Genetic Variation Dissection of Rice Blast Resistance Using an Indica Population
2020-07-06ZhangMengchenWeiZhonghuaYuanXiaopingWangCaihongWangShanNiuXiaojunXuXinXuQunFengYueYuHanyongWangYipingZhuZhiweiZhaiRongrongYangYaolongWeiXinghua
Zhang Mengchen, Wei Zhonghua, Yuan Xiaoping, Wang Caihong, Wang Shan, Niu Xiaojun, Xu Xin, Xu Qun, Feng Yue, Yu Hanyong, Wang Yiping, Zhu Zhiwei, Zhai Rongrong, Yang Yaolong, Wei Xinghua
Letter
Genetic Variation Dissection of Rice Blast Resistance Using anPopulation
Zhang Mengchen1, Wei Zhonghua2, Yuan Xiaoping1, Wang Caihong1, Wang Shan1, Niu Xiaojun1, Xu Xin1, Xu Qun1, Feng Yue1, Yu Hanyong1, Wang Yiping1, Zhu Zhiwei1, Zhai Rongrong3, Yang Yaolong1, Wei Xinghua1
(State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 310006, China; Suihua Branch of Heilongjiang Academy of Agricultural Sciences, Suihua 152000, China; Institute of Crop and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China)
Rice blast disease, caused by fungus pathogen, is one of the most destructive diseases that impact rice farming. In the worldwide, rice harvests lose 10% to 30% of the total production because of blast infection, which is estimated to be enough for feeding 60 million people (Skamnioti and Gurr, 2009). The most cost-effective approach to prevent rice blast disease is by employing host resistance in rice cultivars. To date, more than 84 resistance loci have been identified by various mapping approaches (http://www.ricedata. cn/gene/). Among these, 35genes have been isolated, mostly by positional cloning strategy. Except(Chen et al, 2006),(Fukuoka et al, 2009),(Li et al, 2017) and(Zhao et al, 2018),genes share conserved gene structure and encode nucleotide-binding site (NBS) and leucine-rich repeat (LRR) proteins (Liu et al, 2007). The relatively rapid molecular evolution ofgenes results in abundant alleles and contributes to the adaptability to polymorphic pathogen effectors in nature (Jones and Dangl, 2006). For instance, at least five allelic genes have been identified from blast resistance locus, known as,,,and(Ashikawa et al, 2008; Yuan et al, 2011; Zhai et al, 2011, 2014; Hua et al, 2012). With respectto rice blast disease research, recently several association studies were implemented and various candidate locus were obtained (Wang et al, 2014; Kang et al, 2016). Nevertheless, research based on large-scale association population is still lack. In this study, 1005varieties with 4202 high quality single nucleotide polymorphisms (SNPs) (Lu et al, 2015; Zhang et al, 2017) were inoculated with nineisolates. We, then, conducted a comprehensive dissection on genetic variation of rice blast resistance. The results are expected to deepen our knowledge on rice blast resistance.
To evaluate the population genetic components and reduce its potential influence on genome-wide association study (GWAS), we firstly performed genetic structure analysis. We used ADMIXTURE (Alexander et al, 2009) to estimate ancestry relationship. Results indicated a clear clustering of genetic components when usingas 3. Based this, we then divided the population into three groups (Fig. 1-A). Next, we calculated the pairwise genetic distance and constructed a neighbor-joining tree based on’s 1973 genetic distance (Fig. 1-B). The phylogenetic relationship showed a reasonable identical result compared with groups derived from ADMIXTURE. Then, we carried out principle component analysis (PCA) to further verify our grouping results. The first two principle components that explained the largest genetic variation classified the population into visualized three clusters and a mix group (Fig. 1-C). These results indicated that a weak but clear genetic structure existed in this population.
We evaluated the resistance of each variety against nineisolates (Supplemental Table 1). The descriptive statistical analysis showed diverse distributions of phenotype values (Supplemental Fig. 1). For strains S102, S159 and S182, the frequency distribution showed clear negative skewness. On the contrary, positive skewness distribution was detected with S359 and S362. Based on the resistance degrees to the nine isolates, we calculated the rate of resistant spectrum (RRS) for each variety. The data showed a positive skewed distribution while most varieties possessed RRS less than 40% (Supplemental Fig. 1). According to the standard described in Materials and Method section by INGER (1996) (Supplemental File 1), we identified 61 varieties resistant to at least 8 isolates and 76 varieties resistant to none of the 9 isolates. These 61 and 76 varieties were used to construct the broad spectrum resistant pool and the susceptible pool, respectively (Supplemental Table 2). To understand the relationship between blast resistance and genetic differentiation of the population, we compared the average RRS values of the classified fourgroups. Group 2 showed an average RRS of 62.60% that was significantly higher than the other three groups (Fig. 1-D). This result implied potential correlation existed between rice blast resistance and genetic structure. We then performed statistical analysis using the broad spectrum resistant pool. Interestingly, 80.32% varieties in resistant pool belonged to Group 2 (Fig. 1-E). These results demonstrated the resistance distribution among rice subpopulation was not uniform, and a higher frequency of resistance was found in Group 2.
We carried out trait-marker association using the first three principle components as the covariant under the mixed linear model (MLM). With a threshold of< 0.001, we detected a total of 137 significant SNPs distributed in 10 chromosomes except chromosomes 4 and 5 (Supplemental Table 3 and Supplemental Fig. 2). The highest rank of SNP was seq-rs4195, with a-value of 6.997 × 10-12associated with isolate S359. Among these significant SNPs, 82 were detected on chromosome 12, mostly distributed from 9.9 Mb to 13.1 Mb. Previous research identified a major blast resistance genein this region (Bryan et al, 2000). To evaluate the potential relationship between these signals and, we performed linkage calculation with all significant SNPs detected on chromosome 12. The result indicated a genome region from seq-rs5638 (Position: 9905544) to seq-rs5706 (Position: 13696562) containing SNPs in strong linkage disequilibrium (Supplemental Fig. 3). Since this region located close to the centromere of chromosome 12, we concluded that the local linkage disequilibrium scale was enlarged to at least three megabases and caused a lot of significant signals. Using the other associated SNPs, we then identified 25 genetic loci, namedto, respectively. Among these loci, seven were mapped to known genes,(Hayashi and Yoshida, 2009),(Wang et al, 1999),(Li et al, 2009),(Lee et al, 2009; Deng et al, 2019),(Okuyama et al, 2011),(Zhai et al, 2011) and(Bryan et al, 2000) (Supplemental Table 3). Moreover, 7 of the 25 genomic loci were mapped to previously reported QTLs (Supplemental Table 3). These co- localized genetic loci suggested the accuracy of our association model. The rest 11 loci were defined as newly detected resistance loci as no information has been reported before.
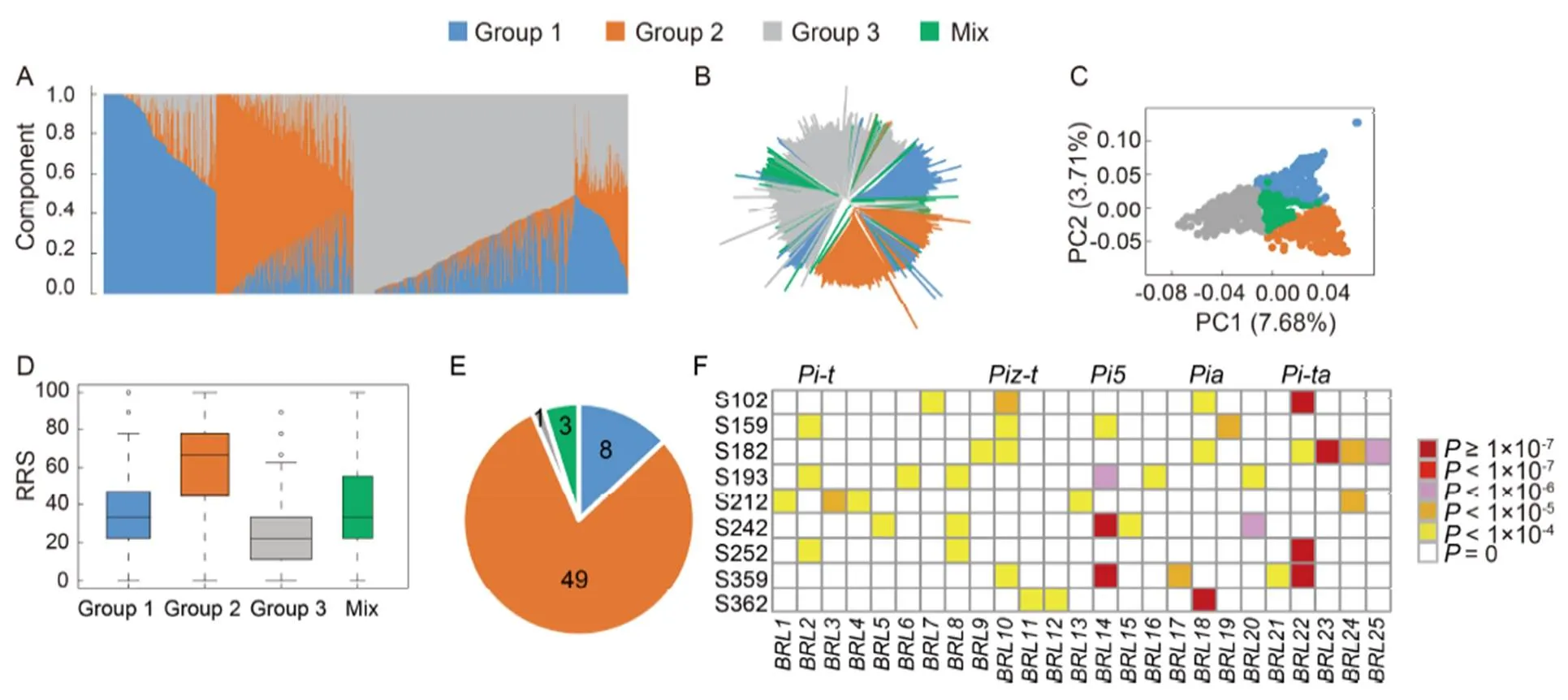
Fig. 1. Genetic structure and resistance level ofpopulation.
A, Model-based population assignment using ADMIXTURE. B, Neighbor-joining tree based on’s 1973 genetic distance. C, Principle component (PC) analysis showed by the first two genetic components. D, Comparison on average rate of resistance spectrum (RRS) of three majorgroups and a mix group. E, Group distribution of the resistant pool (61 varieties). F, Heat map shows the multiple resistance of 25 associated genomic loci.
To evaluate the potential broad resistance of the associated genomic loci, we constructed a heatmap with the GWAS results (Fig. 1-F). As a consequence, 6 of the 25 genomic loci confer resistance to more than 3isolates. Among these six loci, five were cloned genes (,,,and). A novel locuswas associated with resistance to blast strains S193, S242 and S252. A previously detected resistance QTL,(t), was found to cover, demonstrating its real value. To better understand the effect of alleles of the associated genetic loci, we calculated the average phenotype value for all the 25 loci (if there were more than one strain associated with one locus, all of them were analyzed). A set of boxplots comparing average resistance level of each two alleles were displayed in Supplemental Fig. 4. Most loci showed substantial resistance differences except two (with no color filling in the box). These results implied correlation between the resistance level and SNP alleles while superior alleles can be used as genetic markers for resistance selection in rice breeding project.
In summary, we identified 61 highly resistant varieties by screening blast resistance from 1005varieties and found Group 2 was correlated significantly with rice blast resistance, while the average RRS of Group 2 was remarkably higher than those of the other three groups. These varieties must contain diverse resistance genes and can be used as donator in breeding improvement. Through GWAS approach, we detected 25 genetic loci associated with resistance against 9isolates. The co-localized cloned resistance genes indicated the accuracy of our strategy. Genetic loci containingandwere detected with extremely low-values, suggesting major effect of these genes on blast resistance (Supplemental Table 3 and Supplemental Fig. 2). We also screened the resistant spectrum of the associated loci and found,,,,andcontribute to resistance against at least three isolates which suggested their great values in breeding system (Fig. 1-F).
ACKNOWLEDGEMENTs
This work was supported by the Ministry of Science and Technology of China (Grant No. 2017YFD0102002) and National Natural Science Foundation of China (Grant Nos. 31600999 and 31601282).
Supplemental DatA
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/journal/ 16726308; http://www.ricescience.org.
Supplemental File 1. Materials and methods used in this study.
Supplemental Table 1. Virulence profile of nineisolates.
Supplemental Table 2. Information of 1005varieties.
Supplemental Table 3. Blast resistance loci identified by association mapping using 1005varieties.
Supplemental Fig. 1. Phenotypic value distribution of resistance against nineisolates.
Supplemental Fig. 2. Manhattan plots and Quantile-Quantile plots of genome-wild association study.
Supplemental Fig. 3. Linkage analysis of the associated single nucleotide polymorphisms on chromosome 12.
Supplemental Fig. 4. Statistical analysis on phenotypic effects of 25 associated genomic loci.
Alexander D H, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals., 19(9): 1655–1664.
Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M. 2008. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer-specific rice blast resistance., 180(4): 2267–2276.
Bryan G T, Wu K S, Farrall L, Jia Y L, Hershey H P, Mcadams S A, Faulk K N, Donaldson G K, Tarchini R, Valent B. 2000. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene., 12(11): 2033–2046.
Chen X W, Shang J J, Chen D X, Lei C L, Zou Y, Zhai W X, Liu G Z, Xu J C, Ling Z Z, Cao G, Ma B T, Wang Y P, Zhao X F, Li S G, Zhu L H. 2006. A B-lectin receptor kinase gene conferring rice blast resistance., 46(5): 794–804.
Deng Y F, Liu M H, Wang D, Zuo S M, Kang H X, Wang G L. 2019. Origin, distribution and sequence diversity of rice blast resistance locus LABR_64 in rice., 33(1): 20–27. (in Chinese with English abstract)
Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M. 2009. Loss of function of a proline-containing protein confers durable disease resistance in rice., 325: 998–1001.
Hayashi K, Yoshida H. 2009. Refunctionalization of the ancient rice blast disease resistance geneby the recruitment of a retrotransposon as a promoter., 57(3): 413–425.
Hua L X, Wu J Z, Chen C X, Wu W H, He X Y, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, Pan Q H. 2012. The isolation of, an allele at thelocus which confers broad spectrum resistance to rice blast., 125(5): 1047–1055.
INGER. 1996. Standard evaluation system for rice. 4th edn. Manila, the Philippines: International Rice Research Institute (IRRI).
Jones J D, Dangl J L. 2006. The plant immune system., 444: 323–329.
Kang H X, Wang Y, Peng S, Zhang Y L, Xiao Y, Wang D, Qu S H, Li Z Q, Yan S, Wang Z L, Liu W, Ning Y, Korniliev P, Leung H, Mezey J, McCouch S R, Wang G L. 2016. Dissection of the genetic architecture of rice resistance to the blast fungus., 17(6): 959–972.
Lee S K, Song M Y, Seo Y S, Kim H K, Ko S, Cao P J, Suh J P, Yi G, Roh J H, Lee S, An G, Hahn T R, Wang G L, Ronald P, Jeon J S. 2009. Rice-mediated resistance torequires the presence of two coiled-coil-nucleotide-binding- leucine-rich repeat genes., 181(4): 1627–1638.
Li W, Wang B H, Wu J, Lu G D, Hu Y J, Zhang X, Zhang Z G, Zhao Q, Feng Q, Zhang H Y, Wang Z Y, Wang G L, Han B, Wang Z H, Zhou B. 2009. Theavirulence geneencodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene., 22(4): 411–420.
Li W T, Zhu Z W, Chern M, Yin J J, Yang C, Ran L, Cheng M P, He M, Wang K, Wang J, Zhou X G, Zhu X B, Chen Z X, Wang J C, Zhao W, Ma B T, Qin P, Chen W L, Wang Y P, Liu J L, Wang W M, Wu X J, Li P, Wang J R, Zhu L H, Li S G, Chen X W. 2017. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance., 170(1): 114–126.
Liu J L, Liu X L, Dai L Y, Wang G L. 2007. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants., 34(9): 765–776.
Lu Q, Zhang M C, Niu X J, Wang S, Xu Q, Feng Y, Wang C H, Deng H Z, Yuan X P, Yu H Y, Wang Y P, Wei X H. 2015. Genetic variation and association mapping for 12 agronomic traits inrice., 16: 1067.
Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Galam D C, Undan J, Ito A, Sone T, Terauchi R. 2011. A multifaceted genomics approach allows the isolation of the riceblast resistance gene consisting of two adjacent NBS-LRR protein genes., 66(3): 467–479.
Skamnioti P, Gurr S J. 2009. Against the grain: Safeguarding rice from rice blast disease., 27(3): 141–150.
Wang C H, Yang Y Y, Yuan X P, Xu Q, Feng Y, Yu H Y, Wang Y P, Wei X H. 2014. Genome-wide association study of blast resistance inrice., 14(1): 311.
Wang Z X, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. 1999. Thegene for rice blast resistance belongs to the nucleotide binding and leucine- rich repeat class of plant disease resistance genes., 19(1): 55–64.
Yuan B, Zhai C, Wang W J, Zeng X S, Xu X K, Hu H Q, Lin F, Wang L, Pan Q H. 2011. Theresistance toin rice is mediated by a pair of closely linked CC-NBS-LRR genes., 122(5): 1017–1028.
Zhai C, Lin F, Dong Z Q, He X Y, Yuan B, Zeng X S, Wang L, Pan Q H. 2011. The isolation and characterization of, a rice blast resistance gene which emerged after rice domestication., 189(1): 321–334.
Zhai C, Zhang Y, Yao N, Lin F, Liu Z, Dong Z Q, Wang L, Pan Q H. 2014. Function and interaction of the coupled genes responsible forencoded rice blast resistance., 9(6): e98067.
Zhang M C, Lu Q, Wu W, Niu X, Wang C H, Feng Y, Xu Q, Wang S, Yuan X P, Yu H Y, Wang Y P, Wei X H. 2017. Association mapping reveals novel genetic loci contributing to flooding tolerance during germination inrice., 8: 678.
Zhao H, Wang X, Jia Y, Minkenberg B, Wheatley M, Fan J, Jia M H, Famoso A, Edwards J D, Wamishe Y, Valent B, Wang G L, Yang Y. 2018. The rice blast resistance geneencodes an atypical protein required for broad-spectrum disease resistance., 9(1): 2039.
Wei Xinghua (weixinghua@caas.cn);
Yang Yaolong (yangxiao182@126.com)
12 March 2019;
15 May 2019
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2020.05.001
杂志排行
Rice Science的其它文章
- OsPS6 Plays Important Role in Anther Development and Microspore Formation
- Deciphering Rice Lesion Mimic Mutants to Understand Molecular Network Governing Plant Immunity and Growth
- Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis
- RNA-Seq Study Reveals AP2-Domain-Containing Signalling Regulators Involved in Initial Imbibition of Seed Germination in Rice
- RNAi-Mediated Silencing of ITPK Gene Reduces Phytic Acid Content, Alters Transcripts of Phytic Acid Biosynthetic Genes, and Modulates Mineral Distribution in Rice Seeds
- Accumulation of Polyphenolic Compounds and Osmolytes under Dehydration Stress and Their Implication in Redox Regulation in Four Indigenous Aromatic Rice Cultivars
