lmproving grain appearance of erect-panicle japonica rice cultivars by introgression of the null gs9 allele
2021-06-24ZHAODongshengLlUJinyuDlNGAiqiuZHANGTaoRENXinyuZHANGLinLlQianfengFANXiaoleiZHANGChangquanLlUQiaoquan
ZHAO Dong-sheng,LlU Jin-yu,DlNG Ai-qiu,ZHANG TaoREN Xin-yuZHANG LinLl Qian-feng,FAN Xiao-lei,ZHANG Chang-quan,LlU Qiao-quan
1 Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding/Key Laboratory of Plant Functional Genomics of the Ministry of Education,College of Agriculture,Yangzhou University,Yangzhou 225009,P.R.China
2 Co-innovation Center for Modern Production Technology of Grain Crops,Jiangsu Key Laboratory of Crop Genetics and Physiology,Yangzhou University,Yangzhou 225009,P.R.China
Abstract The panicle architecture and grain size of rice affect not only grain yield but also grain quality,especially grain appearance. The erect-panicle (EP) trait controlled by the qpe9-1/dep1 allele has been widely used in high-yielding japonica rice breeding,but usually accompanied with moderate appearance of milled rice. The null gs9 allele shows a good potential for improving grain shape and appearance. However,GS9 and qPE9-1/DEP1 loci are tightly linked,and their interaction is unclear,which obviously restricts their utilization in modern rice breeding. In the present study,comparative analyses of protein and mRNA levels revealed that GS9 and qPE9-1 function independently. Three nearisogenic lines (NILs) carrying various allelic combinations of these two loci,NIL (gs9/qpe9-1),NIL (GS9/qPE9-1) and NIL (gs9/qPE9-1),in the EP japonica cultivar 2661 (GS9/qpe9-1) background were developed for genetic interaction analysis.GS9 and qPE9-1 had additive effects on determining grain size,and the null gs9 allele could decrease grain chalkiness and improve grain appearance without affecting plant and panicle architecture in EP japonica cultivars. Additionally,introgression lines (ILs) developed in another released EP japonica cultivar Wuyujing 27 (WYJ27) background showed the same additive effect and the feasibility of utilizing the gs9 allele to improve grain appearance quality in high-yielding EP cultivars. This study provides an effective strategy for rice breeders to improve rice grain appearance in EP japonica and related cultivars.
Keywords:rice,GS9,qPE9-1/DEP1,genetic interaction,erect panicle,grain appearance
1.lntroduction
Rice (OryzasativaL.) is one of the most important staple cereal crops in the world,and a continuous increase in rice production will be required to meet the demands of the burgeoning human population and increased consumption. However,the grains of high-yielding rice cultivars usually have a poor appearance due to large grain size. Therefore,it is important for rice breeders to develop high-yielding cultivars that have superior grain appearance.
Rice appearance is mainly determined by grain shape and chalkiness (Zhouet al.2020). Grain shape is a complex trait that is controlled by multiple genes with relatively high inheritance. Large grain size results in high yield but poor grain appearance,because large grain size,especially the grain width,is usually associated with much chalkiness due to insufficient starch storage accumulation during grain filling (Zhaoet al.2018). Chalkiness affects the transparency of milled rice and thus its commercial value,and has a negative influence on milling and cooking properties (Harberd 2015). Numerous cloned grain-shape genes have provided important clues on the molecular mechanisms underlying grain shape in rice (Maet al.2002;Liet al.2019;Xueet al.2019). Several important genes that control grain shape,such asGS3,GW8,GL7/GW7,GLW7,andGS9,have great application potential in modern rice breeding (Maoet al.2010;Wanget al.2012;Wang Set al.2015;Wang Yet al.2015;Siet al.2016;Zhaoet al.2018). At present,though many quantitative trait loci (QTLs) have been mapped for rice chalkiness,only one gene has been cloned (Liet al.2014),and the genetic mechanism behind this trait remains unknown.
To achieve breakthroughs for improved rice yield,ideal plant architecture is considered to be a viable approach. The erect-panicle (EP) trait contributes to the preferred plant type injaponicarice cultivars. Plants with erect panicles are able to utilize solar energy effectively,and consequently exhibit dramatic increases in yield potential (Feiet al.2019;Wang Set al.2019). To date,a number of high-yieldingjaponicarice cultivars with dense and erect panicles have been released and occupy a predominant place in China (Xuet al.2016). Currently,several erect panicle genes,DEP1/qPE9-1,DEP2/EP2,EP3,andDEP3,have been cloned (Huanget al.2009;Piaoet al.2009;Zhouet al.2009;Liet al.2010;Zhuet al.2010;Qiaoet al.2011). Among these,the riceqPE9-1/DEP1gene,which controls panicle type and responses to nitrogen,has been widely used injaponicarice breeding (Huanget al.2009;Zhouet al.2009;Sunet al.2014).The dominant allele at theqPE9-1/DEP1locus is a gain-of-function mutation that results in reduced panicle length and erect panicle,and it was the target of artificial selection during domestication,mostly injaponicarice lines (Huanget al.2009;Zhouet al.2009).
The qPE9-1/DEP1 belongs to the G protein family,and the rice G protein contains one Gα subunit (RGA1),one Gβ subunit (RGB1),and five Gγ homologous subunits (RGG1,RGG2,GS3,qPE9-1/DEP1,and GGC2) (Sunet al.2018).RGA1andRGB1show positive regulation on plant growth and grain size (Fujisawaet al.1999;Utsunomiyaet al.2011).RGG1andRGG2are involved in the response to abiotic stress (Yadavet al.2012;Swainet al.2017).RGG2acts as a negative regulator of plant growth and grain size (Miaoet al.2019).GGC2individually or in combination withDEP1increases grain length when complexed with Gβ (Sunet al.2018).GS3functions as a negative regulator of grain size (Maoet al.2010). These findings implicate G protein components in the regulation of grain size and their complex roles in rice.
Although the erect-paniclejaponicacultivars have the potential for high grain yields,they tend to have inferior grain quality due to their relatively larger grain size (Xuet al.2004,2005). Several studies have established that it is practical to breed high-yielding and high-quality rice cultivars by pyramiding grain size QTLs (Wanget al.2012;Wang Set al.2015;Zhaoet al.2018). Thus,it is of considerable importance to identify the genetic interaction effects between grain size QTLs and theqpe9-1allele in EP cultivars,which would facilitate further crop improvement.
However,thegs9allele improves grain shape and appearance,which is consequently used to improve the weakness ofqpe9-1(Zhaoet al.2018).GS9is closely linked with theqPE9-1gene on chromosome 9 that are located approximately 350 kb apart (Fig.1-A),which might result in linkage drag during the introgression of the target chromosomal fragment. More importantly,their interaction is unclear. In this study,the nullgs9allele was introgressed into EPjaponicarice cultivars with theqpe9-1allele and the relationship betweenGS9andqPE9-1was tested. The results show that the independence betweenGS9andqPE9-1,two closely linked and beneficial genes,provides a predictable approach to improve grain appearance in EP cultivars.
2.Materials and methods
2.1.Rice materials
The introgression line N138 carrying thegs9andqPE9-1alleles in the ‘Nipponbare’ background was used as the donor parent (Zhaoet al.2018). Two EPjaponicacultivars grown in the lower Yangtze River region that carry theGS9andqpe9-1alleles,2661 and Wuyujing 27 (WYJ27),were used as recurrent parents. Three nearisogenic lines (NILs) carrying thegs9/qpe9-1,GS9/qPE9-1,andgs9/qPE9-1allelic combinations in the 2661 background were identified from a BC4F2:3generation derived from backcrosses between 2661 and N138 to break the unfavorable linkage between the two target genes (Fig.1;Appendix A). The NILs possessed nearly all the genetic background of the cultivar 2661,with the exception of the introgressed chromosomal segment containing the target genes. Similarly,four other introgression lines (ILs) in the WYJ27 background,namely IL (GS9/qpe9-1) which carries the same alleles as wild type WYJ27,IL (gs9/qpe9-1),IL (GS9/qPE9-1),and IL (gs9/qPE9-1),were selected from the progeny of backcrosses between WYJ27 and N138 (Fig.1;Appendix A). The genotypes and phenotypes of selected rice materials are given in Appendix A.
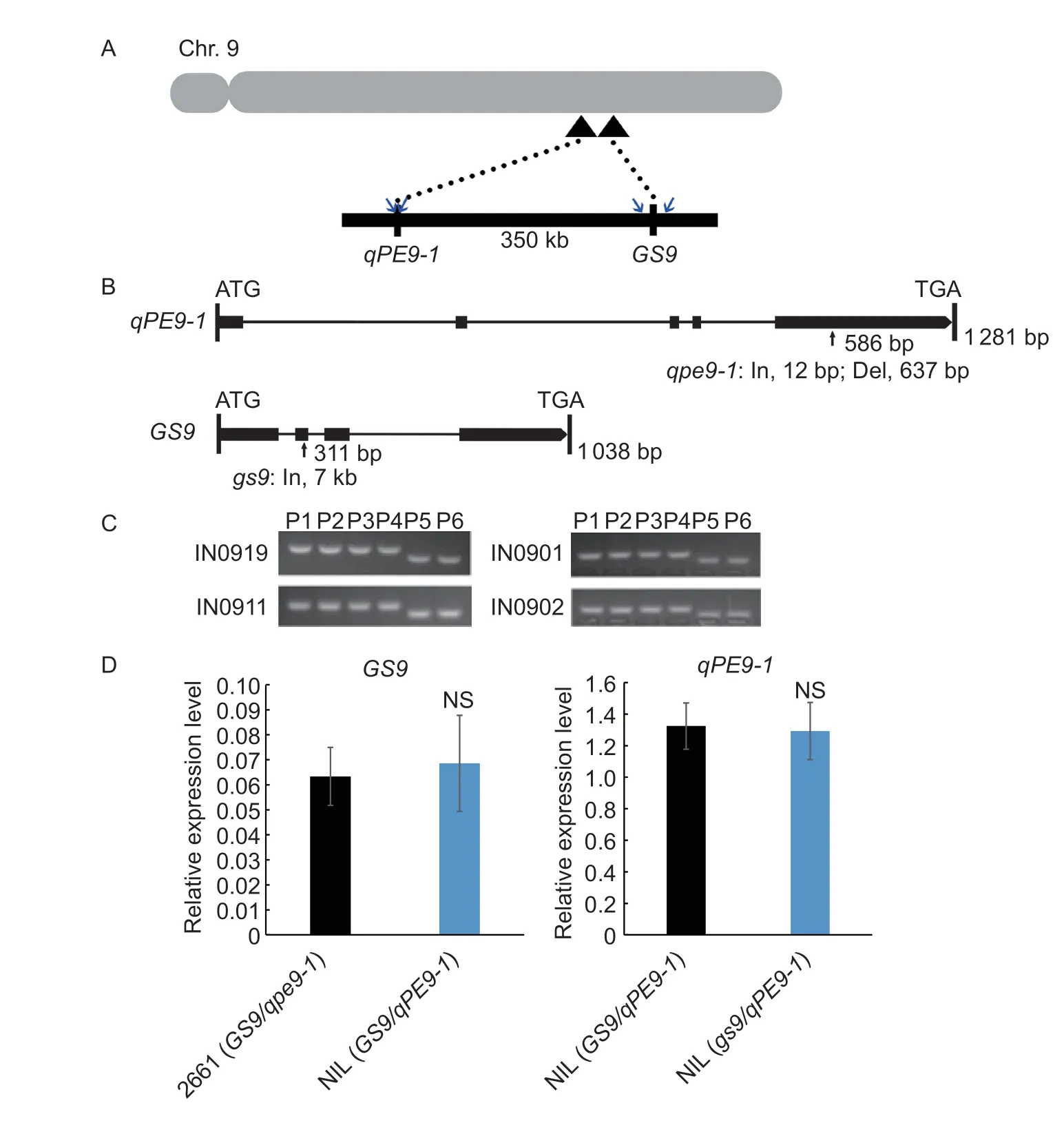
Fig.1 Genetic linkage and allelic variation of GS9 and qPE9-1 genes. A,physical positions of GS9 and qPE9-1. GS9 is closely linked with qPE9-1 on chromosome 9. Four blue arrows indicate the physical position of molecular markers as follows. B,allelic variation in the qPE9-1 and GS9 genes. The qpe9-1 allele encodes a presumably truncated protein due to the InDel4 at position 586 bp. In,insertion mutation;Del,deletion mutation. No transcript from the gs9 allele was detected due to the 7 kb insertion at position 311 bp. C,molecular markers designed to detect the GS9 and qPE9-1 alleles. IN0919 and IN0911 map to loci flanking the GS9 gene with physical distances of 17-kb upstream and 6-kb downstream. IN0901 and IN0902 are located inside the qPE9-1 gene as shown in Fig.1-A. P1-P4,2661,WYJ27,Wuyujing 8,and Yangdao 8,respectively,which are erect-panicle (EP) japonica rice cultivars;P5 and P6 are 9311 and Qingluzan 11,respectively,which are indica rice cultivars with drooping panicles. D,GS9 mRNA levels in inflorescences that differ with respect to the qPE9-1 and qpe9-1 alleles in 2661 and near-isogenic line (NIL (GS9/qPE9-1)). qPE9-1 mRNA levels in inflorescences that differ with respect to the GS9 and gs9 alleles in NIL (GS9/qPE9-1) and NIL (gs9/qPE9-1). Data are given as mean±SD (n=3). NS,no significant difference (P<0.05,t-test).
2.2.Rice phenotyping
All rice plants were grown in an experimental field at Yangzhou University (32.4°N,119.4°E,Yangzhou,Jiangsu) or Lingshui (18.5°N,110.1°E,Hainan) in China using established planting and management practices for rice. Plant height was measured from the ground surface to the tip of the tallest panicle. Panicle length was measured on the main culm from the panicle neck to the panicle tip. Panicle curvature was calculated from the angle between two lines:the line connecting the panicle pedestal and panicle tip,and the elongation line of the stem. Fully filled grain length,width,lengthto-width ratio,and grain chalky were measured using a grain appearance analyzer ScanMaker (Microtek,China),and the 1 000-grain weight was then measured. Grains was de-husked and milled for ECQ measurement. The apparent amylose content (AAC) was measured using the NY/T 593-2013 standard (Zenget al.2017). Viscosity was measured with a rapid visco analyzer (RVA) (Newport Scientific,Australia). The assay of taste and palatability in cooked rice was conducted by an STA1A rice taste analyzer (SATAKE,Japan). Three or five replications were performed for each trait.
2.3.Molecular markers,genomic DNA isolation,and genotyping
Four molecular markers mapped to closely linked loci on chromosome 9 were developed for assisted forward selection of the allelic variation in theGS9andqPE9-1genes (Fig.1). The PCR marker primers were designed using Primer Premier 5.0 based on the sequence differences between parental lines. Molecular marker loci IN0919 and IN0911 are flanking theGS9gene at physical distances of 17 and 6 kb,respectively;the other two marker loci IN0901 and IN0902 are located inside theqPE9-1gene (Fig.1-C;Appendix B). Notably,thegs9allele is a natural mutation of theGS9gene present in introgression segment of the N138 line. Thus,it is straightforward to use IN0919 and IN0901 to examine the introgression of thegs9allele from theindicabackground intojaponicarice. IN0901 and IN0902 were designed based on two specific variable sites between drooping and EP rice cultivars. DNA was isolated from leaf tissue using the CTAB method and then diluted with nucleasefree water for PCR amplification. PCR was carried out using the Taq Master Mix system (Vazyme,China). The amplified products were separated by electrophoresis on a 3% agarose gel.
2.4.RNA extraction and gene expression analysis
Young panicles from 2661 (GS9/qpe9-1),NIL (GS9/qPE9-1) and NIL (gs9/qPE9-1) plants were harvested for RNA extraction. Total RNA was isolated using a Plant RNA Extraction Mini Kit (Tiangen,China). Firststrand cDNA was subsequently synthesized using reverse transcriptase (TaKaRa,Japan) and an oligo dT18primer. Three biological replicates were prepared for each sample. The rice housekeeping geneActin(LOC_Os03g0718100) was used as an internal control.Quantitative real-time (RT) PCR analysis was performed on a Bio-Rad CFX96 Real-Time PCR Detection System using SYBR Green. The specificity of amplified products was confirmed by melting curve analysis,and the relative expression level is presented as 2-ΔΔCTaccording to the ΔCTmethod (Livak and Schmittgen 2001). The specific primers used for amplification ofGS9and the known G protein genes,includingRGA1/Os05g0333200,RGB1/Os03g0669200,RGG1/Os03g0635100,RGG2/Os02g0137800,GS3/Os03g0407400,qPE9-1/DEP1/Os09g0441900,andGGC2/Os08g0456600,are given in Appendix C. In addition,young panicles of NPB-gs9 (gs9) and ‘Nipponbare’ (GS9) plants were harvested for RNA preparation and subsequent RNA sequencing. NPB-gs9 (gs9) was the NIL selected from N138 and ‘Nipponbare’ backcross progeny in previous study. The filtered clean reads were aligned to the rice ‘Nipponbare’ reference genome and genes (http://rice.plantbiology.msu.edu/).RNA sequencing data has been deposited in the NCBI SRA database (accession code SRP132484) (Zhaoet al.2018). The transcript abundances of the known and predicted genes in the G protein pathway were compared between thegs9andGS9allelic variants (Appendix D).
2.5.Yeast two-hybrid assays
Yeast two-hybrid assays were performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech,Japan). The truncated coding region of theGS9/LOC_Os09g27590gene (GS9-C1,without transcription activation activity) was introduced into the pGBKT7 vector used as bait protein. Full-length cDNAs of the known G protein subunit genes (RGA1,RGB1,RGG1,RGG2,GS3,qPE9-1/DEP1,andGGC2) were amplified and cloned into the pGADT7 vector,as prey proteins (RGA1,RGB1,RGG1,RGG2,and GGC2 were provided by Sunet al.(2018)).OFP8was cloned into the pGBKT7 vector and used as the positive control (Zhaoet al.2018). The relevant primers are given in Appendix C.
2.6.Statistical analysis
Values were presented as the mean±standard deviation (SD). The exact sample size of each trait for statistical analysis was listed in figure legends. The SPSS 16.0 Software was used for all statistical tests. Statistical significance was determined by independent samplet-tests for comparison of two groups and by one-way ANOVA for comparison of more than two groups. Differences were considered statistically significant whenP≤0.05.
3.Results
3.1.Function of GS9 independent of qPE9-1
The qPE9-1 protein has been identified as an important member of the G protein family in higher plants,whileGS9acts as transcriptional regulator (Sunet al.2014;Zhaoet al.2018),which both control rice grain size.Therefore,we further confirmed the relationship ofGS9,qPE9-1,and other G protein genes. Three NILs carrying different allelic combinations ofGS9andqPE9-1(gs9/qpe9-1,GS9/qPE9-1andgs9/qPE9-1) were selected in the cultivar 2661 (GS9/qpe9-1) background (Fig.1;Appendix A).GS9allelic variation has no effect onqPE9-1expression,and the reserve is equal (Fig.1-D). In addition,identical transcription levels of other G protein subunit genes,RGA1,RGB1,RGG1,RGG2,GS3,andGGC2,were observed between NIL (gs9/qPE9-1) and NIL (GS9/qPE9-1) (Appendix E). Furthermore,GS9allelic variation did not result in differential expression of whole-genome predicted genes involved in the G protein from RNA sequencing data (Appendix D). Thus,these data indicate thatGS9might not reciprocally affect the transcriptional level ofqPE9-1and other G protein genes.
Using a yeast two-hybrid system,it was found that GS9 did not interactin vivowith qPE9-1 (Appendix F) and other G protein components including RGA1,RGB1,RGG1,RGG2,GS3,and GGC2 (Appendix F). These results revealed thatGS9indeed functioned independently from the G protein pathway,includingqPE9-1,implying the feasibility of usinggs9to improve the appearance of EP cultivars.
3.2.No influence of the null gs9 allele on plant and panicle architecture in EP rice cultivars
Based on their independent functions identified previously,the genetic interaction betweenGS9andqPE9-1were further examined. Compared with the recurrent parent 2661 (GS9/qpe9-1),introgression of the nullgs9allele did not alter plant and panicle architecture in NIL (gs9/qpe9-1) in both Hainan and Yangzhou fields (Fig.2;Appendix G). As expected,when theqpe9-1allele was substituted byqPE9-1,plant height,panicle length,panicle curvature and grain yield per plant in NIL (GS9/qPE9-1) were quite different from the erect panicle in the recurrent parent 2661 (GS9/qpe9-1) in both field locations (Fig.2;Appendix G). There were no significant differences in plant and panicle architecture between theGS9andgs9alleles under bothqpe9-1andqPE9-1conditions,which strongly indicated that there was no genetic interaction effect betweenGS9andqPE9-1.
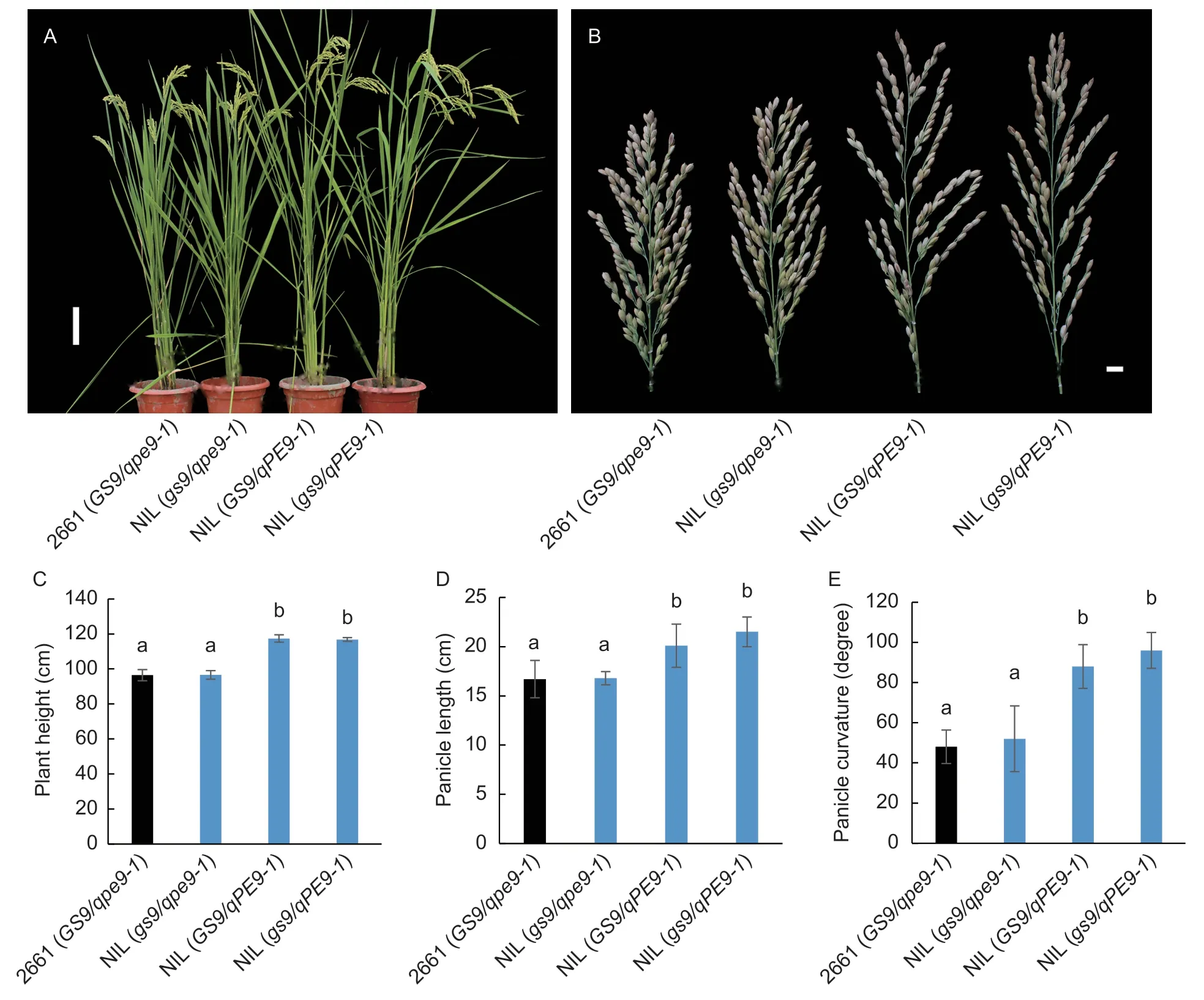
Fig.2 Plant height and panicle architecture of 2661 and three near-isogenic lines (NILs) grown in Yangzhou field,Jiangsu Province,China. A,comparison of plant architecture between 2661 and three NILs carrying different combinations of the GS9 and qPE9-1 alleles. Scar bar=10 cm. B,comparison of panicle architecture between 2661 and the three NILs. Scar bar=1 cm. C-E,plant height,panicle length,and panicle curvature in 2661 and the three NILs. 2661 (GS9/qpe9-1),wild type 2661 carrying the GS9 and qpe9-1 alleles;NIL (gs9/qpe9-1),NIL carrying the gs9 and qpe9-1 alleles;NIL (GS9/qPE9-1),NIL carrying the GS9 and qPE9-1 alleles;NIL (gs9/qPE9-1),NIL carrying the gs9 and qPE9-1 alleles. Data are given as mean±SD (n=5). Different letters indicate significant differences (P<0.05,one-way ANOVA).
In addition,four ILs,carrying theGS9/qpe9-1,gs9/qpe9-1,GS9/qPE9-1,andgs9/qPE9-1allelic combinations in the same segregating progeny of backcrosses between WYJ27 and N138,were selected (Fig.1;Appendix A). As expected,the nullgs9allele did not alter plant and panicle architecture,but the functionalqPE9-1allele had a clear effect on plant height,panicle length,and panicle curvature (Appendices G and H).These results confirmed that the allelic effects of theGS9andqPE9-1loci in different genetic backgrounds. In short,the nullgs9allele had no significant effect on plant and panicle architecture in EPjaponicacultivars.
3.3.The null gs9 allele improves grain shape in EP japonica rice cultivars
The allelic functions ofGS9andqPE9-1on grain shape were further investigated. First,in the Hainan field,compared with 2661,NIL (gs9/qpe9-1) caused a significant increase (6.8%) in grain length and a decrease (-5.1%) in grain width which consequently produced a longer and narrower grain shape (Appendix I). In addition,NIL (GS9/qPE9-1) caused a significant increase (3.4%) in grain length but had no significant effect on grain width (Appendix I). Moreover,when thegs9andqPE9-1alleles were both substituted,NIL (gs9/qPE9-1) led to a 10.1% increase and 3.9% decrease in grain length and width,respectively (Appendix I).
It was also confirmed that the changes of these grain traits during a normal growing season at the Yangzhou field. Similarly,compared with 2661,introgression of thegs9allele caused a significant increase (9.1%) in grain length and a decrease (-4.2%) in grain width,producing a significant increase (13.8%) in grain lengthwidth ratio,yet there was no difference in 1 000-grain weight (Fig.3). Moreover,introgression of theqPE9-1allele caused an increase (4.3%) in grain length and no significant difference in grain width;this resulted in an increase (3.9%) in grain length-width ratio and a significant increase (10.3%) in 1 000-grain weight (Fig.3). Combining thegs9andqPE9-1alleles caused a 12.9% increase in grain length,a 3.4% decrease in grain width,a 16.8% increase in grain length-width ratio,and a 10.0% increase in 1 000-grain weight (Fig.3),which almost equal to the effect of theoretical addition of thegs9andqPE9-1alleles. Meanwhile,the increased grain weight effect of theqPE9-1allele resulted in increased grain yield per plant (Fig.3-F).
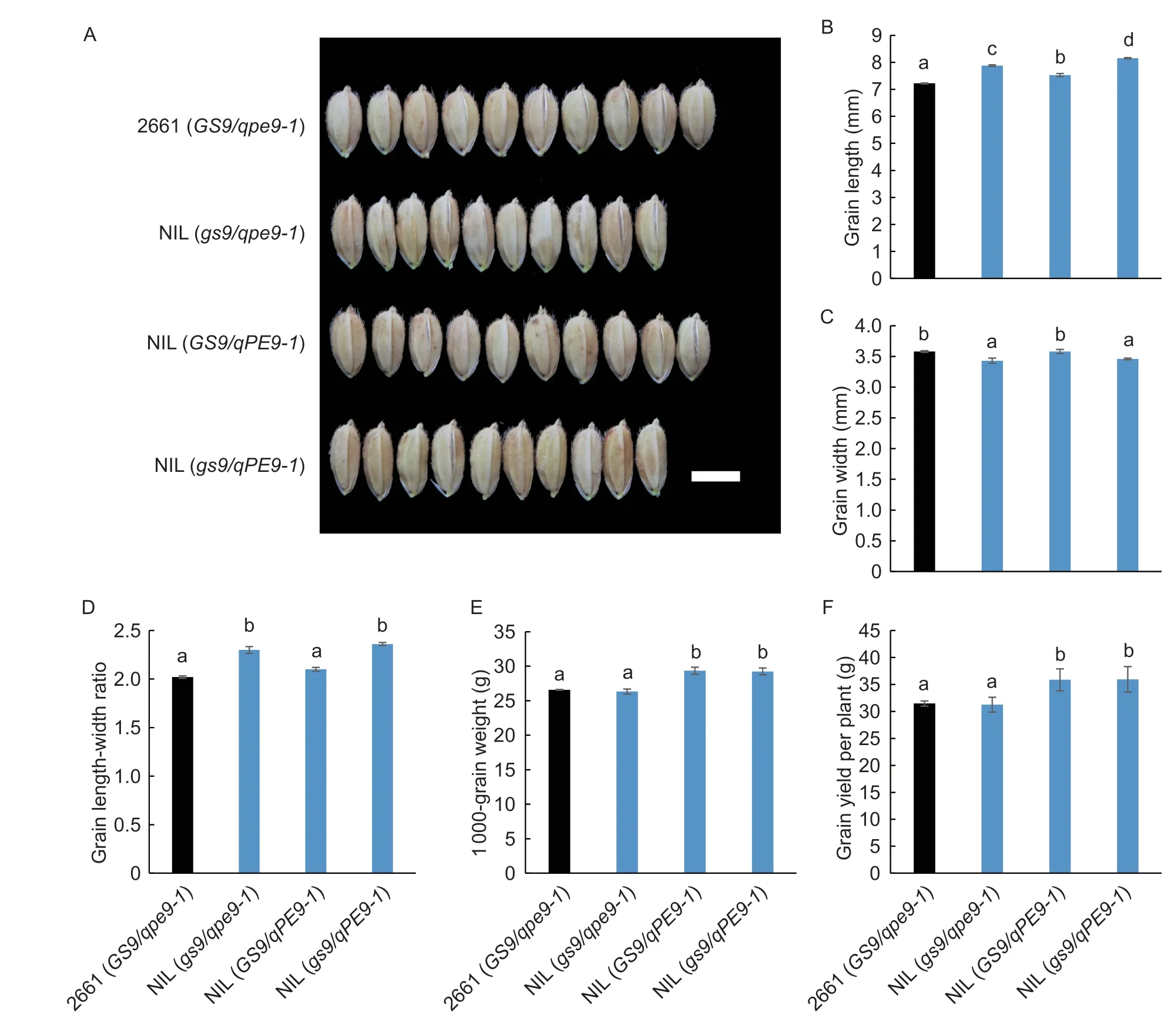
Fig.3 Grain phenotypes in 2661 and three near-isogenic lines (NILs) grown in Yangzhou field,Jiangsu Province,China. A,comparisons of mature grain shape. Scar bar=5 mm. B-F,grain length,grain width,grain length-width ratio,grain weight,and grain yield per plant in 2661 and three NILs. Data are given as mean±SD (n=3). The different lowercase letters indicate significant differences between the means (P<0.05,one-way ANOVA). The genotypes of 2661 and three NILs are given in the legend to Fig.2.
While eitherGS9orqPE9-1has a significant effect on grain shape,there is an obvious additive effect ofGS9andqPE9-1together on grain shape. The nullgs9allele has a significant effect on improving grain shape in EPjaponicarice cultivars.
3.4.The null gs9 allele improves grain appearance of milled rice in EP japonica rice without changing ECQ
AAC and RVA,two important physicochemical properties,affect ECQ. In addition,the palatability value is another factor of ECQ for cooked rice (Zenget al.2017). In this study,AAC,RVA and palatability value were compared among the NILs carrying different combinations of theGS9andqPE9-1alleles. The data showed that there was no significant effect of these two loci,either alone or in combination (Fig.4). As previously reported,GS9orqPE9-1alone had no significant effect on these ECQ indexes.
One final goal of the study is to utilize the nullgs9allele to improve grain appearance of EP cultivars. A slender grain shape usually accompanies better appearance due to a low level of chalkiness (Harberd 2015). Similar with unhusked grain,in 2661 (GS9/qpe9-1) and NIL (GS9/qPE9-1) carrying theGS9allele,grain length-towidth ratio is relatively low. Therefore,an obvious grain chalkiness was present (Fig.4-D);introgression of thegs9allele significantly decreased the percentage of chalky grain and chalkiness degree (Fig.4),indicating an improved appearance in both grain shape and chalkiness (Fig.4). However,in the case ofqPE9-1alone,there was no obvious allelic effect on grain chalkiness,andqPE9-1did not alter the decreased grain chalkiness caused by thegs9allele (Fig.4).
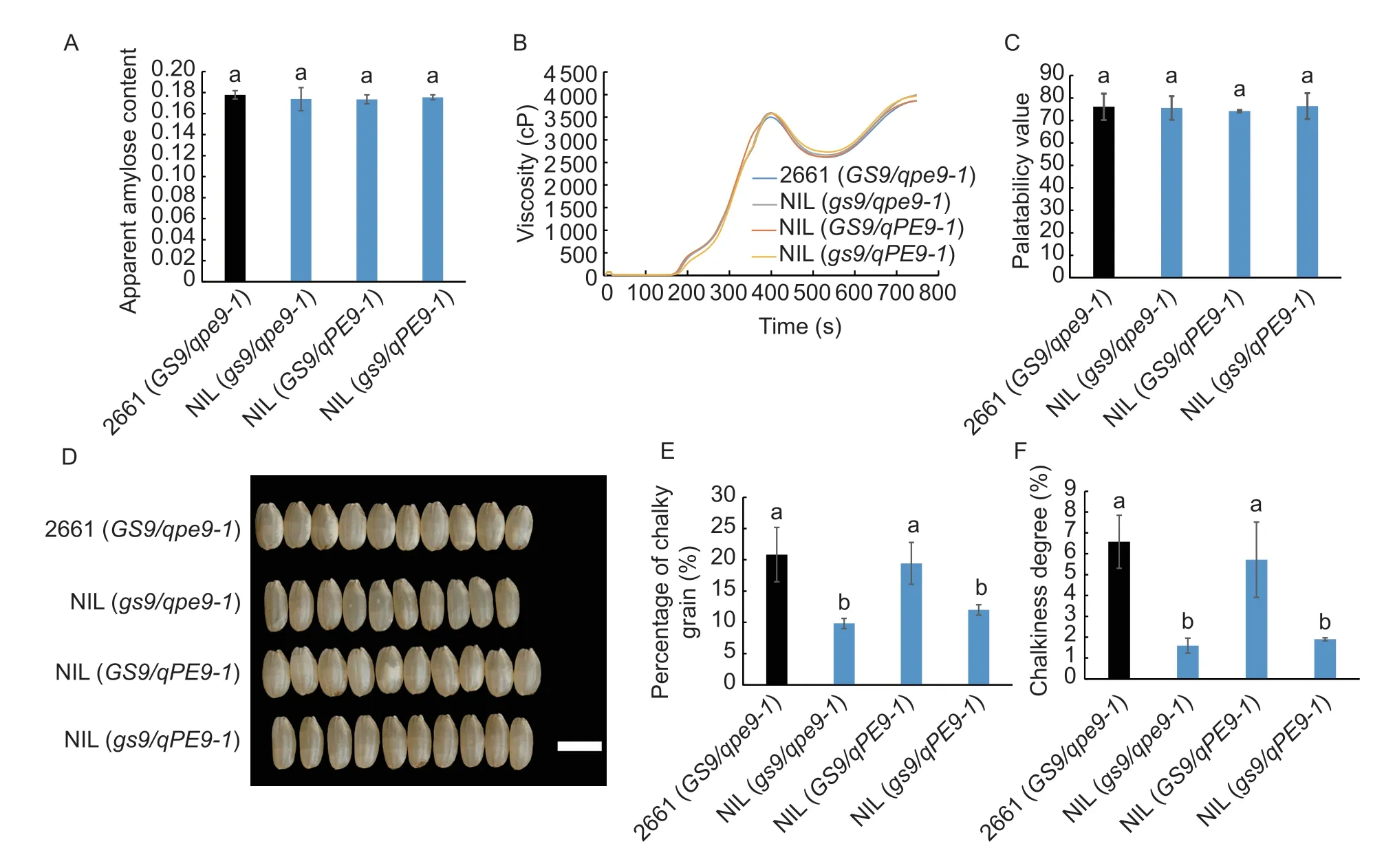
Fig.4 Grain quality in 2661 and three near-isogenic lines (NILs) grown in Yangzhou field,Jiangsu Province,China. A-C,apparent amylose content,rice flour viscosity and palatability value in 2661 and NILs. D,comparison of grain appearance. Decreased chalkiness results from introgression of null gs9 allele. Scar bar=5 mm. E and F,percentage of chalky grain,and chalkiness degree in 2661 and the NILs. Data are given as mean±SD (n=3). The different lowercase letters indicate significant difference between the means (P<0.05,one-way ANOVA). The genotypes of 2661 and three NILs are given in the legend to Fig.2.
Finally,the effect of the nullgs9allele on grain shape and chalkiness was confirmed in the WYJ27 background,another high-yielding commercial cultivar. For grain shape,compared with IL (GS9/qpe9-1),the mature grains of IL (gs9/qpe9-1) were significantly longer and narrower due to increased grain length and decreased grain width,resulting in a 8.2 and 8.6% increase in the grain lengthwidth ratio at Hainan and Yangzhou field,respectively (Fig.5;Appendix J). Meanwhile,a similar change was observed in milled rice (Fig.5-B). For grain chalkiness,introgression of thegs9allele significantly decreased the percentage of chalky grain and chalkiness degree (Fig.5).Additionally,thegs9allele did not change ECQ (Appendix J). These results confirmedgs9’s ability to improve grain appearance in different genetic backgrounds.
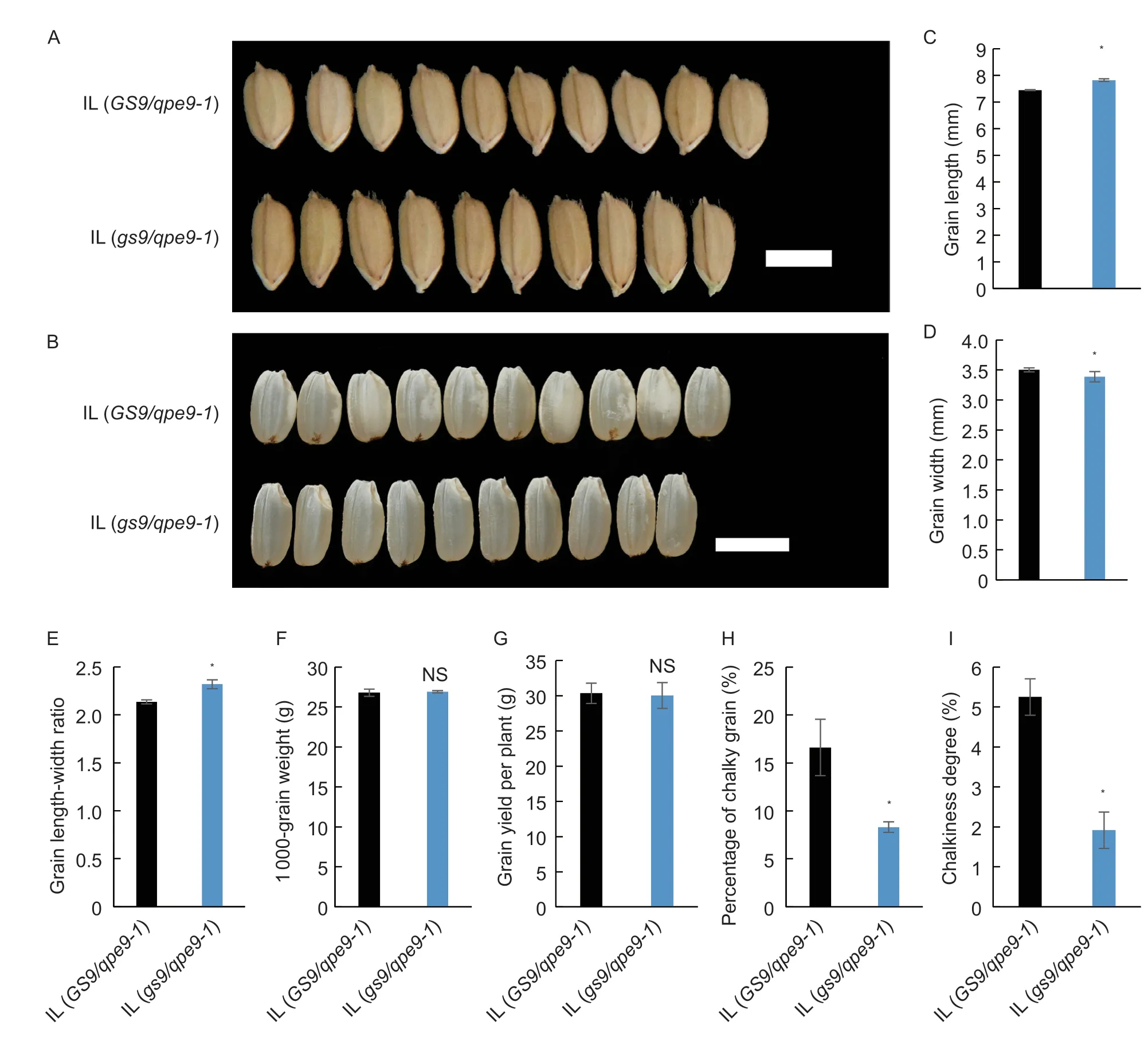
Fig.5 Grain shape and appearance in WYJ27 introgression lines (ILs) grown in Yangzhou field,Jiangsu Province,China. The two ILs,IL (GS9/qpe9-1) and IL (gs9/qpe9-1),were selected from the same segregating population and were homozygous for the GS9 and gs9 alleles,respectively,in the WYJ27 background. A,mature grain shape. Scar bar=5 mm. B,grain appearance. gs9 allele also decreased chalkiness in the WYJ27 background. Scar bar=5 mm. C-I,grain length,grain width,grain length-width ratio,grain weight,grain yield per plant,percentage of chalky grain,and chalkiness degree. Data are given as mean±SD (n=3).* and NS indicate significant and non-significant differences (P<0.05,t-test),respectively.
In summary,introgression of thegs9allele can significantly improve grain appearance by changing grain shape and decreasing chalkiness in EPjaponicacultivars without affecting ECQ. These results suggest that it is feasible to use thegs9allele to improve grain appearance in EP cultivars.
4.Discussion
The EPjaponicarice cultivars generally have mediocre grain quality,including processing quality,appearance,and ECQ. Thus,future breeding projects utilizingdep1/qpe9-1should address these inadequacies,especially grain appearance (Xuet al.2004,2016).GS9orqPE9-1alone had no signifcant effect on ECQ indexes (Yiet al.2011;Zhaoet al.2018;Wang Yet al.2019),which still had no change in combination ofGS9andqPE9-1(Fig.4). Grain shape is an important aspect for appearance improvement (Harberd 2015). The effect ofDEP1/qPE9-1on grain shape is clear,yet there was no consensus on the effect of thedep1/qpe9-1allele on grain yield (Huanget al.2009;Zhouet al.2009). Moreover,the effect ofdep1/qpe9-1on grain quality is controversial,which might be due to different cultivars’ genetic backgrounds (Wang Yet al.2019). Importantly,introducing the nullgs9allele into EPjaponicarice cultivars significantly improves grain shape and appearance without altering grain yield and ECQ. However,neither theqPE9-1norqpe9-1alleles affected the improved grain appearance caused by the nullgs9allele (Figs.4 and 5). The nullgs9is a rare allele in bothindicaandjaponicarice cultivars,while a high proportion of Chinese high-yielding rice cultivars carry thedep1/qpe9-1alleles. Therefore,gs9shows a good potential to improve the breeding of EP cultivars.
Rational design by pyramiding multiple complex traits is a powerful strategy for breeding excellent cultivars to meet the challenges of future crop breeding (Zenget al.2017). The essence of rational design is genetic interaction. Therefore,identification of improved genes and ready incorporation of these genes into rice varieties will become increasingly important in feeding the increasing human population. Using this strategy,a few excellent lines were bred (Zenget al.2017).
Grain shape controlled by several signaling pathways is an important breakthrough point to improve grain appearance in EPjaponicacultivars. Compared with recent advances in the understanding of molecular networks (Liet al.2019),understanding the genetic effects of combinations of grain size genes only progressed moderately. So,what is the effect of the genetic interaction between them? Genetic analysis has revealed mutual interactions betweenqSW5andGS3(Yanet al.2011). PyramidingGW2andqSW5/GW5showed a significant increase in grain width compared to plants carrying only one of two major QTLs,which suggests that there are no significant QTL interactions between them (Yinget al.2012).GS3andqGL3had additive effects on the regulation of rice grain length revealed by genetic analysis (Gaoet al.2015). Notably,little is known about the effect of introducing grain shape or appearance genes into EP cultivars.GS9had no direct relationship withqPE9-1gene in both protein and transcriptional level (Fig.1;Appendix F).
In summary,after the introgression of the nullgs9allele into EPjaponicarice cultivars,the NIL (gs9/qpe9-1) and IL (gs9/qpe9-1) lines exhibited improved grain appearance with no alteration in grain yield and ECQ compared with their recurrent parental lines. At present,studies have identified and characterized many genes governing grain chalkiness (Chalk5) (Liet al.2014),grain shape (GW8,GL7/GW7andGLW7) (Wanget al.2012,Wang Set al.2015;Wang Yet al.2015;Siet al.2016),and ECQ (Tianet al.2009;Zhanget al.2019). Thus,it is possible to further improve the grain quality of these high-yielding EP rice lines by reasonably pyramiding multiple favorable alleles for grain quality-related traits.
5.Conclusion
The panicle architecture and grain size of rice affect not only grain yield but also grain quality,especially grain appearance. The genetic effect between two closely linked lociGS9andqPE9-1/DEP1is unclear. The introgression lines carrying various allelic combinations of these two loci were developed in two cultivar backgrounds for genetic and molecular interaction analysis.GS9andqPE9-1had additive effects on grain size. The nullgs9allele could decrease grain chalkiness and improve grain appearance without affecting plant and panicle architecture in EPjaponicacultivars.GS9andqPE9-1functioned independently. This study provides an effective strategy for rice breeders to improve rice grain appearance in EPjaponicaand related cultivars.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31971914),the National Key Research and Development Program of China (2016YFD0100501),the Key Research and Development Program of Jiangsu Province,China (BE2018357),the Science Fund for Distinguished Young Scholars of Jiangsu Province,China (BK20200045),the Jiangsu Agricultural Science and Technology Innovation Fund (CX(18)1001),the Jiangsu PAPD Talent Project,and the Yong Elite Scientists Sponsorship Program by China Association for Science and Technology (2018QNRC001).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Effects of plant density and nitrogen rate on cotton yield and nitrogen use in cotton stubble retaining fields
- Adoption of small-scale irrigation technologies and its impact on land productivity:Evidence from Rwanda
- Lignin metabolism regulates lodging resistance of maize hybrids under varying planting density
- Natural nematicidal active compounds:Recent research progress and outlook
- Comparative transcriptome analysis of different nitrogen responses in low-nitrogen sensitive and tolerant maize genotypes
- ldentifcation of blast-resistance loci through genome-wide association analysis in foxtail millet (Setaria italica (L.) Beauv.)
