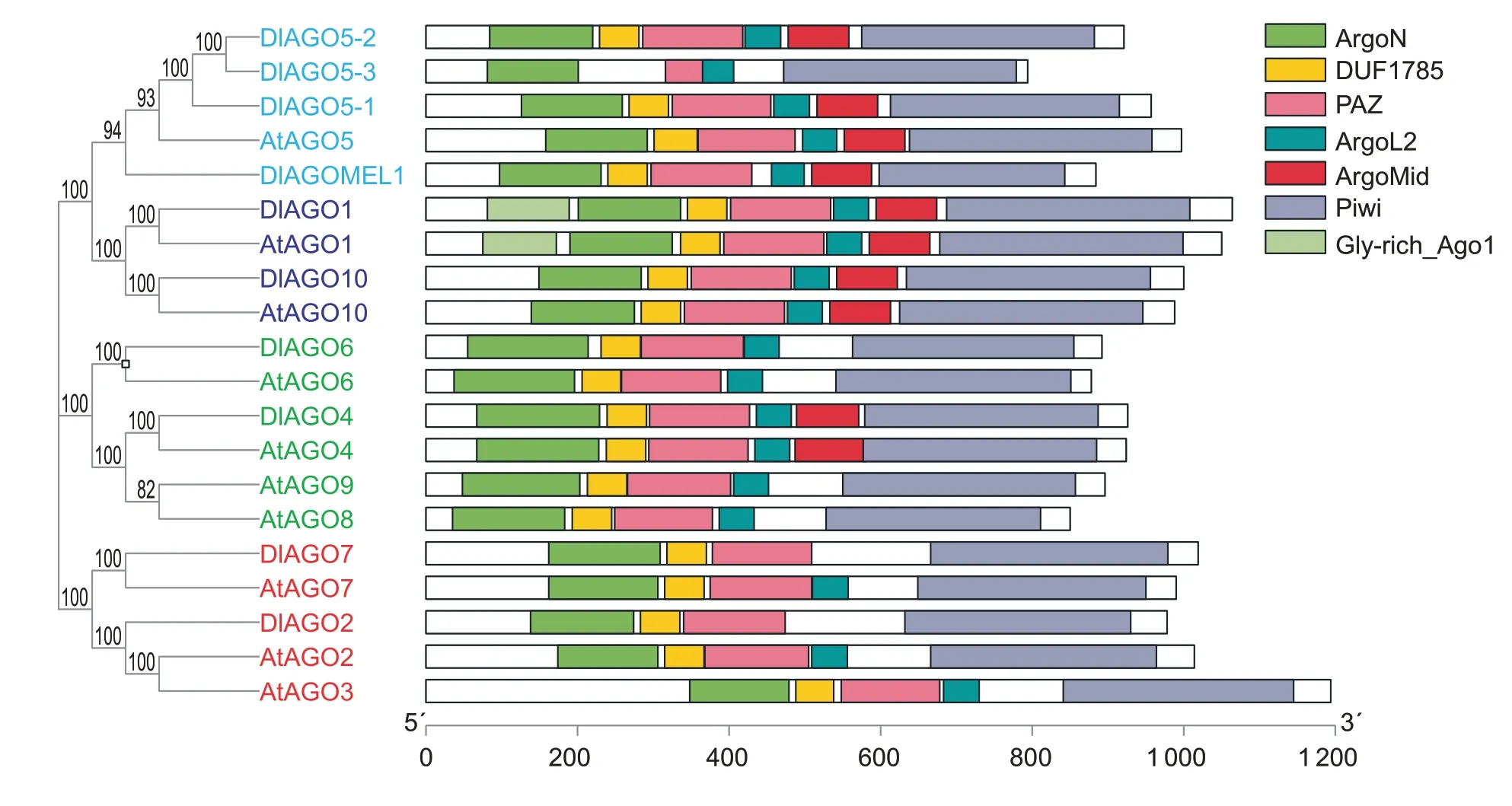
Fig.1 Analysis of conserved domains of proteins of DlAGO and AtAGO. AtAGOs represent the conserved domain of AGO proteins from Arabidopsis thaliana,DlAGOs represent the conserved domain of AGO proteins from longan.
The Piwi domain is similar to RNase H enzymes,and can specifically bind to the siRNA 5´ end and cleave target RNAs. The domain has an active site usually containing an aspartate-aspartate-histidine (DDH) motif and a conserved histidine at position 798 (H798),which was found in AtAGO1.The Piwi domain of all the longan AGO proteins and AGO1 inArabidopsiswere aligned using DNAMAN6.0,in order to find whether the Piwi domains of DlAGO proteins possess the conserved catalytic residues (Fig.2). Four DlAGO proteins (DlAGO1,DlAGO5-1,DlAGO7,and DlAGO10) contain conserved DDH/H798 residues. D760,D845 H986,and H798 were not conserved in the remaining six AGO proteins of longan,the histidine residue (H) at position 986 of DlAGO2 was replaced by glutamine (Q) and became DDQ/H,while in DlAGOMEL1 it was missing and became DD-/H. The histidine residues (H) at position 798 of DlAGO4 and DlAGO5-2 were replaced by proline residues (P),while the histidine residue (H) at position 798 of DlAGO6 was replaced by a serine residue (S) and became DDH/S. Aspartic acids (D) at position 760 of DlAGO5-2 and DlAGO5-3 were replaced by alanine (A),becoming ADH/P and ADH/H,respectively. The diversity of DDH/H798 motif in the Piwi domain also reflects the diversity of the protein’s function (Table 2). InC.sinensis,the DDH/H motif was present in CsAGO1,CsAGO7 and CsAGO10,DDD/H motif for CsAGO2a,DDH/P motif for CsAGO4a and CsAGO4b;a DDH/S motif in CsAGO6,DDY/H and DDY/P motifs for CsAGO5a and CsAGO5b,respectively,and a D-H/T motif was observed in CsAGO6-like protein (Agustínet al.2019). Therefore,the DDH/H motif of Piwi in longan and sweet orange had both similarities and differences.
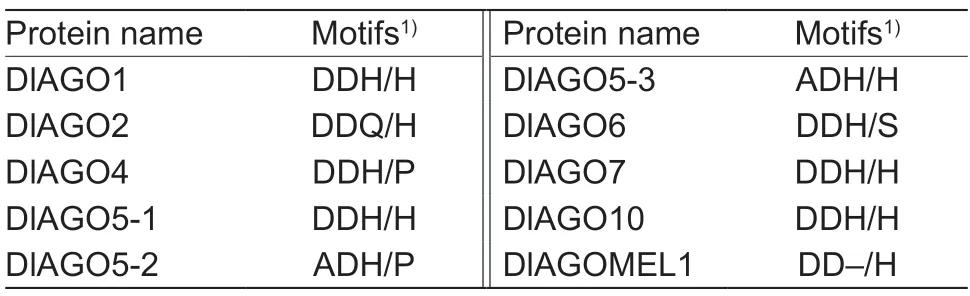
Table 2 Argonaute proteins with catalytic residue(s) in Piwi domains of longan
3.2.Phylogenetic analysis and classification of the longan AGOs
To explore the phylogenetic relationships between dicot and monocot AGOs,an unrooted neighbor-joining tree using full length protein sequences from longan,Arabidopsis,orange,poplar,grapevine and rice was built (Fig.3). The phylogenetic analysis revealed that AGO proteins can be clustered into four clades,AGO1/10,MEL1/AGO5,AGO2/AGO7,and AGO4/AGO6. DlAGO1 and DlAGO10 are included in the AGO1/10 clade,and AGO1/10 can coordinate regulation of the maintenance of stem and root meristem,and the establishment of leaf polarity in combination with miR165/166 inArabidopsis(Jiet al.2011). Meanwhile in the MEL1/AGO5 clade,four genes (DlAGO5-1,DlAGO5-2,DlAGO5-3 and DlAGOMEL1) are included,and inArabidopsisand rice,it was found that AGO5/MEL1 participated in regulating cell division during meiosis in gametes (Nonomuraet al.2007;Komiyaet al.2014;Brosseau and Moffett 2015). The AGO2/AGO7 clade contained DlAGO2 and DlAGO7 in longan,but AGO2 and AGO7 may have different functions,i.e.,while AGO2 is involved in antiviral infection,AGO7 is involved in the formation of tasiRNA in combination with miR390 (Quet al.2008;Scholthofet al.2011). DlAGO4 and DlAGO6 belonged to the AGO4/AGO6 clade,and AGO4/AGO6 play important roles in the RdDM pathway,which can bind to 24-nt siRNA (Qiet al.2006;Duanet al.2015). Further analysis found that the longan AGO proteins displayed a high similarity with CsAGOsin orange,and the AGO distribution was similar between monocots and dicots.

Fig.3 Phylogenetic analysis of the DlAGO proteins and five other plants. Dl,Dimocarpus longan;At,Arabidopsis thaliana;Os,Oryza sativa;Vv,Vitis vinifera;Cs, Citrus sinensis;Pt, Populus turanga. The different colored areas indicate different clades of AGO family.
3.3.Chromosomal location of longan AGO genes
AGOgenomic distribution was analyzed by localizing the genes on longan chromosomes (Fig.4). The 10DlAGOgenes were distributed on eight chromosomes;DlAGO4andDlAGO5-1were located on chromosome 12,whileDlAGO5-2andDlAGO5-3were located on chromosome 4,and chromosomes 1,8,10,13,14,and 15 each contained a singleAGOgene,but chromosomes 2,3,5,6,7,9,and 11 did not include anyDlAGOs. Further analysis revealed that theAGOgenes of longan had not undergone tandem duplication,and the result showed theAGOfamily members may have different modes of expression regulation.
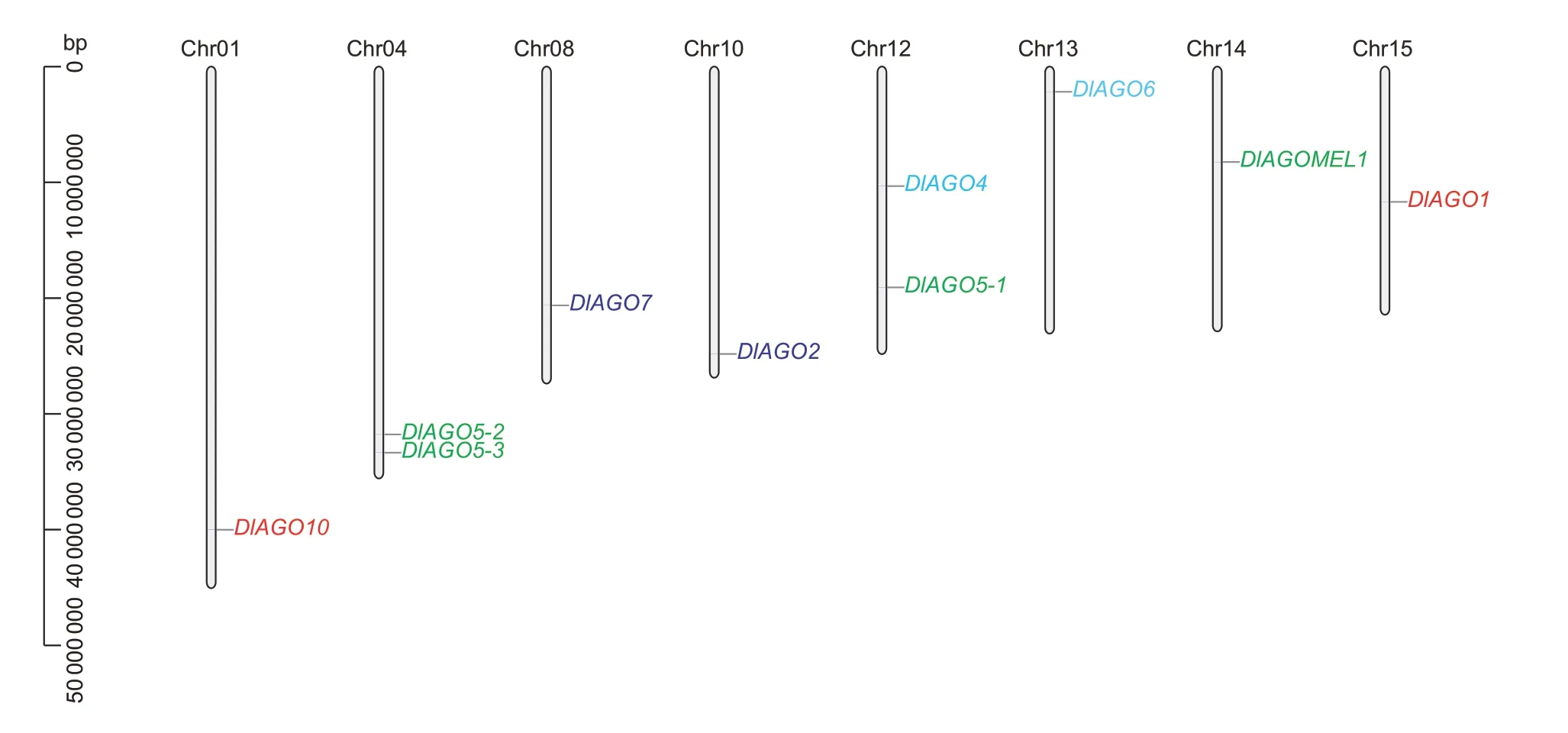
Fig.4 Genomic distribution of AGO genes on the chromosomes of longan. Chromosome numbers are shown at the top of each chromosome,and the different clades are indicated by different colors.
3.4.Conserved motif and gene structure analysis of the longan AGO genes
Through analysis of the conserved motifs and exon-intron organization of all identifiedDlAGOs,we can gain insights into their evolution. The results showed that different genes ofDlAGOsusually have different structures. In order to analyze the conserved motifs among the DlAGO proteins,the MEME suite of tools was used. A total of 10 motifs were identified and used to analyze the characteristics of the DlAGO proteins (Fig.5-A;Appendix E). Ten motifs were present in seven DlAGO proteins,but motifs 7,8,3 and 6 were missing in DlAGO2,DlAGO5-1,and DlAGOMEL1,respectively. Through further analysis,it was found that the number and location of motifs in the AGO1/AGO10 and AGO4/AGO6 clades were similar,but the MEL1/AGO5 clade was quite different.
In order to further understand the biological functions of longanAGOs,theAGOgene structures were analyzed (Fig.5-B). The results showed that six of the ten genes had complete sequences,andDlAGO5-1andDlAGO10genes had no 3´ UTR,whileDlAGO5-2andDlAGO5-3had no UTR.From the gene structure analysis,it can be found that the number of introns ofAGOfamily members varied greatly,from 2 to 23. Interestingly,the clade members had roughly the same number of introns. For example,AGO2/AGO7clade members had two introns,whileAGO1/AGO10andAGO4/AGO6clades had about 22 introns,but the number of introns inMEL1/AGO5clade members was quite different,from 19 (DlAGO5-3andDlAGOMEL1) to 22 (DlAGO5-1).The close similarity of conserved motif composition and gene structure within the same clade indicated that the phylogenetic classification was reliable.
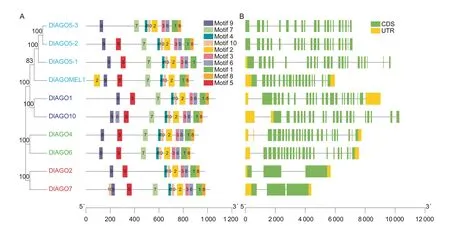
Fig.5 Architecture of conserved protein motifs and gene structures in the AGO family members of longan. The motifs,numbers 1-10,are displayed in different colored boxes. Green boxes indicate exons,and black lines indicate introns.
3.5.Expression profiling of DlAGOs during early stages of SE by RNA-seq
To assess whether theAGOgene family members affected early SE in longan,we analyzed the transcript levels in the longan transcriptome dataset using FPKM values,including EC,IcpEC,and GE of the“HHZ”cultivar (Fig.6).The expression profiling ofAGOgenes during longan early SE can be roughly divided into five patterns,three genes (DlAGO2,DlAGO5-1,DlAGO7) were up-regulated in EC stage,only one gene (DlAGO1) showed highest expression in ICpEC stage,two genes (DlAGO4andDlAGO10) were upregulation in GE stage,whereasDlAGO6andDlAGOMEL1were up-regulated in EC and IcpEC stages,butDlAGO5-3gene was not expressed in the three stages. Interestingly,DlAGO1andDlAGO4were highly expressed in the three stages,butDlAGO7,DlAGO5-2andDlAGO5-3were only expressed at low levels or not at all. So,we presume thatAGOgenes may affect the development of early stage of longan SE,but different members have different functions.
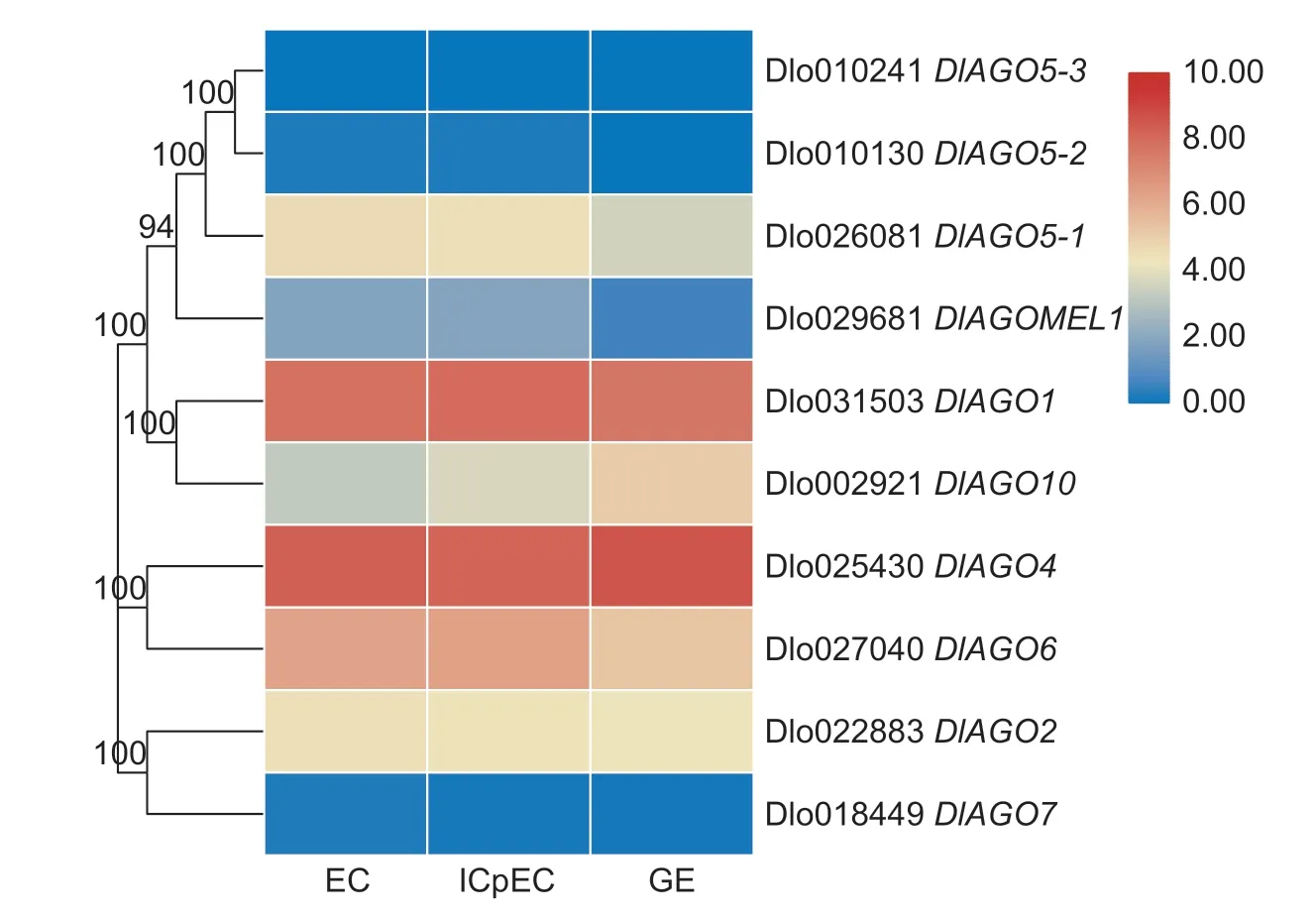
Fig.6 Hierachical clustering of expression profiles of AGO genes in the early stage of longan embryogenic cultures. EC,embryogenic callus;ICpEC,incomplete compact pro-embryogenic cultures;GE,globular embryos.
3.6.AGO protein interactions among specific proteins in longan
To confirm the potential functions of the DlAGO proteins,a protein-protein interaction network was constructed using STRING 10.5 Software based on anArabidopsisassociation model (Fig.7). All the proteins participated in gene silencing by RNA,posttranscriptional regulation of gene expression,and regulation of transcription,while most proteins are involved in the defense response to viruses,except AT5G21030,KTF1 and NRPD1A. Seven of the 12 proteins have the function of posttranscriptional gene silencing but only three proteins take part in DNA methylation,which are AGO4,AGO6,and KTF1,meanwhile,AGO4 and AGO6 are involved in the RNA-directed DNA methylation.
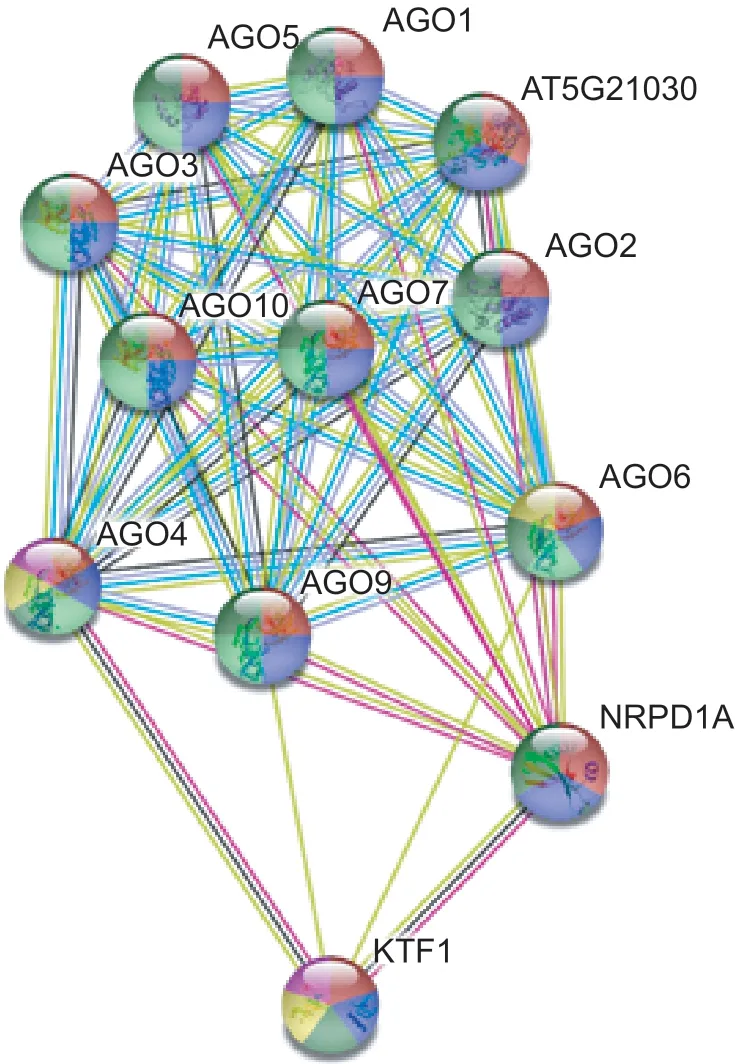
Fig.7 Potential protein-protein interaction network of DlAGO proteins. The result was based on an Arabidopsis association model.
3.7.Expression patterns of longan AGO genes during the early stage of SE,ZE,and in different tissues
To investigate the transcriptional regulation ofAGOgenes during longan embryogenesis,the transcriptional profiles during SE were analyzed by qRT-PCR (Fig.8). The results showed that five of eightAGOgenes had the same expression trends as the data from RNA-seq,but the other three genes had different expression trends. From the qRTPCR dates,we can find that four genes (DlAGO2,DlAGO4,DlAGO7,andDlAGO10) had the highest expression levels in the GE stage,whileDlAGO5-1,DlAGO6,andDlAGOMEL1were highly expressed in EC stage,and onlyDlAGO1gene had its highest expression in ICpEC stage.
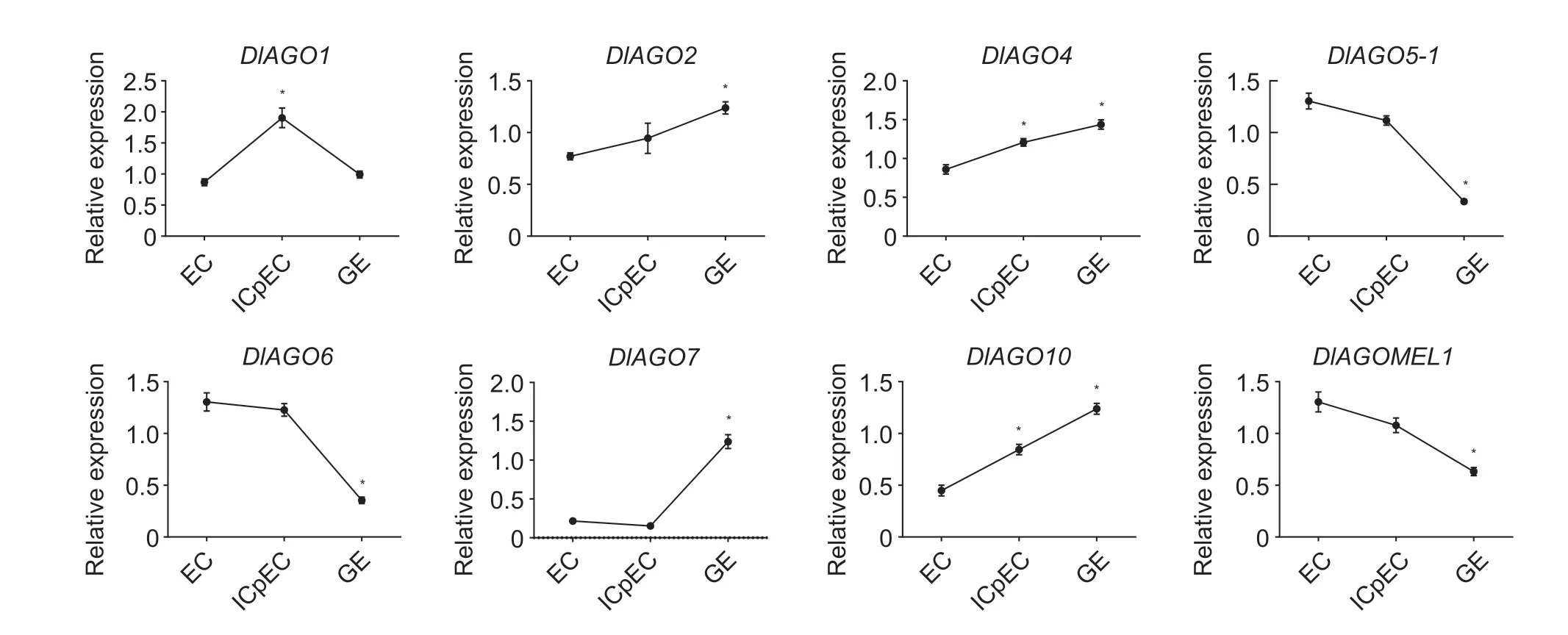
Fig.8 Expression analysis of AGO genes in the embryogenic cultures of longan by qRT-PCR. EC,embryogenic callus;ICpEC,incomplete compact pro-embryogenic callus;GE,globular callus. Reference genes EF-1α,Fe-SOD,and eIF4α. Data are mean±SD (n=3). * indicates significant difference at 0.05.
Furthermore,the expression levels of longanAGOgenes were detected in the zygotic embryo development stages of“HHZ”longan (Fig.9). Four genes expression levels were the highest in S5 stage,and three genes showed a peak in S3 stage,whileDlAGOMEL1gene showed high levels in S2 stage. These results indicated thatDlAGOgenes showed a similar expression pattern in longan SE and ZE and played an important role during longan ZE,especially at the early and middle embryogenesis stages.
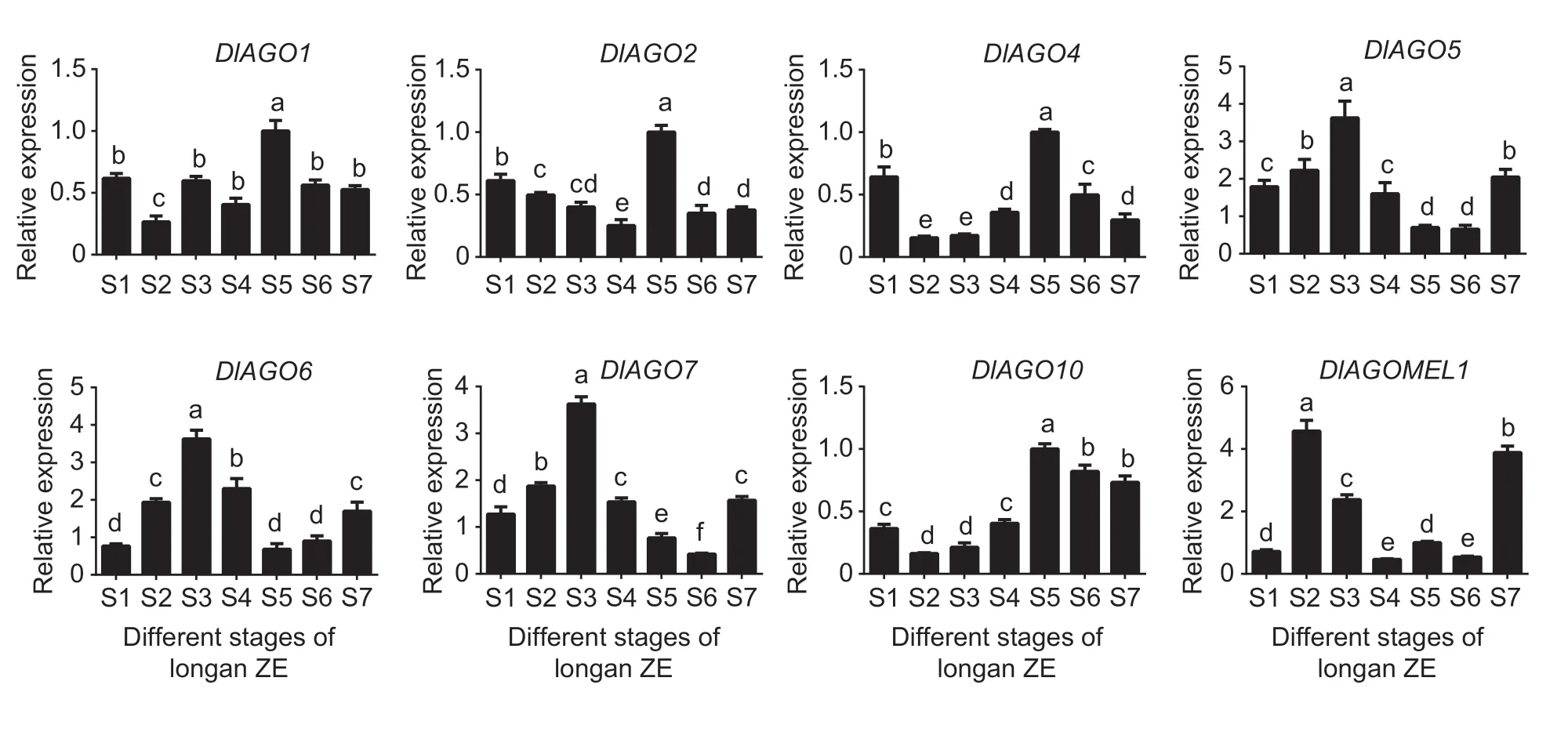
Fig.9 Relative expression of DlAGO genes at different stages of longan zygotic embryo (ZE). Different development stages of ZE,when the young fruits emerge the cotyledon embryo stage was marked as S1,then the zygotic embryos were collected every four days,marked as S2,S3,S4,S5,S6,and S7,respectively. Reference genes are UBQ and EF-1α. Data are mean±SD (n=3). Lower-case letters indicate significant differences at 0.05.
The expression ofDlAGOgenes in“HHZ”longan tissues,including roots,stems,leaves,leaf buds,flower buds,inflorescence,alabastrum,male flowers,filaments,anthers,female flowers,young fruits,pulp,ripe fruits,and seeds,was analyzed using qRT-PCR technology,and differential expression patterns were observed (Fig.10).The results showed thatDlAGO1gene was highly expressed in all tissues and relatively high in male flowers and seeds. Five genes showed the highest expression in seeds,includingDlAGO2,DlAGO4,DlAGO7,DlAGO10,andDlAGOMEL1. WhileDlAGO6was a flower budspecific gene,DlAGO5-1showed the high expression in stems and young fruits,followed by leaves and leaf buds. The results indicated the functional conservation and diversity ofAGOsin longan.
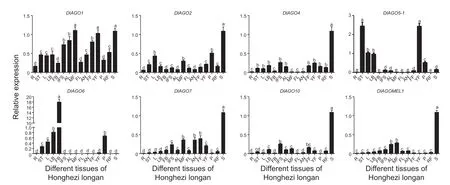
Fig.10 Relative expressions of DlAGO genes in different tissues of Honghezi longan. R,root;ST,stem;L,leaf;LB,leaf bud;FB,flower bud;IFS,inflorescence;AL,alabastrum;MF,male flower;FL,filament;AN,anther;FF,female flower;YF,young fruit;P,pulp;RF,ripe fruit;S,seed. Reference genes are UBQ and EF-1α. Data are mean±SD (n=3). Lower-case letters indicate significant differences at 0.05.
3.8.Expression patterns of longan AGO genes in response to exogenous hormones
To understand whetherDlAGOsare hormone responsive genes,we analyzed the transcription ofDlAGOsunder different concentrations of 2,4-D,kinetin (KT),MeJA,and SA for 24 h. Among the treatments,exogenous 2,4-D inhibited the expression of fourDlAGOgenes (DlAGO2,DlAGO4,DlAGO7,andDlAGO10),while certain concentrations of 2,4-D promoted the expression of the other fourDlAGOs,and the expression ofDlAGO1,DlAGO5-1andDlAGOMEL1had a peak with the treatment of 1.0 mg L-12,4-D (Fig.11-A). Interestingly,KT treatment had an active effect onDlAGOstranscript levels.DlAGO7andDlAGOMEL1had the highest expression levels when the concentrations of KT were respectively 1.0 and 2.0 mg L-1,while the remaining sixDlAGOswere upregulated dramatically by the 0.25 mg L-1KT concentration (Fig.11-B). The relative expression ofDlAGOsunder different concertations of MeJA showed that MeJA inhibited expression of four genes (DlAGO2,DlAGO4,DlAGO5-1andDlAGO6) compared with the control,but a certain concentration of MeJA could enhance the transcript levels of the other four genes (Fig.11-C). Relatively low concentrations of SA can improve the expression ofDlAGOs,but high concentrations could have negative effects on them (Fig.11-D).
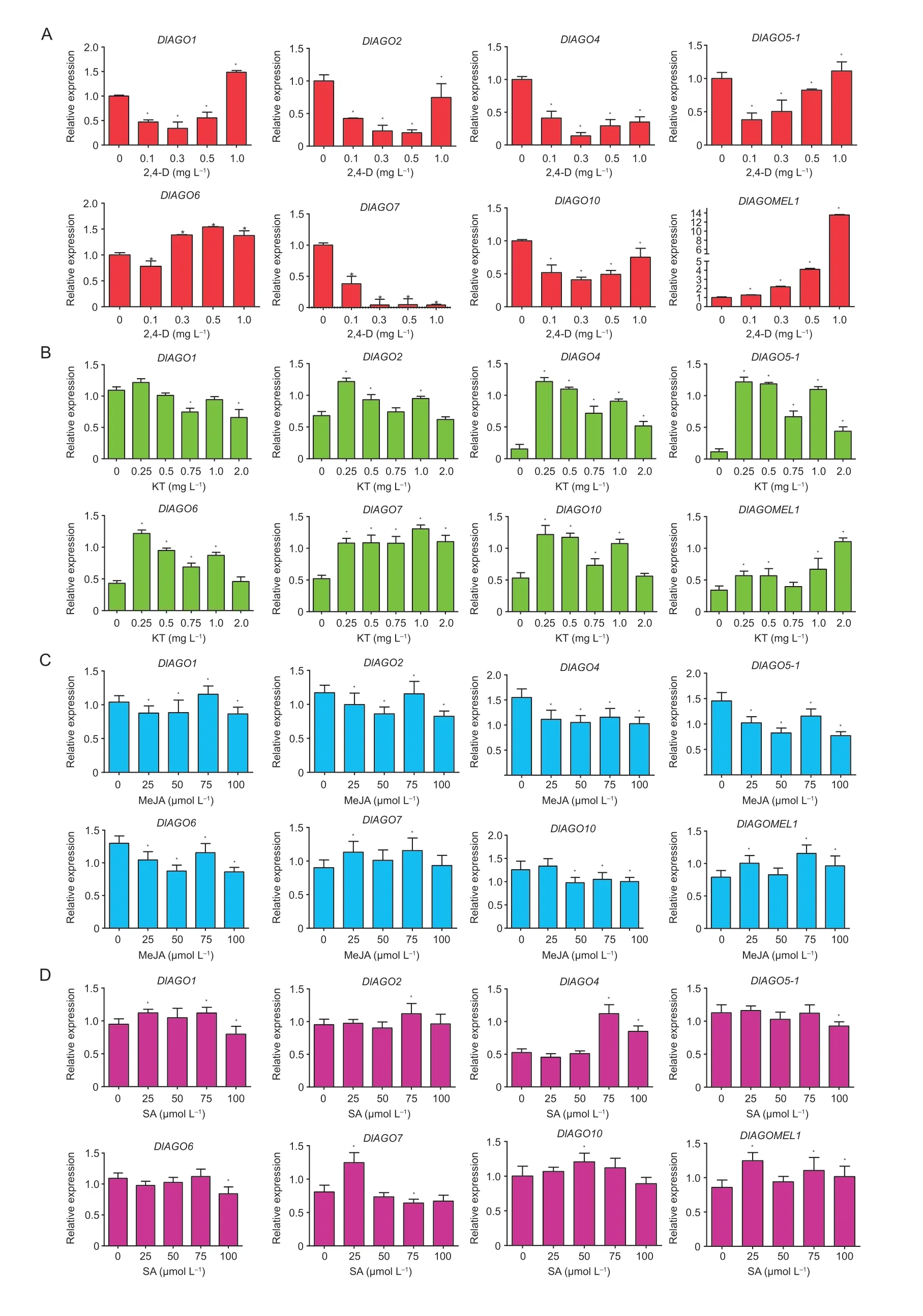
Fig.11 Relative expression of DlAGO transcripts in response to exogenous hormones. Embryogenic callus (EC) was treated with nutrient solution in the presence of the listed hormones at the indicated concentrations (MS,0 as control). A,treatment with 2,4-D (0.1,0.3,0.5,and 1.0 mg L-1). B,treatment with KT (0.25,0.5,0.75,1.0,and 2.0 mg L-1). C,treatment with MeJA (25,50,75,and 100 μmol L-1).D,treatment with SA (25,50,75,and 100 μmol L-1). The expression levels of DlAGOs after longan EC treatment with 2,4-D were normalized to Fe-SOD,and the DlAGOs expression levels after longan EC treatments with KT,MeJA,and SA were normalized to EF-1α,Fe-SOD,and eIF4α. Data are mean±SD (n=3). * indicates significant difference at 0.05.
These results indicated that the transcription machinery ofDlAGOgenes did react differently in responding to different exogenous hormones in longan EC,and different members had different hormone regulation effects.
3.9.The effects of light and abiotic stress on longan AGO genes expression
In order to evaluate the abiotic stress-responsiveness of the longanAGOfamily members,we investigated the transcript levels in longan EC exposed to 150 mmol L-1NaCl,150 mmol L-1mannitol,10% PEG-4000,and 10 μmol L-1ABA,and then sampled at six different points (0,1,2,4,8,and 12 h) at 25°C,110 r min-1and dark conditions for 12 h. In addition,longan EC was treated under different light quality conditions for 24 h to determine whetherDlAGOsresponded to light. The results showed that allDlAGOsresponded to abiotic stress and light.
After 150 mmol L-1NaCl treatment,it was found that six (DlAGO1,DlAGO2,DlAGO4,DlAGO5-1,DlAGO10,andDlAGOMEL1) of eightDlAGOswere initially up-regulated and then down-regulated,and had relative peaks at 4 h after treatment. Unlike the expression patterns of the above genes,DlAGO7was up-regulated,and had the highest expression at 1 h treatment. In addition,DlAGO6was down-regulated relative to the control treatment (Fig.12-A).
For mannitol stress,the results of the relative expression levels showed that compared to the control,DlAGO6was down-regulated,but the otherDlAGOswere up-regulated to varying degrees by this treatment.DlAGO4,DlAGO5-1andDlAGO7increased slightly up to 2 h after treatment and were down-regulated thereafter,while the expression ofDlAGO2andDlAGOMEL1were up-regulated significantly up to treatment for 4 h and then declined gradually,andDlAGO10had the highest expression after treatment for 8 h. Interestingly,the treatment can improve the relative expression level of theDlAGO1gene,which increased slightly up to 12 h after treatment (Fig.12-B).
After PEG-4000 treatment,the expression analysis showed that allDlAGOswere induced in response to it.DlAGO5-1was down-regulated,whereas the treatment can improve the expression of the otherDlAGOsat different points. The expression ofDlAGO2,DlAGO4,DlAGO10andDlAGOMEL1were up-regulated up to 2 h and then followed by a decrease gradually at 4,8 and 12 h,whereasDlAGO7andDlAGO6showed peaks at 1 and 4 h treatment,respectively. Unlike the above genes,the expression level ofDlAGO1gene showed a pattern of first rising,then falling slightly and finally rising sharply,meanwhile it had the highest expression at 12 h treatment (Fig.12-C).
To determine the responsiveness ofDlAGOsto ABA treatment,we investigated the transcript levels by qRTPCR. We found that allAGOgenes responded to the ABA treatment with time-dependent patterns.DlAGO2,DlAGO4andDlAGO10were up-regulated notably up to 1 h after the ABA treatment and then decreased at the other times.DlAGO7andDlAGOMEL1showed the highest expression at 2 h and then declined. Meanwhile,ABA treatment had an active effect on the expression ofDlAGO1and the highest expression level was at 12 h. But beyond that,DlAGO5-1andDlAGO6were strikingly downregulated by the ABA treatment compared to the control (Fig.12-D).
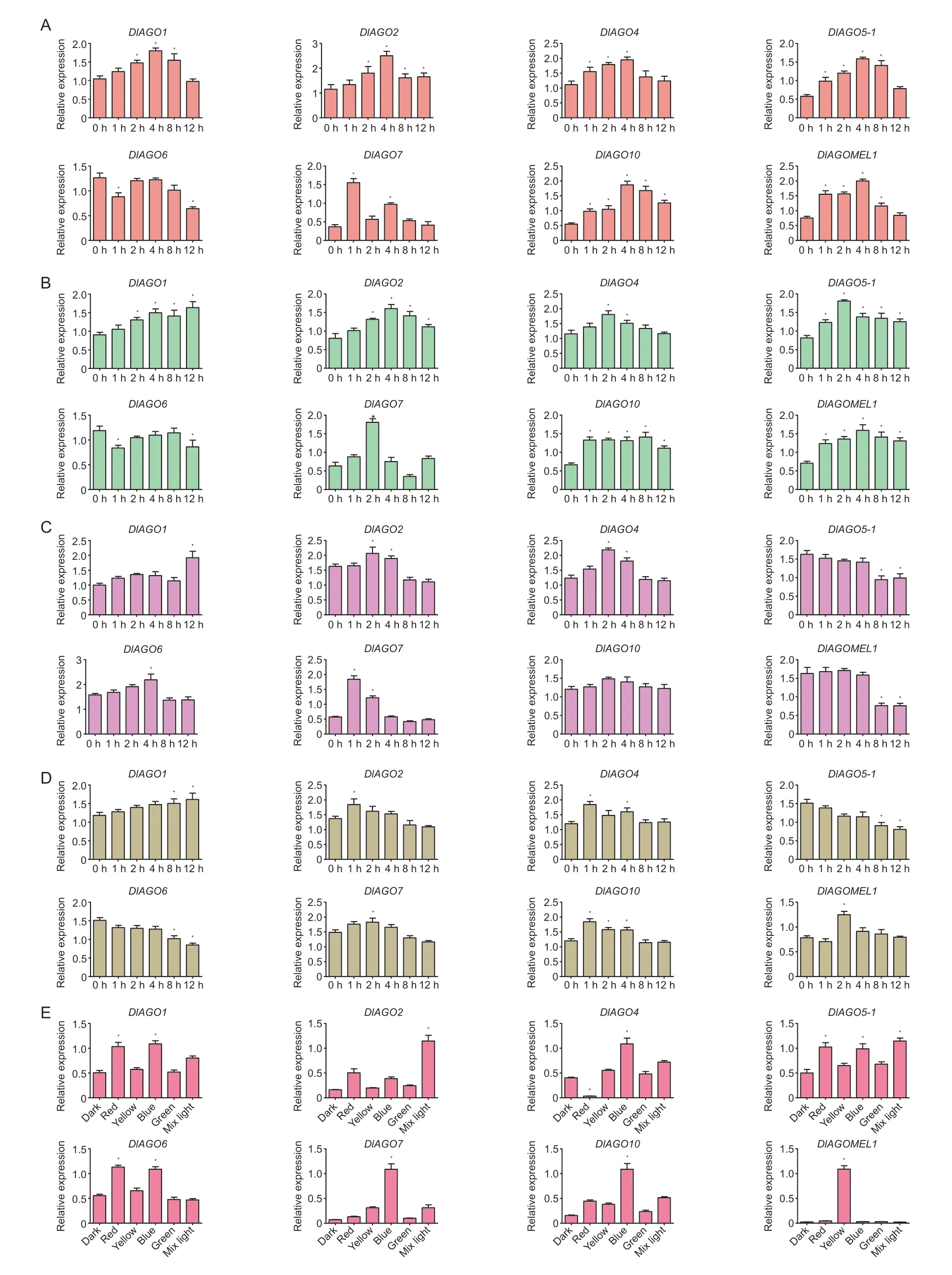
Fig.12 DlAGOs expression in response to abiotic stress and light treatment. A-D,embryogenic callus (EC) was treated with nutrient solution containing 150 mmol L-1 NaCl,150 mmol L-1 mannitol,10% PEG-4000 or 10 μmol L-1 ABA for different times,respectively. E,different light qualities (dark,red,green,blue,and white) for 24 h. Reference genes are EF-1α,eIF4α,and Fe-SOD. Data are mean±SD (n=3). * indicates significant difference at 0.05.
The results indicated thatDlAGOsmRNA differently responded to salinity,mannitol,PEG-4000,and ABA stress.These stresses did promoteDlAGO1,DlAGO2,DlAGO4,DlAGO7,DlAGO10,andDlAGOMEL1. Salinity and mannitol stress had a positive impact toDlAGO5-1and inhibitedDlAGO6,while PEG-4000 and ABA stress had a negative impact onDlAGO5-1,butDlAGO6was down-regulated in ABA treatment and up-regulated in PEG-4000 stress.
As Fig.12-E shows,blue light can significantly promote the expression ofDlAGO1,DlAGO4,DlAGO7andDlAGO10genes. The relative expression levels ofDlAGO2andDlAGO5-1genes increased significantly under mixed light treatment. Both blue and red light can significantly promote the expression ofDlAGO6gene. The expression ofDlAGOMEL1wasup-regulated significantly by yellow light. So,DlAGOsare light-responsive genes,the accumulation ofDlAGOsmRNA is stimulated by various light qualities and they may be involved in different light regulation pathways.
4.Discussion
4.1. AGO genes may play a key role in longan embryogenesis and developmental processes
Somatic embryogenesis is involved in dedifferentiation and redifferentiation through reconstruction of somatic cells to generate embryogenic cells. More and more evidence has proven that the plant somatic embryogenesis is controlled not only by specific genes but also by epigenetic regulation,especially DNA methylation (Jahnke and Scholten 2009;Zemachet al.2010;Songet al.2015). A certain level of DNA methylation enables more genes to be actively transcribed in the embryogenic callus,thus facilitating the development of plant embryos (Lambéet al.1997;Zavattieriet al.2010;Teyssieraet al.2013). In studies on mosses and rice,24-nt long miRNA can mediate DNA methylation,and this process is influenced by related proteins participating in the miRNA synthesis pathway (Hao and Deng 2002;Chellappanet al.2010;Khraiweshet al.2010). MiRNAs,especially 24-nt long miRNAs in longan,played an important role in longan SE (Huet al.2014). AGO comprises the core component of RNA-induced silencing complexes (RISC),and is involved in miRNA synthesis,so it also plays an important role in somatic embryogenesis. In studies of loblolly pine,carrot,and orange,PtAGO9L,C-Ago1,CsAGO4a,CsAGO6andCsAGO10were specifically expressed during somatic embryogenesis (Ohet al.2008;Takahata 2008;Agustínet al.2019),and the likely role ofAtAGO10was shown to be involved in embryo development (Moussian 1998;Tuckeret al.2008). In our study,the expression of longanAGOgenes showed different patterns in early SE,and they showed similar expression during longan ZE processes,especially high expression at the early or middle embryo stages.
AGOgenes are also involved in the development of plant organs. InArabidopsis,AtAGO1was expressed especially in leaves,roots,flowers and siliques,butago1mutantwas a dwarf (Bohmertet al.1998). InS.miltiorrhiza,theAGOsexpression of flowers,leaves,stems and roots are different and the members of each subgroup have the same expression pattern (Shao and Lu 2013). In grapevines,VvAGOsare expressed in different organs,butVvAGO10awas only expressed in the stem (Zhaoet al.2015). AllCsAGOsin cucumber,especiallyCsAGO1c,CsAGO1dandCsAGO7showed rich expression in leaves and tendrils compared to the other organs (Ganet al.2017).CaAGO1b,CaAGO5andCaAGO10bexhibited higher transcript levels in flower;CaAGO2andCaAGO4ashowed high expression in fruit and leaves,respectively (Qinet al.2018).AGO1aandAGO2played a key role in the flowering process and flower development in rice (Kapooret al.2008).In our study,DlAGO2,DlAGO4,DlAGO7,DlAGO10,andDlAGOMEL1showed special expression in seeds,DlAGO5-1was expressed in stem and young fruits,DlAGO6showed the highest expression in flower buds,andDlAGO1may participate in all the longan organs. We can conclude thatDlAGOsexhibits diverse expression patterns in different organs and play different roles in the formation of organs. The different expression patterns ofAGOsin various plant species exhibit considerable variations which likely contributes to a diverse set of functions in different species of plants.
4.2.AGO genes were involved in exogenous hormone signaling pathways during longan SE
Exogenous hormones can be used as a signal of embryogenesis,plant organ differentiation and responses to stress. Certain concentrations of 2,4-D could affect somatic embryogenesis of carrot and grapevine (Borkirdet al.1986;Yanget al.2013). During longan SE,embryogenic induction was accompanied by high levels of 2,4-D and the level of endogenous IAA in early SE was higher,decreasing after the GE stage until the late stages,while 1 mg L-12,4-D can make the EC growth vigorous and maintained long term (Lai and Chen 2002). SA and MeJA can act as endogenous signals stimulating plant defense responses (Vicente and Plasencia 2011).AtAGO1is responsive to hormones (Liuet al.2018),and six of 19OsAGOsshowed specific up/down-regulation in response to Gibberellin A3 (GA3),KT,or 1-naphthaleneacetic acid (NAA) treatments (Yanget al.2013). Among theAGOgenes in pepper,CaAGO10aandCaAGO10bwere significantly induced by ABA andCaAGO1aexpression was up-regulated by SA (Qinet al.2018). Our results suggested that exogenous 2,4-D inhibitedDlAGO2,DlAGO4,DlAGO7,andDlAGO10,but promotedDlAGO1,DlAGO5-1,DlAGO6,andDlAGOMEL1,whereas,the transcripts ofDlAGOswere promoted by KT in longan EC. Some of theAGOsin longan were significantly induced and others were repressed by MeJA,while the transcript levels ofDlAGOswere increased by SA. These results showed thatDlAGOsparticipate in different hormone signaling pathways during longan SE. However,at present the relationships betweenAGOsand these hormones (KT,2,4-D,SA,and MeJA,) in plant embryos are still unknown,so,further study is needed onDlAGOgenes in the regulation of hormone signaling pathways in longan SE.
4.3. DlAGOs may participate in responses to abiotic stress
Some studies reported that plantAGOgenes also respond to abiotic stresses.ZmAGO1amight play an important role in response to environmental changes,and allZmAGOsshowed different expression modes under various abiotic stress treatments (Zhaiet al.2019). InSetaria italica,19AGOshave differential expression patterns at different time points of stresses (Yadavet al.2015). TheMdAGOsare mostly involved in the responses to NaCl,PEG,heat,and low-temperature stresses (Xu R Ret al.2016). Also,stress was considered as an embryogenic stimulus in the induction of somatic embryogenesis (Shiet al.2009). Interestingly,we found here that certain treatment times of NaCl and mannitol could increase the accumulation of seven of eightDlAGOs(DlAGO1,DlAGO2,DlAGO4,DlAGO5-1,DlAGO7,DlAGO10,andDlAGOMEL1),but decrease the expression ofDlAGO6. Additionally,the transcript levels of most ofDlAGOs(DlAGO1,DlAGO2,DlAGO4,DlAGO6,DlAGO7,DlAGO10,andDlAGOMEL1) could be up-regulated and that ofDlAGO5-1was down-regulated by PEG-4000 treatment. Therefore,it seems thatDlAGOsplay direct and important roles in responses to abiotic stresses such as osmotic,salinity,and drought stress.
Some studies have revealed that ABA can promote the maturation of plant somatic embryos (Finkelsteinet al.1985;Fernando and Gamage 2000;Langhansováet al.2004). During SE,ABA plays the key role in normal morphological development of embryos,which can improve somatic embryo quality. Our results indicated that ABA has a positive effect on the accumulation of mostDlAGOs,and only inhibits the transcript level ofDlAGO6. All these results indicate thatDlAGOsparticipate in the abiotic stress responses during longan SE.
5.Conclusion
RNA silencing regulates many developmental events in plants,andAGOproteins play an important role in this conserved mechanism. So,it is critical to identify theAGOgenes family using the longan genome database and investigate their temporal and spatial expression patterns. In our study,a total of 10DlAGOgenes were identified in longan EC. Phylogenetic,chromosomal location,and gene structure analyses were carried out.The results showed that they were divided into four clades,distributed on chromosomes 1,4,8,10,12,13,14 and 15,while the number of introns ofAGOfamily members varied greatly,from 2 to 23. In addition,the expression profiles ofDlAGOgenes under hormone (2,4-D,KT,MeJA,SA),light,and abiotic stress (NaCl,mannitol,PEG,ABA) were detected by qRT-PCR. The results revealed significant roles ofDlAGOsin longan embryogenesis and will provide a new insight into the RNAi-mediated gene silencing in longan EC.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31572088 and 31672127),the Science and Technology Plan Major Projects of Fujian Province of China (2015NZ00021),the New Century Excellent Talents Support Program in Fujian Province University of China (20151104),and the Special Fund for Scientific and Technological Innovation of Fujian Agricultural and Basic Research of China (CXZX2017314).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Lignin metabolism regulates lodging resistance of maize hybrids under varying planting density
- Adoption of small-scale irrigation technologies and its impact on land productivity:Evidence from Rwanda
- Comparison of grain yield and quality of different types of japonica rice cultivars in the northern Jiangsu plain,China
- Natural nematicidal active compounds:Recent research progress and outlook
- lmproving grain appearance of erect-panicle japonica rice cultivars by introgression of the null gs9 allele
- Comparative transcriptome analysis of different nitrogen responses in low-nitrogen sensitive and tolerant maize genotypes
