Increased CMTM4 mRNA expression predicts a poor prognosis in patients with hepatocellular carcinoma
2021-01-07HeQiZhouJianHaoLiLiWenLiuJiaMinLouZhiGangRen
He-Qi Zhou , Jian-Hao Li , Li-Wen Liu , Jia-Min Lou , Zhi-Gang Ren ,*
a Department of Infectious Diseases, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450052, China
b Key Laboratory of Clinical Medicine, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450052, China
Hepatocellular carcinoma (HCC) is one of the most common human malignancies and main cause of cancer mortality worldwide [1] . Conventional treatment for HCC consists of hepatic resection, liver transplantation and radiofrequency ablation [ 2 , 3 ]. Despite improvements in clinical treatment, the 5-year survival rate of advanced HCC patients remains low. The exploration of novel therapeutic targets and the identification of prognostic biomarkers for HCC are vital and essential to improve clinical outcomes. CKLF-like MARVEL transmembrane domaincontaining member 4 (CMTM4), mapped to chromosome 16q22.1, is the most conserved member of theCMTMfamily. TheCMTMfamily comprises 9 genes:CMTM1-8andCKLF. Proteins fromCMTMfamily are involved in the immune system [4] , the male reproductive system [5] , angiogenesis regulation, and tumorigenesis.CMTM4/6protect programmed death-1 (PD-1) ligand 1 (PD-L1) protein from ubiquitination and suppress tumor specific T cell response via PD-L1 regulation, presenting critical immune checkpoints and potential therapeutic targets for anti-tumor immunity [4] . At the protein level, high CMTM6 expression was associated with worse patient survival in head and neck squamous cell carcinoma(HNSCC) [6] , while the reduced CMTM6 protein predicted poor prognosis for HCC patients [7] . At the transcriptome level, a negative correlation was found between CMTM6 expression and survival time of patients with glioma [8] . The mRNA levels ofCMTMfamily members appears diverse in different tumors [9] . Recently,Bei et al. [10] investigated the clinical significance of the CMTM4 protein in HCC based on 75 pairs of specimens collected from HCC patients. However, the expression and clinical value ofCMTM4mRNA are not well investigated. In this study, we investigateCMTM4mRNA expression and its role in HCC diagnosis and prognosis using data from the Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) database. Moreover, we explored the molecular mechanism ofCMTM4in HCC, which may help explain the unfavorable survival of HCC.
CMTM4mRNA expression data of HCC were obtained from TCGA ( http://cancergenome.nih.gov/ ) and GEO ( https://www.ncbi.nlm.nih.gov/geo/ ). The general work algorithm of GEO data is summarized ( Table 1 ). In the TCGA database, level 3 data ofCMTM4mRNA expression of 369 HCC tissues and 50 normal tissues were collected, and 334 cancer samples with complete clinical data were used to explore the relationship betweenCMTM4mRNA expression and clinicopathological features. Results revealed thatCMTM4mRNA expression was significantly upregulated in tumor samples compared to adjacent non-tumor samples ( Fig. 1 A).After retrieval in the GEO database, a total of ten microarrays ofCMTM4expression profiles were included. Seven microarrays(GSE6764, GSE45436, GSE54238, GSE62232, GSE77314, GSE102083,and GSE76297) showed that the expression ofCMTM4mRNA in HCC tissues was higher than that of normal liver tissues( Fig. 1 B). Furthermore, we found thatCMTM4mRNA expression was much higher in patients with advanced TNM stage ( Fig. 1 C).Kaplan-Meier survival curves demonstrated that patients with highCMTM4mRNA expression had shorter overall survival (OS) and progression-free survival (PFS) than those with lowCMTM4expression ( Fig. 1 D and E). X-tile was used to determine the best cutoff points for low or highCMTM4 mRNA levels [11] . Pearson correlation analysis showed that theCMTM4mRNA level was positively correlated with marker of proliferation Ki-67 (MKI67) ( Fig. 1 F).
Most ofCMTMfamily members have been reported as tumor suppressor genes and their down-expressions are associated with poor survival in malignancies.CMTM4,CMTM5andCMTM8showed a cancer-suppressing role in most solid tumors, such as renal carcinoma, glioma and digestive system tumors [ 9 , 10 , 12 , 13 ].Bei et al. [10] demonstrated that CMTM4 protein expression was reduced in HCC. However, a high level of mRNA does not necessarily guarantee a high level of protein [14] . We speculated that post-translation regulation or posttranscriptional regulation might exist in protein synthesis, leading to the degradation or translational silencing of the target mRNA. Moreover, the mRNA levels ofCMTMfamily members in different tumors appear diverse. Delic et al. [9] found that high mRNA expression ofCMTM2,CMTM3andCMTM6in glioblastomas compared to normal tissues, and higher mRNA expressions ofCMTM3andCMTM6revealed shorter OS of glioma patients. Together, these results indicate that the levels ofCMTM4mRNA are frequently increased in HCC tissues and correlated with an unfavorable prognosis of HCC.
Clinicopathologic analysis indicated that mRNA expression ofCMTM4was associated with TNM stage, race and sex ( Table 2 ).Males had a higherCMTM4 mRNA expression than females for its high expression in the testis and its key role in spermatogenesis and sperm quality [5] . Further univariate and multivariate analyses were completed and results revealed that highCMTM4mRNA expression together with high TNM stage were independently associated with poor PFS ( Table 3 ), while only TNM stage was a prognostic marker of OS in HCC patients ( Table 4 ). ROC curves were conducted to verify the diagnostic value ofCMTM4mRNA expression in distinguishing HCC in eight independent HCC cohorts. The AUCs of different cohorts were as follows: the TCGA cohort, AUC = 0.704(P<0.001, Fig. 2 A); GSE6764, AUC = 0.816 (P<0.001, Fig. 2 B);GSE45436, AUC = 0.921 (P<0.001, Fig. 2 C); GSE54238, AUC =0.710 (P= 0.007, Fig. 2 D); GSE62232, AUC = 0.854 (P<0.001,Fig. 2 E); GSE76297, AUC = 0.887 (P<0.001, Fig. 2 F); GSE77314,AUC = 0.758 (P<0.001, Fig. 2 G); and GSE102083, AUC = 0.745(P<0.001, Fig. 2 H). Taken together, theCMTM4mRNA level was of promising value in the diagnosis and prognosis of HCC patients.

Table 1 Characteristics of CMTM4 mRNA expression profiling datasets obtained from GEO

Table 2 The relationship between CMTM4 expression and clinicopathological features of liver cancer
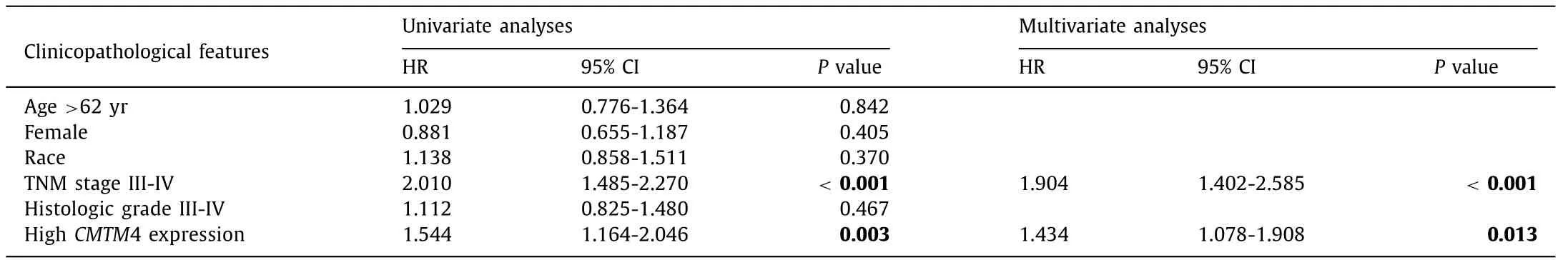
Table 3 Univariate and multivariate analyses of progression-free survival of liver cancer
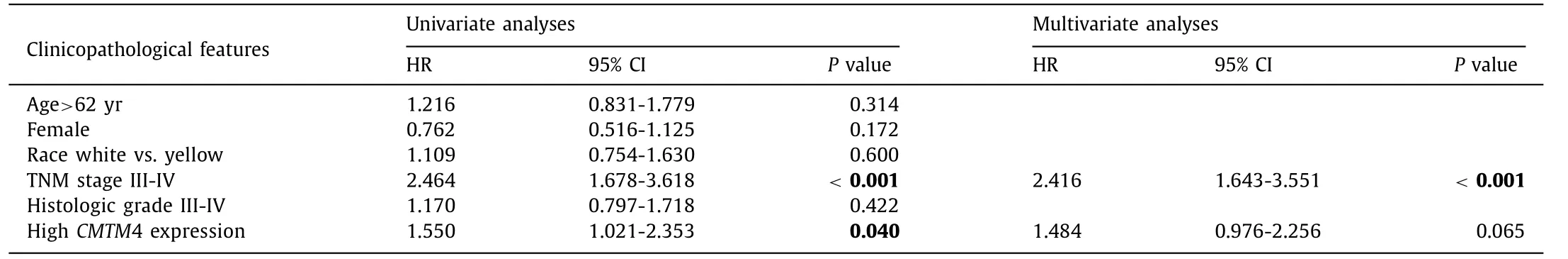
Table 4 Univariate and multivariate analyses of overall survival of liver cancer
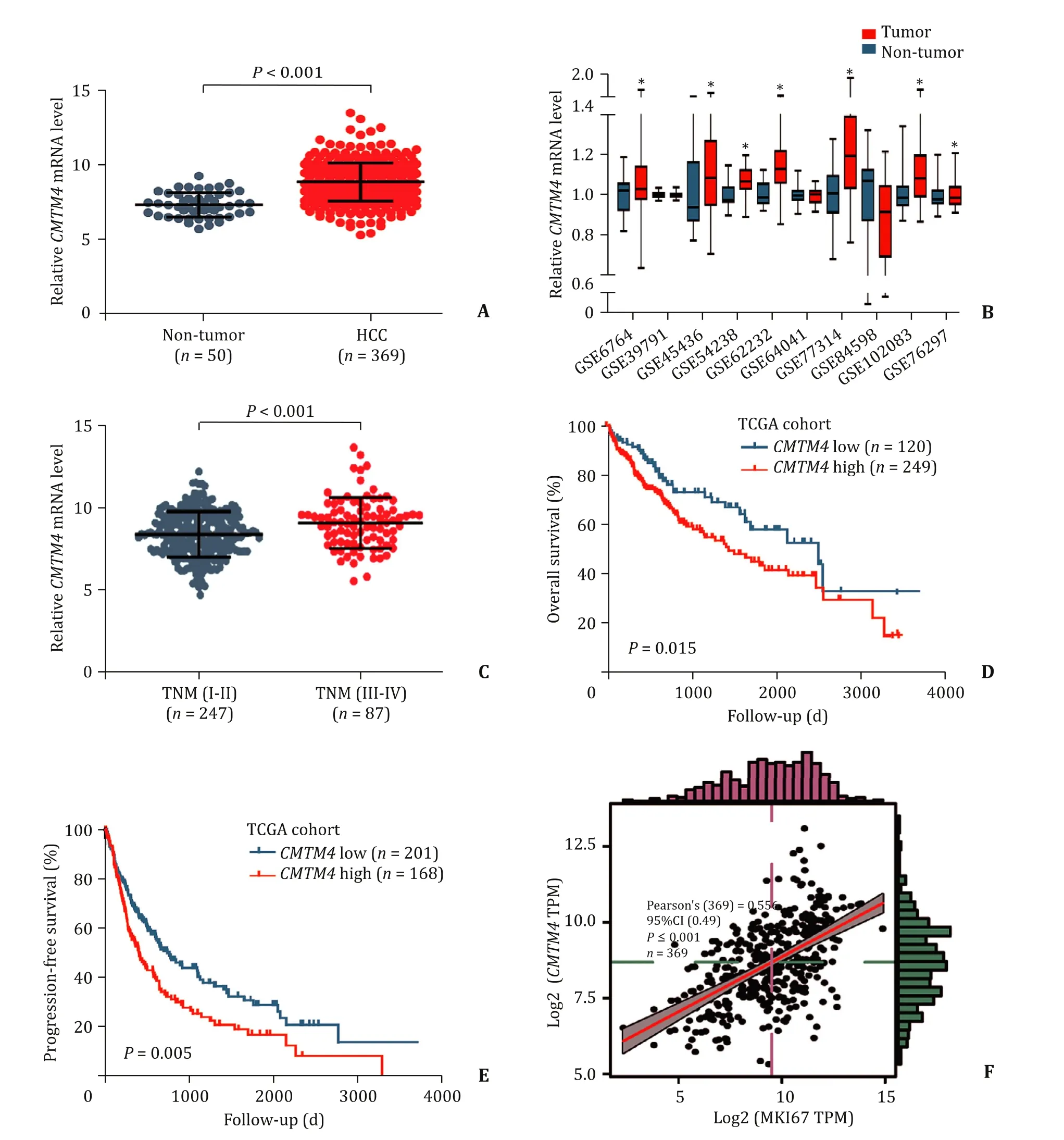
Fig. 1. CMTM4 mRNA was overexpressed in HCC tissues and negatively correlated with survival of patients with HCC. A : CMTM4 mRNA expression was up-regulated in HCC tissues according to TCGA data analysis; B : CMTM4 mRNA was up-regulated in HCC tissues in most GEO data sets; C : Expression of CMTM4 mRNA was higher in advanced TNM stage; D & E : Kaplan-Meier method and Log-rank test of overall survival and progression-free survival for HCC patients with low CMTM4 expression versus high CMTM 4 expression; F : Correlation between MKI67 and CMTM4 . CMTM4: CKLF-like MARVEL transmembrane domain-containing member 4; HCC: hepatocellular carcinoma; TCGA: The Cancer Genome Atlas; GEO: Gene Expression Omnibus; MKI67: marker of proliferation Ki-67.
To further investigate the association between theCMTM4gene and tumors, we used the gene set variation analysis (GSVA) package to explore biological processes between the low and high expression groups according to the gene sets of defined biological processes. Results suggested that the samples with high mRNA expression were enriched in immune system processes and cell junctions ( Fig. 3 A). One paper argued thatCMTM4functioned as a positive backup regulator of PD-L1 expression that enhanced the ability of PD-L1-expressing tumor cells to influence the antitumor effect of T cells [4] . Given the association ofCMTM4mRNA expression with immune system processes, we next found the correlation between immune cells andCMTM4expression in HCC. Results showed thatCMTM4mRNA level had negative correlations with cytotoxic cells (r= -0.40,P<0.001), dendritic cells (DCs) (r= -0.42,P<0.001), T cells (r= -0.19,P= 0.04) and CD8 + T cells (r= -0.16,P= 0.02) in HCC based on TCGA database ( Fig. 3 B). The tumorinfiltrating immune cells are involved in shaping the evolution and progression of HCC, and the HCC immune landscape serves as a prognostic factor [15] . In addition, viral status (HBV, HCV or negative) has no contribution to immune cell composition in HCC [16] , which makes the results more convincing. These results indicate thatCMTM4may play an important role in the immune microenvironment and the progression of HCC.
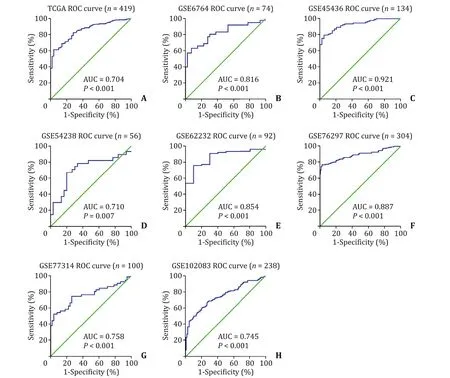
Fig. 2. Receiver operating characteristic (ROC) analysis was used to determine the sensitivity and specificity of CMTM4 mRNA level using area under the ROC curve (AUC)analysis. CMTM4: CKLF-like MARVEL transmembrane domain-containing member 4; TCGA: The Cancer Genome Atlas.
In conclusion, our findings indicate thatCMTM4mRNA expression may serve as an important prognostic and diagnostic marker of HCC.CMTM4negatively influences immune cell infiltration in HCC tissues, which may provide new options for the treatment of HCC.
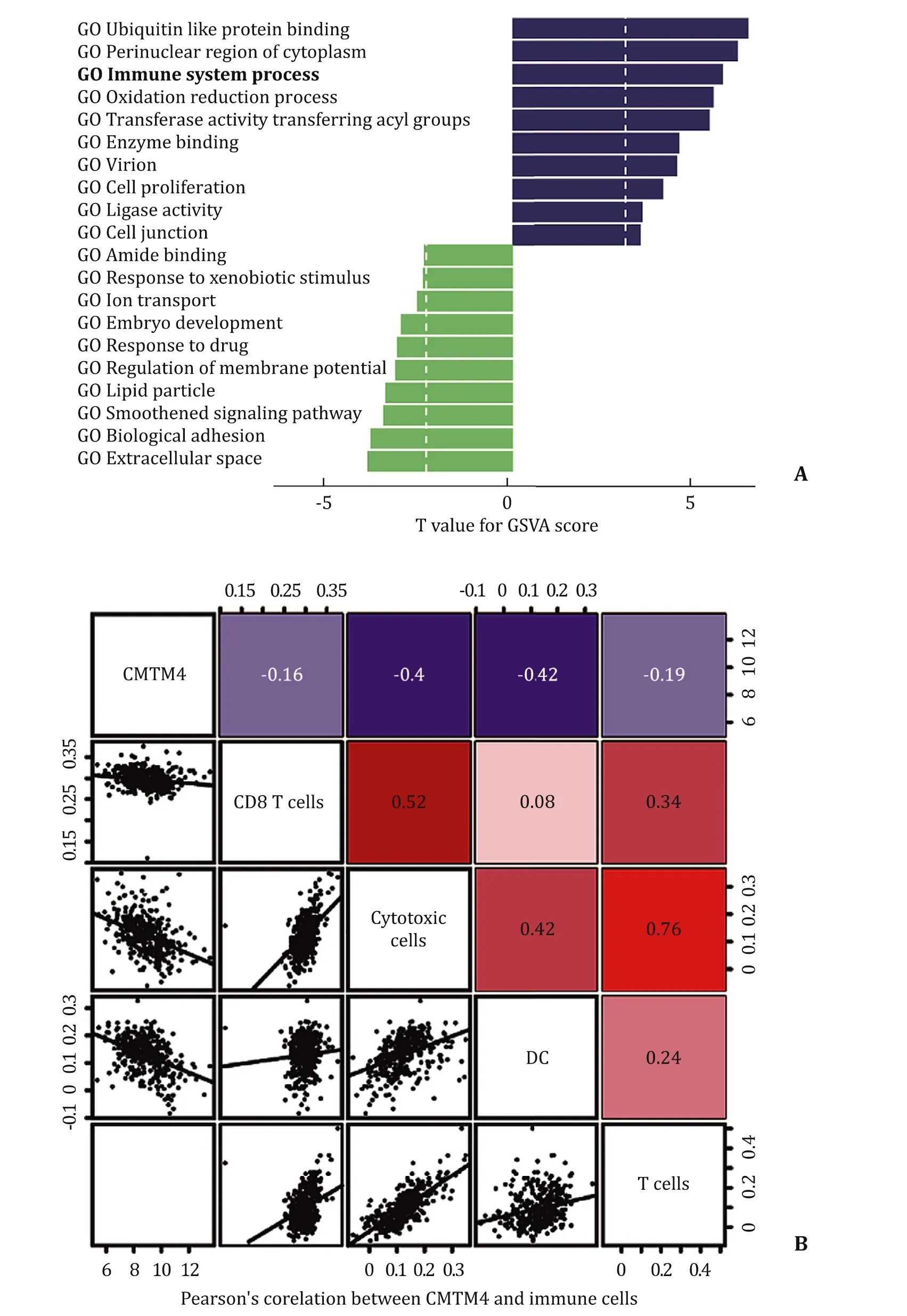
Fig. 3. CMTM4-related biological signatures in HCC. A : The top 10 CMTM4-related GO terms via GSVA; B : Pearson’s correlation coefficients between CMTM4 and CD8 + T cells, cytotoxic cells, DCs and T cells ( P < 0.05). DC: dendritic cell.
Acknowledgments
We thank Drs. Zu-Jiang Yu and Ran-Ran Sun from the First Affiliated Hospital of Zhengzhou University for giving many helpful suggestions for the design of the research.
CRediT authorship contribution statement
He-Qi Zhou:Data curation, Formal analysis, Writing - original draft, Writing - review & editing.Jian-Hao Li:Investigation, Resources.Li-Wen Liu:Methodology.Jia-Min Lou:Formal analysis,Software, Visualization.Zhi-Gang Ren:Conceptualization, Funding acquisition, Writing - review & editing.
Funding
This study was supported by grants from the National S&T Major Project of China (2018ZX10301201-008) and Henan Province Science and Technology Project (2020-387).
Ethical approval
This study was approved by the Institutional Review Board of the First Affiliated Hospital of Zhengzhou University, Zhengzhou,China. All participants signed written informed consents upon enrollment.
Competing interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Non-operative management of pancreatic trauma in adults
- Serum non-high-density lipoprotein cholesterol level is increased in Chinese patients with nonalcoholic fatty liver disease
- Folfirinox chemotherapy prolongs stent patency in patients with malignant biliary obstruction due to unresectable pancreatic cancer
- Role of phosphorylated Smad3 signal components in intraductal papillary mucinous neoplasm of pancreas ✩
- Diagnostic accuracy of administrative database for bile duct cancer by ICD-10 code in a tertiary institute in Korea
- Long noncoding RNA HAND2-AS1 re duce d the viability of hepatocellular carcinoma via targeting microRNA-300/SOCS5 axis
