Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis
2020-07-06YuanShutingXuChunjueYanWeiChangZhenyiDengXingwangChenZhufengWuJianxinTangXiaoyan
Yuan Shuting, Xu Chunjue, Yan Wei, Chang Zhenyi, Deng Xingwang, Chen Zhufeng, Wu Jianxin, Tang Xiaoyan
Research Paper
Alternative Splicing ofDefines C-Terminal Domain Essential for Protein Function in Meiosis
Yuan Shuting1, 2, 3, #, Xu Chunjue2, #, Yan Wei1, 2, Chang Zhenyi1, 2, Deng Xingwang2, 3, Chen Zhufeng2, Wu Jianxin1, Tang Xiaoyan1, 2
(Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Sciences, South China Normal University, Guangzhou 510631, China; Shenzhen Institute of Molecular Crop Design, Shenzhen 518107, China; School of Life Sciences, Southern University of Science and Technology, Shenzhen 518055, China; These authors contributed equally to this work)
Alternative splicing can generate multiple mRNAs that differ in their untranslated regions or coding sequences, and these differences might affect mRNA stability or result in different protein isoforms with diverse functions and/or localizations. In this study, we isolated a sterile mutant in rice with abnormal meiosis of microspore mother cells and megaspore mother cells that carried a point mutation ingene. Cloning ofcDNAs revealed three transcript variants, named as,and, respectively, which were derived from alternative splicing of the last intron. Proteins derived from the three transcripts were mostly identical except the difference in the very C-terminal domain. The three transcripts exhibited similar expression patterns in various tissues, but the expression level ofwas the highest. Specific knockout ofled to sterility, while knockout ofandtogether did not change the plant fertility. Overexpression ofandcDNAs in-specific mutantdid not complement the plant fertility. Yeast two-hybrid assay showed that OsRAD1.1, but not OsRAD1.2 and OsRAD1.3, interacted with the three other meiosis proteins OsHUS1, OsRAD9 and OsRAD17, suggesting that the C-terminal domain of OsRAD1.1 is critical for the protein function.
;; alternative splicing; meiosis; protein interaction; fertility; sterility
Alternative splicing is an important post-transcriptional regulation of genes that greatly increases the diversity of mRNAs and proteins in higher eukaryotes and complex organs (Pan et al, 2008; Kalsotra and Cooper, 2011). Alternative splicing can generate two or more mRNA isoforms from a single gene that differ in their untranslated regions (UTRs) or coding sequences, leading to exon skipping and intron retention (Chen and Manley, 2009). The different mRNA isoforms might have different stabilities or translation efficiencies, and some alternative mRNA isoforms can be translated into different protein isoforms with diverse functions and/or localizations (Black, 2003; Kelemen et al, 2013). Recent studies indicated that alternative splicing is subjected to developmental and physiological regulations (Naftelberg et al, 2015; Szakonyi and Duque, 2018), and a big number of genes have been identified with a function in regulating alternative splicing programs during the plant development and in response to physiological changes (Laloum et al, 2018). However, the functional significance remains unknown for most of the identified alternative splicing events (Chaudhary et al, 2019). This is partly due to the fact that not all alternative splicing events necessarily result in the production of functional proteins (Chaudhary et al, 2019). To make the matter more complicated, although certain alternatively spliced transcripts are non-coding, they may still have a function to modulate other RNAs by competing with them for their regulators (Chaudhary et al, 2019). The complexity often makes it difficult to the resolve functional significance of splicing variants.
Radiation sensitive 1 (RAD1) is an evolutionarily conserved protein that plays a critical role in DNA damage response (Ohashi and Tsurimoto, 2017). RAD1 and two other proteins, RAD9 and HUS1, which are alsoinvolved in DNA damage responseform a heterotrimeric protein complex (RAD9-HUS1-RAD1 or 9-1-1 complex)(Doré et al, 2009; Xu et al, 2009). In response to DNA damage, the 9-1-1 complex is recruited by protein RAD17 to the damaged site on chromatin (Griffith et al, 2002; Zou et al, 2002; Bermudez et al, 2003; Navadgi- Patil and Burgers, 2009). The 9-1-1 complex plays an essential role in sensing the aberrant DNA structures and initiating the checkpoint signaling pathways for DNA damage repair (Zhou and Elledge, 2000; Melo and Toczyski, 2002).
In addition to the role in DNA damage response during genotoxic stress, genetic analyses in yeast (Grushcow et al, 1999; Shinohara et al, 2003), worms (Jaramillo-Lambert et al, 2010), flies (Ubi et al, 2007; Peretz et al, 2009) and mouse (Lyndaker et al, 2013; Vasileva et al, 2013) indicated that the9-1-1 complex also plays an essential role in promoting double-strand DNA break (DSB) repair and homologous recombination during meiosis. Meiosis is proceeded by one round of DNA replication during the S phase followed by two rounds of nuclear division (meiosis I and meiosis II), resulting in four haploid nuclei (Wang and Copenhaver, 2018). Like mitosis, meiosis I and II are divided into four phases: prophase, metaphase, telophase and anaphase. During early prophase I, unique meiotic chromosome interactions occur, including pairing, synapsis and recombination. In most organisms, meiotic recombination is initiated by the programmed formation of DSBs. The processing of DSB producessingle-strand DNA ends, which search either homologous chromosomes or sister chromatids as templates for DNA repair (Wang and Copenhaver, 2018). Genetic studies indicated that mutations of the 9-1-1 components lead to meiosis defects and severely reduce fertility in mouse (Lyndaker et al, 2013; Vasileva et al, 2013). The 9-1-1 complex also plays a critical role for DSB repair in higher plants. It has been shown that mutants of the 9-1-1 components inand rice exhibit defect in meiotic DSBs repair and reduced fertility (Heitzeberg et al, 2004; Che et al, 2014; Hu et al, 2016, 2018).
In this study, we isolated a rice mutant defective in meiosis carrying a point mutation in. We found thatencodes three alternative transcripts with differences only at the 3-end. Overexpression and specific knockout of the three mRNA isoforms showed that, but notand,had a role in regulating rice fertility. Further yeast-two-hybrid assay showed that OsRAD1.1, but not OsRAD1.2 and OsRAD1.3, interacted with OsHUS1, OsRAD9 and OsRAD17, respectively. These results suggested that the small peptide fragment at the very C-terminus is critical for the protein function.
MATERIALS AND METHODS
Plant materials and growth conditions
Themutant, a complete sterile mutant, was isolated from thevariety Huanghuazhan (HHZ) mutant library induced by ethylmethanesulfonate (EMS) (Chen et al, 2014). Themutant was derived from the same mutant library (Chang et al, 2016). All the plant materials were grown in the paddy field in Shenzhen, China under natural conditions with regular management.
Characterization of mutant phenotype
The plant morphology was photographed with a digital camera. To analyze the pollen fertility, mature pollen was stained with 1% I2-KI and photographed with a Nikon AZ100 microscope (Nikon, Japan). To test the female fertility, theandmutants were cross-pollinated with the HHZ pollen at the same time, and themutant served as a control.
Cytological analysis of anther and embryo sac
The spikelets of HHZ and themutant at various developmental stages were fixed and then resin-embedded as previously described (Chang et al, 2016). The resin-embedded samples were sliced using Leica EM-UC6 ultramicrotome. Semi-thin sections (1 μm) were stained with 0.5% toluidine blue and photographed under a bright field using an Axio Imager A1 microscope (Zeiss, Germany).
Mature anthers were fixed in FAA solution (38% formaldehyde 5 mL, acetic acid 5 mL, 50% alcohol 90 mL) for scanning electron microscopy as described by Chang et al (2016). The samples were observed using a scanning electron microscope (S-3400N, Hitachi, Japan) with an acceleration voltage of 10 kV.
To observe meiosis, the anthers at meiotic stage were fixed in Carnoy’s fluid, and then, rehydrated with gradient alcohol (75%, 50% and 30%) and ddH2O as previously described (Wang et al, 2016). Then, the anthers were stained in 1 μg/mL 4,6-diamidino-2- phenylindole (DAPI) working solution in dark for 1 h. The meiotic behavior was observed with the AZ100 microscope (Nikon, Japan) under UV light excitation. The DAPI working solution (1 mL) contained 1 μL of DAPI stock solution (1 mg/mL), 40 μL of 25 mmol/L EDTA, 1 μL of 1.06 g/mL Triton X-100, and 958 μL of 0.1 mol/L phosphate buffer saline (PBS) buffer.
To observe the embryo sac development, the ovaries of wild type (WT) HHZ andmutant at different developmental stages were fixed in 70% FAA solution (70% ethanol : formalin : acetic acid = 18 : 1 : 1), stained and observed with a laser confocal scanning microscope as described by Wang et al (2016).
Molecular cloning of OsRAD1 and high resolution melting (HRM) analysis
Themutant was backcrossed with the WT HHZ to obtain F1generation, and the F1was selfed to produce F2generation. Thirty sterile plants in the F2population were selected for genomic DNA extraction. The DNA was mixed in equal amounts and sequenced using the Illumina Hi-Seq platform. A total of 18.26 Gb genomic sequence data were obtained. The data were analyzed using the simultaneous identification of multiple causal mutations (SIMM) method to identify the mutant gene as described by Yan et al (2017). The linkage relationship between the candidate mutation site and the mutant phenotype in the F2population was validated using HRM analysis (Lochlainn et al, 2011) with a primer set OsRAD1-P1 (Supplemental Table 1).
Cloning of OsRAD1 cDNAs
Total RNA was extracted from anthers at stages 5–6 using the Trizol reagent (Invitrogen, Waltham, USA) and then reverse-transcribed with SuperScript™ III Reverse Transcriptase (Invitrogen, Waltham, USA), according to the manufacture’s instruction. Primer sets OsRAD1-P2 and OsRAD1-P3 (Supplemental Table 1) were used for cDNA cloning.
Plasmid construction and rice transformation
The 10.0 kb HHZ genomic DNA fragment for, including the genic region and 1512 bp upstream and 1928 bp downstream region, was PCR- amplified with primer set OsRAD1-P4 (Supplemental Table 1). The PCR product was digested withI andIII, and cloned into the binary vector pCAMBIA1300 to generate the construct OsRAD1pro:: OsRAD1 for mutant complementation. The cDNAs ofwere PCR-amplified with a primer set OsRAD1-P2 and OsRAD1-P3 (Supplemental Table 1). The PCR products were digested withI andHI, and cloned into the vector Ubi-intron to generate the constructs UBIpro::OsRAD1.1, UBIpro:: OsRAD1.2 and UBIpro::OsRAD1.3. To create mutant alleles ofinvariety Wuyungeng 7 (WYG), the CRISPR/Cas9 genome targeting method described by Ma et al (2015) was used to generate the constructs CR-1, CR-2 and CR-3. The target site sequences are listed in Supplemental Table 1.
Constructs were introduced intoAGL10 strain, and then, transformed into rice calli. The OsRAD1pro::OsRAD1 was introduced into the progeny of OsRAD1 heterozygote plants. UBIpro::OsRAD1.1, UBIpro::OsRAD1.2 and UBIpro:: OsRAD1.3 were introduced into the progeny of cr-2 heterozygous plants. CR-1, CR-2 and CR-3 were introduced into WYG. The positive transgenic lines were determined by PCR, with a primer set OsRAD1-P5 for UBIpro::OsRAD1.1, UBIpro::OsRAD1.2 and UBIpro:: OsRAD1.3 constructs, a primer set SP-LR for CR-1, CR-2 and CR-3, and a primer set OsRAD1-P6 for OsRAD1pro::OsRAD1. To identify the background genotype of OsRAD1pro::OsRAD1 transgenic plants, genomic DNA and a primer set OsRAD1-P7 flanking the OsRAD1 genomic DNA were used for PCR- amplification, and the PCR product was further subjected to HRM analysis with primer set OsRAD1-P1. For transgenic lines of CR-1, CR-2 and CR-3, DNAs were amplified and sequenced using primer sets OsRAD1-P8, OsRAD1-P9 and OsRAD1-P10, respectively. All the primers are listed in Supplemental Table 1. To identify the background genotype of UBIpro:: OsRAD1.1, UBIpro::OsRAD1.2 and UBIpro::OsRAD1.3 transgenic plants, genomic DNA samples were examined by HRM analysis using a primer set OsRAD1-P11 (Supplemental Table 1) for themutation.
Protein alignment and steric structure prediction
The full-length amino acid sequence of OsRAD1.1 was used as a query in BLASTP search of homologs from NCBI. These proteins were aligned using ClustalW with default parameters. The tertiary structures of the proteins were predicted using Phyre2(http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id= index), and then, analyzed by the PyMOL software.
Quantitative real-time PCR (qRT-PCR)
Rice tissues were collected at the flowering stage. The stages of anthers were classified as previously described (Zhang D B et al, 2011). Total RNA was extracted using TransZol reagent (TransGen, Beijing, China) and then reverse-transcribed with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China), according to the manufacturer protocols. Quantitative real-time PCR (qRT-PCR) was performed using Applied Biosystems 7500 Real-Time PCR System and SYBR Premix Ex™ II (TaKaRa, Dalian, China), following the manufacturer’s instructions. Each experiment was repeated three times, each time with three replicates.was used as the normalization reference. The relative expression levels were measured using the comparative CT method. Primer sequences used are listed in Supplemental Table 1.
Yeast two-hybrid assay
The yeast two-hybrid assays were performed using the Matchmaker Gold Yeast Two-Hybrid System (TaKaRa, Dalian, China). The full length coding regions of,,,(the mutant gene of the EMS mutant plant),,andwere cloned into pGBKT7 and pGADT7 to generate BD and AD recombinants. Double dropout selecting medium (SD-Leu-Trp) was used to confirm the successful transformation, and quadruple dropout selection medium (SD-Ade- His-Leu-Trp) was used to verify the interaction.
RESULTS
Isolation and phenotypic characterization of m10690 mutant
Compared with the WT plant,was not significantly abnormal during the vegetative growth. However,panicles showed incomplete heading, resulting in slightly shorter plant height than that of the WT (Fig. 1-A and -B). The mutant anther was thinner and smaller (Fig. 1-D). The WT pollen grains were spherical and stained dark by I2-KI, whereas the mutant pollen grains were irregular in shape and were lightly stained (Fig. 1-E and -F). To determine whether the female fertility was normal, we cross-pollinated the mutant with WT pollen., a male sterility mutant with normal female fertility (Chang et al, 2016), was also cross-pollinated as a control. The seed-setting rate ofwas only 9.5%, while that ofwas 82.1% (Fig. 1-C), indicating that the female fertility of the mutant was also compromised. Whenwas backcrossed with the WT, all of the F1plants were fertile, and the F2population displayed an approximately 3 : 1 segregation of fertile to sterile (186 : 58), indicating that the sterile phenotype was controlled by a single recessive gene.
Anatomic characterization of m10690 mutant anthers
To determine the cause of pollen abortion, we compared the WT andanthers at different developmental stages using semi-thin sections. In rice, anther development is divided into 14 stages based on cytological features (Zhang D B et al, 2011). In the WT anther, pollen mother cells (PMCs) were adhered to the tapetum and began meiosis at stage 7, and the middle layer changed into an obscure band structure (Fig. 2-A). At stage 8a, PMCs finished the first meiotic division and formed the ellipsoidal dyad, and the tapetal cells became vacuolated and shrunken with darkly stained cytoplasm (Fig. 2-C). At stage 8b, the PMCs completed the second meiosis and formed the tetrad of four haploid microspores. The middle layer disappeared, and the tapetum became more condensed and vacuolated (Fig. 2-E). At stage 9, the microspores were released from the tetrad, and a thin layer of exine was deposited on the spherical microspore while the tapetum continued to degrade (Fig. 2-G). Until stage 9, we observed no significant difference between the WT andanthers (Fig. 2-A to -H).
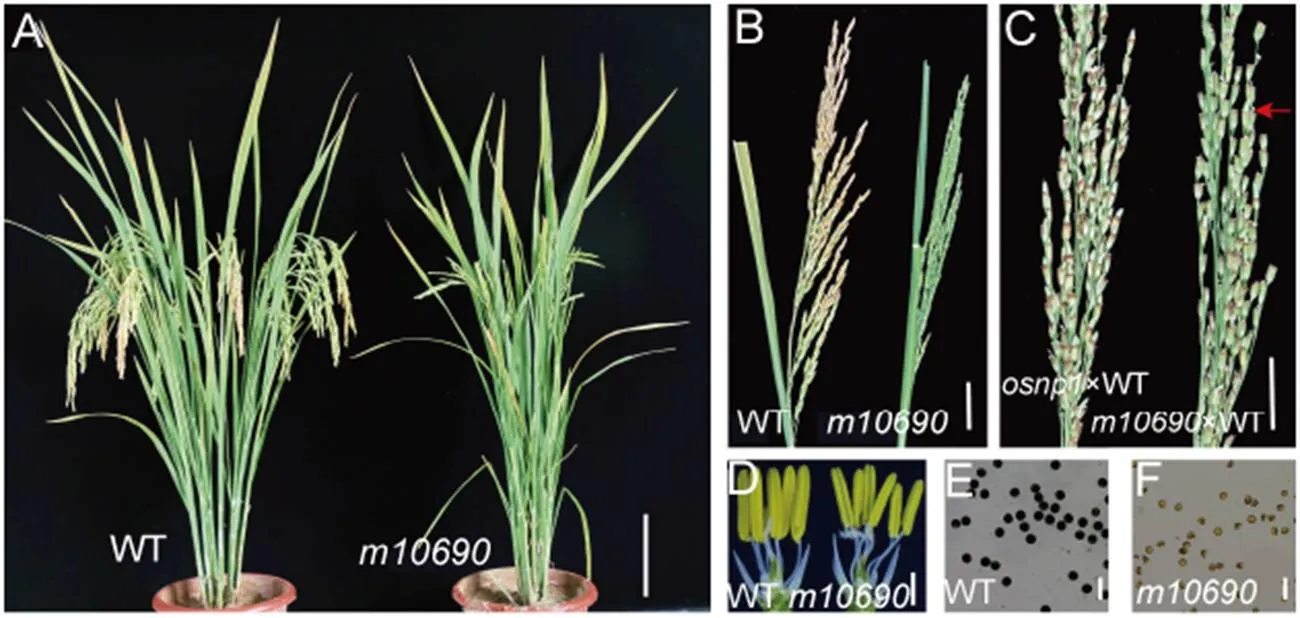
Fig. 1. Phenotypes of Huanghuazhan wild type (WT) andmutant plants.
A, WT andmutant plants after heading. Scale bar, 10 cm. B, Seed-setting in WT and the mutant. Scale bar, 3 cm. C, Seed-setting ofandplants at 20 d after pollinated with the WT pollen. Red arrow indicates the seed-setting in the mutant. Scale bar, 2 cm. D, WT andspikelets with the palea and lemma removed to show the anthers. Scale bar, 1 mm. E, WT pollen grains stained with I2-KI. Scale bar, 100 μm. F,pollen grains stained with I2-KI. Scale bar, 100 μm.
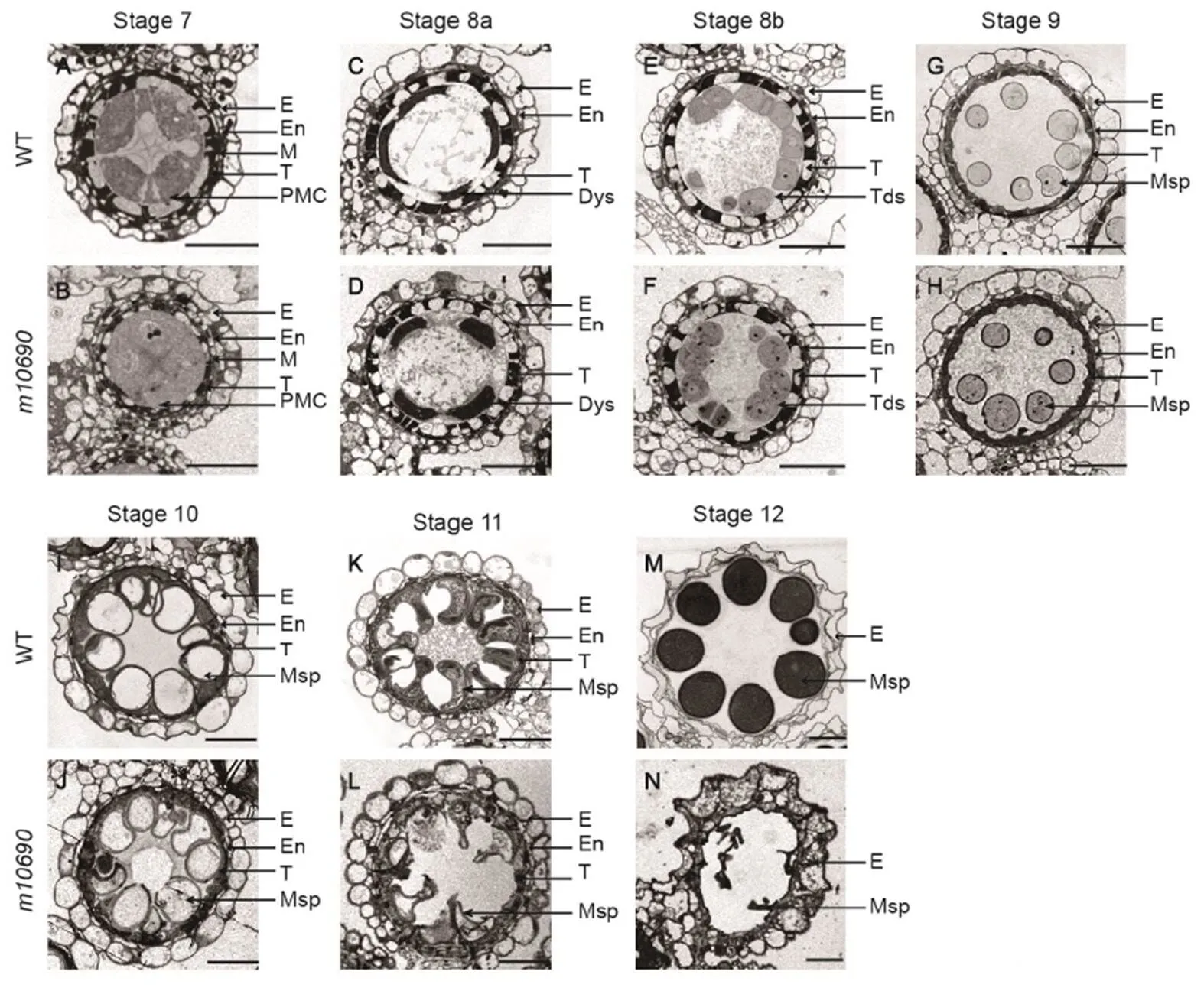
Fig. 2. Transverse sections of anther in Huanghuazhan wild type (WT) andplants.
WT anther sections are shown in A, C, E, G, I, K and M. Themutant anther sections are shown in B, D, F, H, J, L and N.
E, Epidermis; En, Endothecium; M, Middle layer; T, Tapetal layer; PMC, Pollen mother cell; Dys, Dyads; Tds, Tetrads; MP, Mature pollen; Msp, Microspores. Scale bars, 20 μm.
At stage 10, the WT microspores were highly vacuolated and round in shape (Fig. 2-I). The mutant microspores were also large and round in shape, but the center of vacuole appeared to contain small vesicles, implying disruption of the microspore cytoplasm (Fig. 2-J). At stage 11, the WT microspores formed bicellular pollen after the first mitosis and changed into the shape of crescent, while the tapetum cells almost completely degenerated into debris (Fig. 2-K). At this stage, disruption of mutant microspores was more pronounced, andtapetum layer looked more swollen than the WT tapetum (Fig. 2-L). At stage 12, the microspores underwent the second mitosis, and the mature pollen grain contained three nuclei and a large amount of storage substance (Fig. 2-M). But the mutant microspores completely disrupted with only the collapsed pollen exine left (Fig. 2-N).
We further compared the pollen grains, the cuticle on epidermis surface, and the Ubisch bodies on the inner surface of the tapetal cells between the WT and themutant anthers at stage 12 using a scanning electron microscope. In agreement with the transverse sections, the WT pollen was spherical, whereas the mutant pollen was collapsed (Supplemental Fig. 1-A and -B). The Ubisch bodies on the inner surface of the tapetum cell were similar in shape and size, but the density was slightly higher in the WT than in(Supplemental Fig. 1-C and -D). The anther surface cuticle structures were similar betweenand WT (Supplemental Fig. 1-E and -F).
Abnormal meiosis of spore mother cells in m10690 mutant
Meiosis mutants often display male sterility and reduced female fertility (Luo et al, 2014). We therefore examined if the meiosis process was normal during the microspore development in themutant by DAPI staining. In the WT, chromosomes dispersed in microspore mother cells at prophase I(Fig. 3-A) and arranged neatly on the equatorial plate at metaphase I (Fig. 3-B). Then, the chromosomes moved to the cell poles, resulting in separation of the homologous chromosomes and subsequent formation of dyads (Fig. 3-C and -D). The dyads continued the second meiosis and eventually formed tetrads (Fig. 3-E and -F). No significant defect was observed induring prophase I and metaphase I (Fig. 3-G and -H). However, at anaphase I when the chromosomes moved to the cell poles, DNA fragments appeared on the equatorial plate of the mutant (Fig. 3-I). Themutant can form dyads (Fig. 3-J). However, during the second meiosis, DNA fragmentation showed up again at anaphase II (Fig. 3-K), and eventually abnormal tetrads were formed (Fig. 3-L).
We examined themutant embryo sacs at different developmental stages with a laser confocal scanning. In the WT, the megaspore mother cell was clearly visible at the megasporocyte stage (Fig. 4-A), which underwent meiosis to form dyad and tetrad, and the four megaspores were linearly arranged in the tetrad (Fig. 4-B and -C). Then, the three megaspores near the ovule hole degenerated, and the megaspore near the chalaza end developed into the functional megaspore (Fig. 4-D). The functional megaspore elongated, and the vacuole expanded to form a mono-nucleate embryo sac (Fig. 4-E). The mono- nucleate embryo sac underwent three mitotic divisions to form two-nucleate embryo sac (Fig. 4-F), four- nucleate embryo sac (Fig. 4-G), and eight-nucleate embryo sac successively (Fig. 4-H and -I), which eventually formed an embryo sac structure of eight nuclei and seven cells (Fig. 4-J). Inmutant, the megaspore mother cells looked normal (Fig. 4-K) and formed normal dyad (Fig. 4-L). However, the cells in the tetrads were deformed (Fig. 4-M) and unable to go through normal mitosis (Fig. 4-N). Eventually, the mature embryo sacs were shriveled without a typical egg apparatus structure (Fig. 4-O).
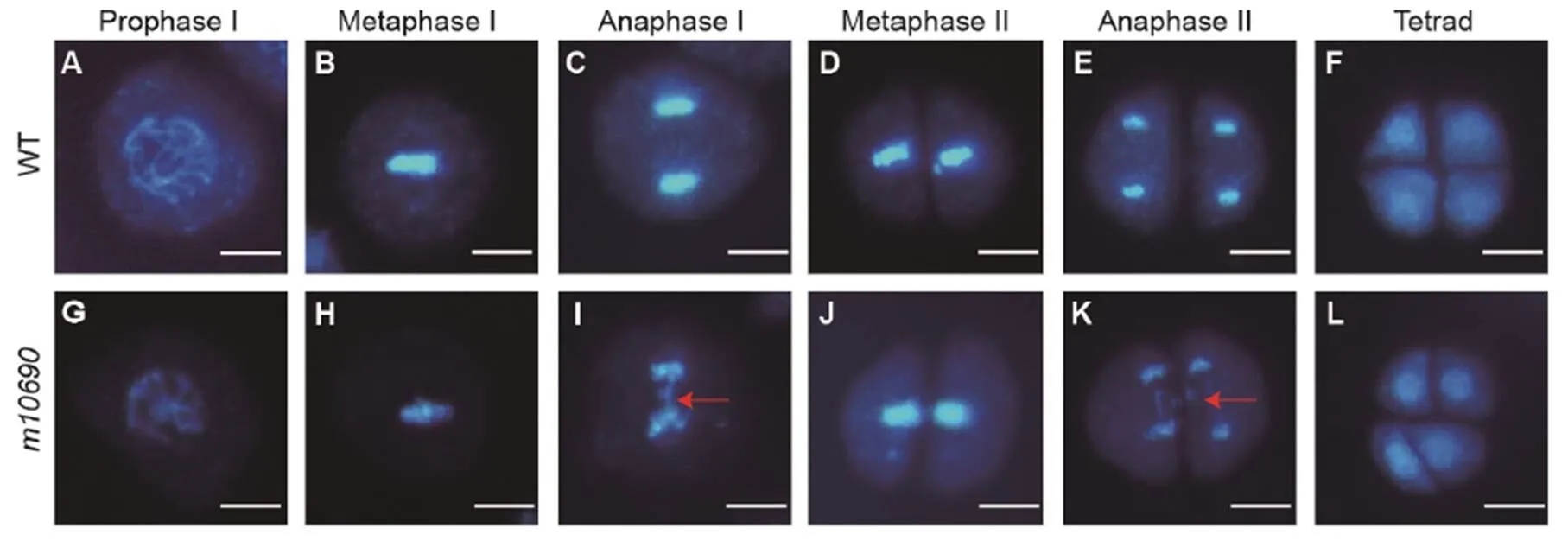
Fig. 3. Chromosome behavior during meiosis in pollen mother cells of Huanghuazhan wild type (WT) and.
A‒F, Different meiosis stages for the WT. G‒L, Different meiosis stages for themutant. Red arrows indicate broken chromosome fragments. Scale bars, 10 μm.
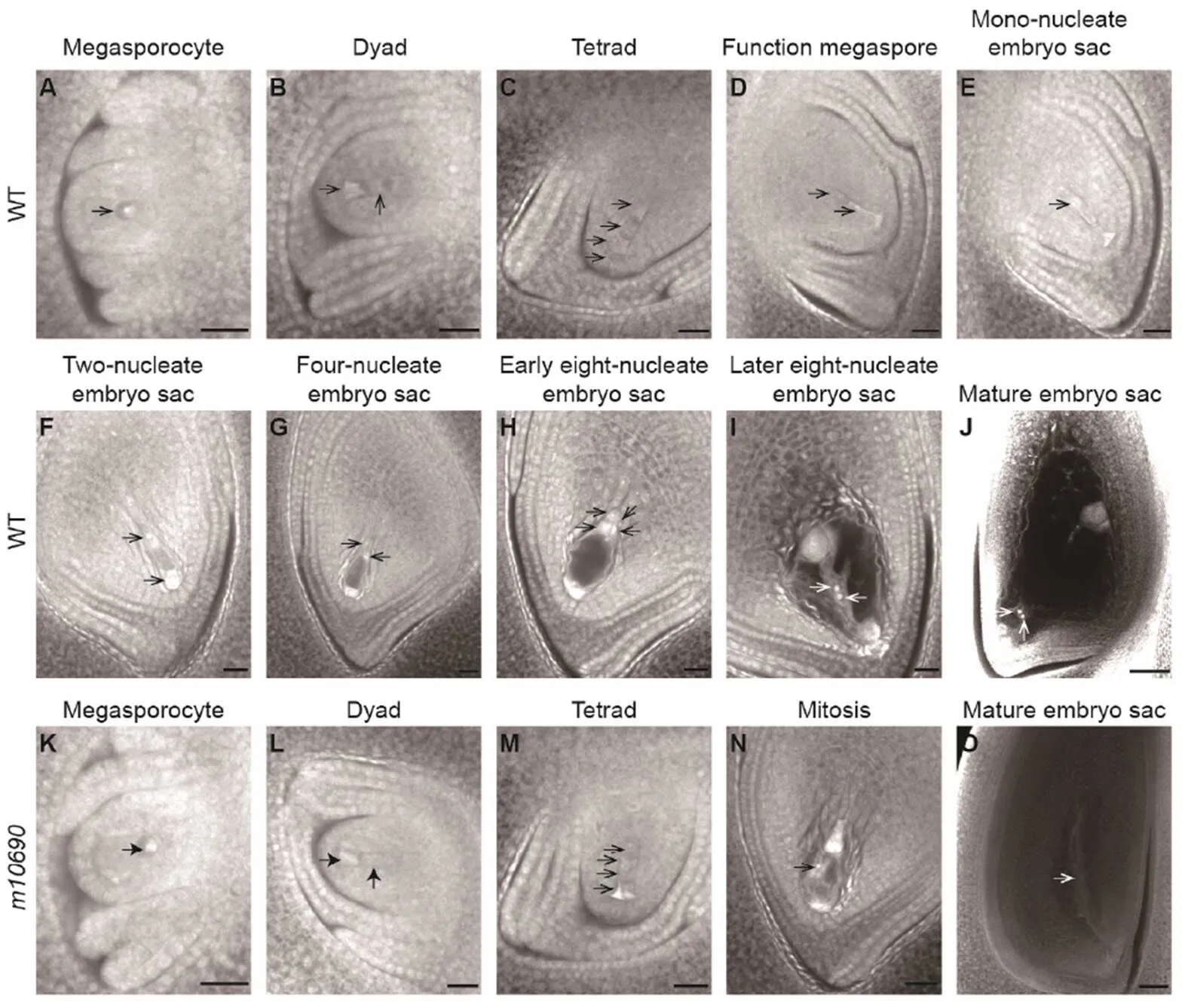
Fig. 4. Development of embryo sac in Huanghuazhan wild type (WT) andplants.
A‒J, The WT embryo sacs. K‒O, Themutant embryo sacs. Arrows indicate the megaspores at various developmental stages. Scale bars, 20 μm.
We further compared the expression of a few key genes acting in meiosis by qRT-PCR in the WT andmutant anthers (Supplemental Fig. 2). The expression levels of(Nonomura et al, 2007), which is involved in meiosis initiation, and(Wang et al, 2010), which encodes a synaptic complex central element protein, were decreased in the mutant. The expression levels of(Nonomura et al, 2004) and(Yuan et al, 2009), which participate in homologous pairing, were increased in the mutant. However, the expression of(Nonomura et al, 2006), another gene acting in homologous pairing, had no difference between the WT andmutant (Supplemental Fig. 2). These results indicated that the mutation caused abnormal meiosis of microspore and megaspore mother cells, leading to male sterility and compromised female fertility.
Cloning of the mutant gene and characterization of its transcripts
To identify the mutant gene, total DNAs from 30 backcrossed F2individuals of the sterile phenotype were bulk-sequenced. The sequence data were processed using the SIMM pipeline (Yan et al, 2017). The analysis identified a candidate region on chromosome 6 with a single base mutation (TAT to TAG) in(Fig. 5-A and -B).
HRM assay was performed to verify the linkage between the candidate mutation and the sterile phenotype in the backcrossed F2population. All the sterile plants (47) showed the homozygous mutant genotype, while the fertile plants displayed approximately 2 : 1 ratio of heterozygous to homozygous wild-type genotypes (119 : 69), indicating that the mutation inwas co-segregated with the sterile phenotype.
To confirm the mutation, a 10.0 kb genomic fragment for(Fig. 5-B), including the genic region and 1512 bp upstream and 1928 bp downstream regions, was transformed into themutant plant. The pollen fertility and seed-setting phenotype of the transgenic plants were restored to normal (Supplemental Fig. 3). These results demonstrated that the sterile phenotype was caused by the single nucleotide mutation in. The protein encoded byis highly homologous to RAD1 proteins in other organisms (Supplemental Fig. 3), and thus, the gene was named as.
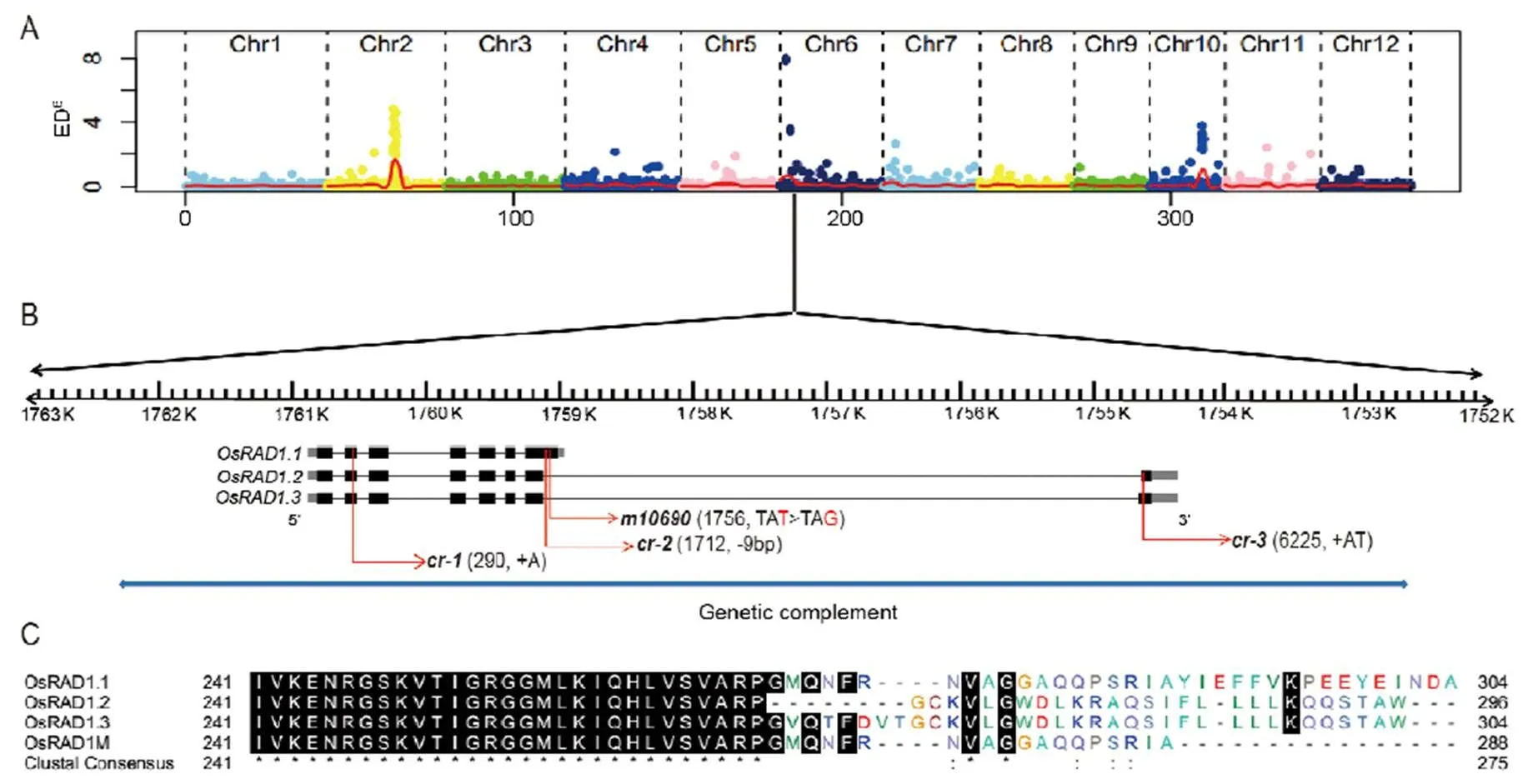
Fig. 5. Simultaneous identification of multiple causal mutations (SIMM) analysis ofmutant and gene structure of.
A, Mapping of thegene based on the distribution of Euclidian Distance (ED6) values of SNPs along the rice chromosomes. B,gene structure. The black boxes indicate exons, the gray boxes indicate 5′-UTR and 3′-UTR, the black lines indicate introns and the red arrows indicate the mutation sites in the originalmutant and three other mutants generated by CRISPR knockout. The blue line represents the DNA fragment for transgenic complementation. C, Alignment of the C-terminal sequences of OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1M.
Cloning of cDNAs encoded byindicated that the WT anthers at the meiosis stage contained three different transcripts, named as,and, respectively. The splicing patterns of the three transcripts were shown in Fig. 5-B. The three transcripts differed only at the 3-ends (Fig. 5-B). Transcripthad seven exons and encoded a protein of 304 amino acids. Transcriptsandboth had eight exons and encoded proteins of 296 amino acids and 304 amino acids, respectively. The difference of the three proteins was limited to the very C-terminal portion (Fig. 5-C). The mutation was on the 7th exon of(Fig. 5-B), causing premature termination of the protein without the C-terminal 16 amino acids (OsRAD1M, Fig. 5-C). This mutation was in the last intron ofand(Fig. 5-B) and presumably did not alter the corresponding transcripts and encoded proteins.
Expression of OsRAD1 alternative transcripts
Besides defects in male and female fertility,mutant showed no clear defects in other tissues. To determine iffunction was limited to the male and female reproductive tissues, we analyzed the expression patterns oftranscripts in different tissues, including root, stem, leaf, glume, palea, lemma and pistil at flowering and anthers at the developmental stages 6 to 12 by qRT-PCR. We first used primers derived from exon 5 and exon 6 (Supplemental Fig. 4), which were shared by all the three transcripts, for qRT-PCR. The results showed thatwas expressed at lower levels in root, stem and leaf tissues than in glume, palea, lemma and pistil at the flowering stage (Fig. 6-A). The expression levels ofalso varied in anthers at different developmental stages (Fig. 6-A).
Next, we investigated if the three alternative transcripts have different expression patterns by qRT- PCR using primers specific to individual transcript (Supplemental Fig. 4). Thetranscript level was 8–10 fold higher than the combinedandtranscripts in all the floral organs, including glume, palea, lemma, anther and pistil (Fig. 6-B and -C). In root and stem, however, thetranscript level was close to the level of combinedandtranscripts (Fig. 6-B and -C). These results suggested thatplayed the major role in floral organs.
Because the mutant showed defects mainly in male and female fertility, we thus characterized the expression of the three transcripts in more details in anthers and pistils at different developmental stages (Fig. 6-D to -F). Despite some differences in the expression patterns,was still the most abundant transcript in anthers and pistils at all the developmental stages. The level oftranscript was slightly higher than(Fig. 6-E and -F).
Functional characterization of three alternative transcripts
Based on the location of the mutation site in, we presumed that the mutation only affected the function of transcriptbut notandin the mutant plant. To determine ifandplay a role in regulating the plant fertility, we constructed three CRISPR knockout vectors. CR-1 targeted the second exon to disrupt all the three transcripts. CR-2targeted the unique region ofin the 7th exon to disrupt. CR-3targeted the common region in the 8th exon ofandto disruptandtogetherWe obtained three homozygous mutants in WYG background.mutant had an ‘A’ insertion in the second exon that disrupted the open reading frames of all the three transcripts (Fig. 5-B). This mutant was sterile with aborted pollen (Fig. 7-B), just like the original mutant of HHZ.mutant specifically deleted 9 bp in the 7th exon of(Fig. 5-B), and this mutant was sterile without functional pollen grains (Fig. 7-C).mutant had 2 bp insertion in the 8th exon ofand(Fig. 5-B), and this mutant showed normal seed-setting and pollen (Fig. 7-D). These results suggested thatplayed major role in executing the meiosis function, andandwere dispensable.
There were two possible explanations for the lack of detectable function ofandin plant fertility. One possibility was that the proteins encoded by the two transcripts were nonfunctional. The other was that OsRAD1.2 and OsRAD1.3 were functional proteins but their expression levels were too low to affect the phenotype. To differentiate these two possibilities, we constructed two overexpression vectors, Ubipro::OsRAD1.2 and Ubipro::OsRAD1.3, and introduced them into, the-specific mutant plant. Neithernorrestored the fertility ofmutant (Fig. 7-E and -F), even though the transcripts were expressed at high levels (Fig. 7-H), indicating thatandwere not functionally equivalent to. Overexpression ofandin the wild type plants did not change the seed-setting and pollen fertility either (Fig. 7-G and -H), suggesting that thetranscripts ofandwere probably nonfunctional.
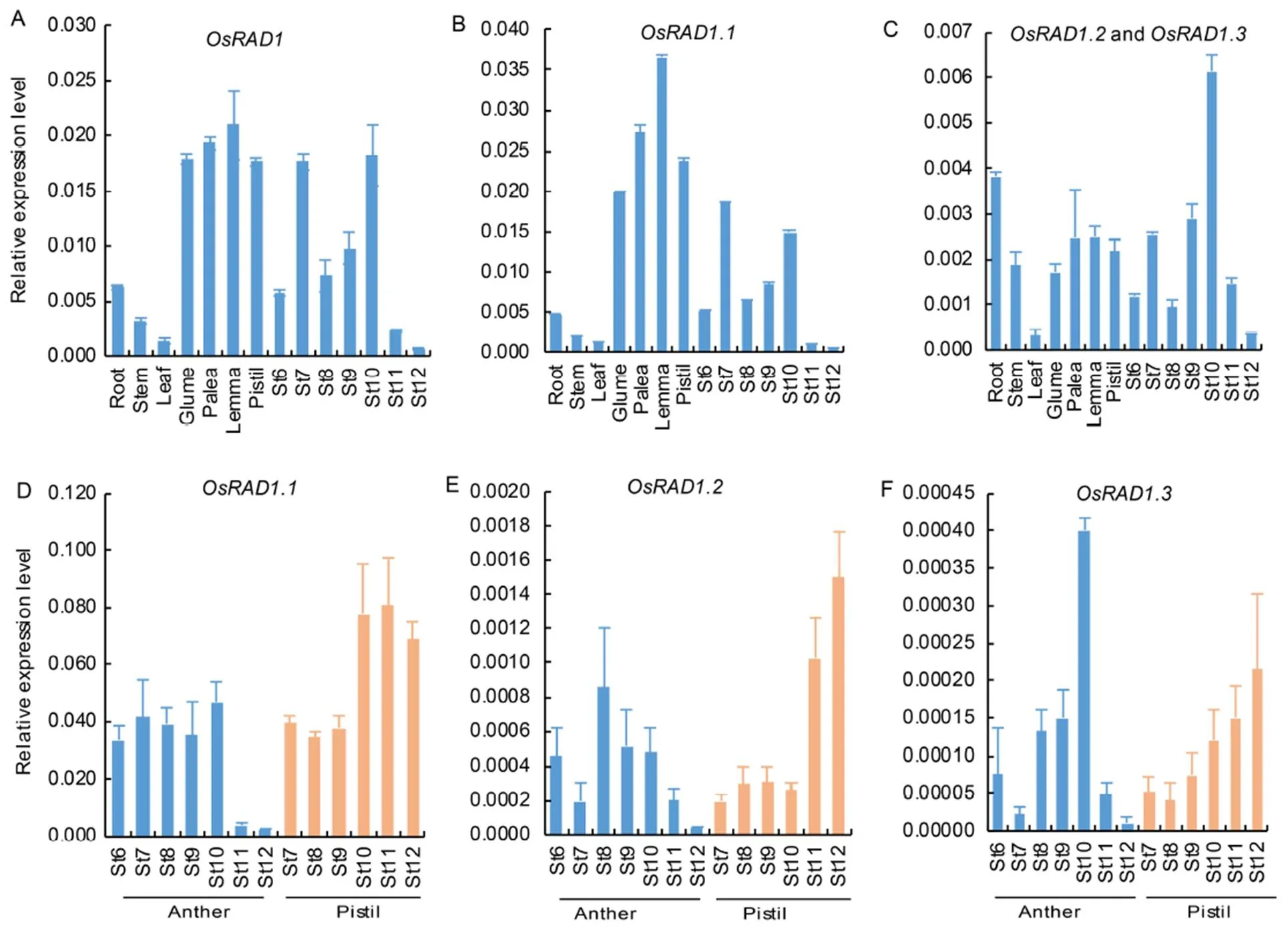
Fig. 6. Analysis of expression patterns of.
A, Expression levels ofat different tissues and anther developmental stages. B, Expression levels ofat different tissues and anther developmental stages. C, Expression levels ofplusat different tissues and anther developmental stages. D, Expression levels oftranscripts in anthers and pistils at different developmental stages. E, Expression levels oftranscripts in anthers and pistils at different developmental stages. F, Expression levels oftranscripts in anthers and pistils at different developmental stages.
St6 to St12 are different developmental stages of anthers. Pistils and other tissues in A‒C were harvested from plants at the flowering stage. Anthers and pistils in D‒F were collected at St6 to St12.was used as the internal control. Data are Mean ± SD (= 3).

Fig. 7. Phenotypes of CRISPR mutant alleles and transgenic plantsoverexpressingplus.
A, Wuyungeng 7. B,. C,. D,. E,overexpressing. F,overexpressing. G, Wuyungeng 7 over- expressing. H, Wuyungeng 7 overexpressing. The seed-setting and pollen grains stained with I2-KI are shown. White scale bars, 5 cm. Red scale bars, 100 μm. I, Relative expression levels ofandin nontransgenic (N) and transgenic (P) plants of Wuyungeng 7 (WT) andbackgrounds.was used as the internal control. Data are shown as Mean ± SD (= 3).
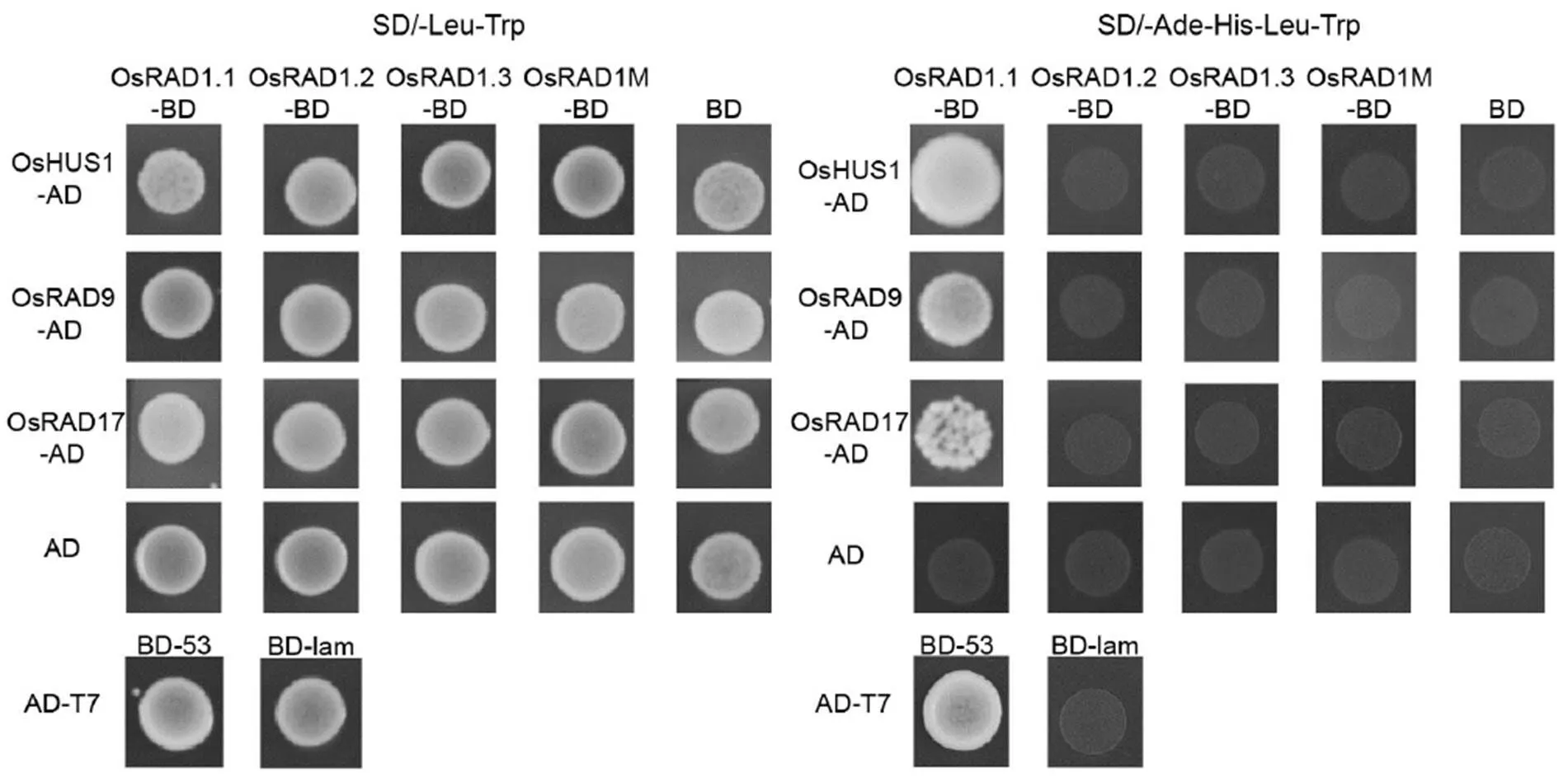
Fig. 8. Yeast two-hybrid assay of OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1M interaction with RAD9, HUS1 and RAD17.
SD/-Leu-Trp was used to test the co-transformation efficiency. The interactions were verified by the growth of yeast on selective medium SD/-Ade-His-Leu-Trp. The interaction of AD-T7 and BD-53 was included as the positive control, the interaction of AD-T7 and BD-lam was included as the negative control, and the interaction of BD and AD empty vectors was also included as the negative control.
OsRAD1.1, but not OsRAD1.2 and OsRAD1.3, is capable of interaction with partner proteins
Alignment of the proteins derived from the three alternative transcripts ofwith their homologous proteins indicated that the C-terminal sequence of OsRAD1.1was highly conserved with RAD1 proteins in other organisms (Supplemental Fig. 5). As RAD1 was known to interact with RAD9, HUS1 and RAD17 in other organisms, we tested if the C-terminal difference can impact the interaction of OsRAD1 isoforms with other proteins. We cloned the cDNAs of,andfrom rice and conducted the yeast two-hybrid assay to test their interactions with OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1.1 mutant lacking 16 amino acid residues at the C-terminal (OsRAD1M), respectively. As shown in Fig. 8, OsRAD1.1, but not OsRAD1.2, OsRAD1.3 and OsRAD1M, interacted with OsHUS1, OsRAD9 and OsRAD17.
DISCUSSION
RAD1 is an evolutionarily conserved protein, and can form the 9-1-1 complex with RAD9 and HUS1. The 9-1-1 complex is involved in DNA damage response, and also plays important roles in DSB repair and homologous recombination during meiosis (Doré et al, 2009; Xu et al, 2009). In, RAD1 is homologous with RAD17, and the loss function of RAD17 leads to meiotic defects, including aberrant synapsis, elevated ectopic recombination level and delayed DSB repair (Grushcow et al, 1999; Shinohara et al, 2003). In human, RAD1 is required for the DNA damage checkpoint mechanisms of both mitosis and meiosis (Freire et al, 1998; Udell et al, 1998). Homozygous deletion of RAD1 in mouse results in embryo-lethal (Han et al, 2010). The mouse embryonic stem cells, with a target deletion of RAD1, are sensitive to ultraviolet light, gamma rays and hydroxyurea. The cells also have defects at G2/M and S/M checkpoints. Moreover, RAD1 plays an important role in homologous recombinant repair of embryonic stem cells (Zhang C B et al, 2011). In, the mutant of 9-1-1 RAD9 is sensitive to DNA-damaging agents and shows defects in general repair of DSBs (Heitzeberg et al, 2004). Previous reports revealed that the9-1-1 complex functions in suppressing nonhomologous end-joining, therefore facilitating accurate meiotic recombination (Che et al, 2014; Hu et al, 2016, 2018). In this study, we showed thatplayed an important role in meiosis.has three alternative transcripts that differ in the 3-coding regions. By gene expression, specific knockout, overexpression and yeast two-hybrid assays, we demonstrated thatwas the functional isoform, andandwere nonfunctional for meiosis. These results indicated that C-terminal domain of OsRAD1.1 is important for the protein function.
The sequences of predicted proteins forandtranscripts are mostly identical to OsRAD1.1 except the very C-terminal 34 amino acid residues. Although this small fragment of OsRAD1.1 contains several residues that are highly conserved in RAD1 proteins from the other organisms, particularly in plants, nobody ever reported that this domain is functionally significant to the RAD1 protein. Therefore, we doubted that lack of phenotypic defect in knockout mutants ofandtranscripts was due to the low expression levels of the two transcripts, which was probably trivial to the gene function. To test this possibility, we overexpressed theandtranscripts in themutant plant. Unexpectedly, the overexpression ofandtranscripts did not restore the fertility of themutant. These results suggested that the small C-terminal domain is critical to the RAD1 protein function. In support of this observation, the original EMS mutant, which was truncated only the very C-terminal 16 amino acid residues of OsRAD1.1, was also infertile. In addition, the yeast two-hybrid analysis indicated that OsRAD1.1, but not OsRAD1.2, OsRAD1.3 and OsRAD1M lacking the C-terminal 16 residues, interacted with OsRAD9, OsHUS1 and OsRAD17, further indicating that the C-terminal domain of OsRAD1.1 is essential for the protein function. These results emphasized the critical role of the C-terminal domain for the RAD1 protein function.
The protein structure of RAD1 in the 9-1-1 complex has been well established previously and can be used for protein steric structure prediction (Venclovas and Thelen, 2000). To understand how the C-terminal domain contributes to the protein function, we conducted a protein structure modeling analysis of OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1M. OsRAD1.2, OsRAD1.3 and OsRAD1M proteins exhibited significant differences from OsRAD1.1 in the steric structure (Supplemental Fig. 6), suggesting that the C-terminal peptide of OsRAD1.1 is critical to maintain the protein tertiary structure. It is expected that the changes in the steric structure in OsRAD1.2, OsRAD1.3 and OsRAD1M would impair the protein interaction with other proteins and thus disrupt the protein function.
Whileandare apparently different fromat the functional level, why does the plant produce the two alternatively spliced forms? Do they have a biological function, or are they simply products of a cryptic intron without biological function? We attempted to answer the questions by overexpressing the two transcripts in the WT plants, but we did not see that the elevated transcripts ofandin the transgenic plants had a clear impact on the plant fertility. Based on these observations, we speculated thatandprobably have no particular function. It was probably that an accidental occurrence of the splicing site downstream of the cryptic 7th intron gaveandtranscripts the chance to be produced. Nonetheless, we cannot rule out the possibility that the productions ofandwere evolutionary selected for certain regulatory role that we have not found.
Acknowledgements
This work was supported by grants from Natural Science Foundation of Guangdong Province (Grant Nos. B030308008, 2017A030310500 and A03013104),National Key Research and Development Plan Program(Grant Nos. 2016YFD0101801 and 2016YFD0100406), Shenzhen Commission on Innovation and Technology Programs (Grant No. JCYJ20160229204920363), Guangzhou Science and Technology Innovation Commission (Grant No. 201804010034) and National Natural Science Foundation of China (Grant No. 31500254). We thank the Microscope Center in Life Science School of Sun Yat-sen University for using the facilities for microscopic analysis, and Liu Yaoguang for the CRISPR/Cas9 system.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/ journal/16726308; http://www.ricescience.org.
Supplemental Table 1. Primers used in this study.
Supplemental Fig. 1. Scanning electron microscopy analysis of pollen grains, inner surface of tapetum, and outer surface of anther wall in the wild type andmutant.
Supplemental Fig. 2. Expression of meiosis genes acting at prophase I in the wild type andmutant anthers.
Supplemental Fig. 3. Transgenic complementation of themutant.
Supplemental Fig. 4. Positions of primers for qRT-PCR.
Supplemental Fig. 5. Alignment of OsRAD1.1, OsRAD1.2, OsRAD1.3 and OsRAD1.1M with RAD1 proteins from other plant species.
Supplemental Fig. 6. Alignment of the predicated steric structures of OsRAD1.1 with OsRAD1.2, OsRAD1.3 and OsRAD1M.
Bermudez V P, Lindsey-Boltz L A, Cesare A J, Maniwa Y, Griffith J D, Hurwitz J, Sancar A. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex., 100(4): 1633–1638.
Black D L. 2003. Mechanisms of alternative pre-messenger RNA splicing., 72(1): 291–336.
Chang Z Y, Chen Z F, Wang N, Xie G, Lu J W, Yan W, Zhou J L, Tang X Y, Deng X W. 2016. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene., 113(49): 14145–14150.
Chaudhary S, Jabre I, Reddy A S N, Staiger D, Syed N H. 2019. Perspective on alternative splicing and proteome complexity in plants., 24(6): 496–506.
Che L X, Wang K J, Tang D, Liu Q Q, Chen X J, Li Y F, Hu Q, Shen Y, Yu H X, Gu M H, Cheng Z K. 2014. OsHUS1 facilitates accurate meiotic recombination in rice., 10(6): e1004405.
Chen M, Manley J L. 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches., 10(11): 741–754.
Chen Z F, Lu J W, Lu Q Q, Wang N, Wang C X, Xie G, Zhou X Y, Tang X Y. 2014. Screening and analysis of male sterile mutants derived from elitecultivar Huanghuazhan., 41(19): 1–4. (in Chinese)
Doré A S, Kilkenny M L, Rzechorzek N J, Pearl L H. 2009. Crystal structure of the Rad9-Rad1-Hus1 DNA damage checkpoint complex-implications for clamp loading and regulation., 34(6): 735–745.
Freire R, Murguía J R, Tarsounas M, Lowndes N F, Moens P B, Jackson S P. 1998. Human and mouse homologs ofSchizosa- ccharomyces pombe rad1and: Linkage to checkpoint control and mammalian meiosis., 12(16): 2560–2573.
Griffith J D, Lindsey-Boltz L A, Sancar A. 2002. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy., 277(18): 15233–15236.
Grushcow J M, Holzen T M, Park K J, Weinert T, Lichten M, Bishop D K. 1999.checkpoint genes,andare required for normal meiotic recombination partner choice., 153(2): 607–620.
Han L, Hu Z S, Liu Y H, Wang X Y, Hopkins K M, Lieberman H B, Hang H Y. 2010. Mousedeletion enhances susceptibility for skin tumor development., 9(1): 67.
Heitzeberg F, Chen I P, Hartung F, Orel N, Angelis K J, Puchta H. 2004. The Rad17 homologue ofis involved in the regulation of DNA damage repair and homologous recombination., 38(6): 954–968.
Hu Q, Tang D, Wang H J, Shen Y, Chen X J, Ji J H, Du G J, Li Y F, Cheng Z K. 2016. The exonuclease homolog OsRAD1 promotes accurate meiotic double-strand break repair by suppressing nonhomologous end joining., 172(2): 1105–1116.
Hu Q, Zhang C, Xue Z H, Ma L J, Liu W, Shen Y, Ma B J, Cheng Z K. 2018. OsRAD17 is required for meiotic double-strand break repair and plays a redundant role with OsZIP4 in synaptonemal complex assembly., 9: 1236.
Jaramillo-Lambert A, Harigaya Y, Vitt J, Villeneuve A, Engebrecht J. 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells., 20(23): 2078–2089.
Kalsotra A, Cooper T A. 2011. Functional consequences of developmentally regulated alternative splicing., 12(10): 715–729.
Kelemen O, Convertini P, Zhang Z Y, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing., 514(1): 1–30.
Laloum T, Martín G, Duque P. 2018. Alternative splicing control of abiotic stress responses., 23(2): 140–150.
Lochlainn S Ó, Amoah S, Graham N S, Alamer K, Rios J J, Kurup S, Stoute A, Hammond J P, Østergaard L, King G J, White P J, Broadley M R. 2011. High resolution melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes., 7(1): 43.
Luo Q, Li Y F, Shen Y, Cheng Z K. 2014. Ten years of gene discovery for meiotic event control in rice., 41(3): 125–137.
Lyndaker A M, Lim P X, Mleczko J M, Diggins C E, Holloway J K, Holmes R J, Kan R, Schlafer D H, Freire R, Cohen P E, Weiss R S. 2013. Conditional inactivation of the DNA damage response genein mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance., 9(2): e1003320.
Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G. 2015. A robust CRISPR/Cas9 system for convenient, high- efficiency multiplex genome editing in monocot and dicot plants., 8(8): 1274–1284.
Melo J, Toczyski D. 2002. A unified view of the DNA-damage checkpoint., 14(2): 237–245.
Naftelberg S, Schor I E, Ast G, Kornblihtt A R. 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure., 84(1): 165–198.
Navadgi-Patil V M, Burgers P M. 2009. A tale of two tails: Activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1., 8(9): 996–1003.
Nonomura K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2004. The novel geneof rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis.,16(4): 1008–1020.
Nonomura K, Nakano M, Eiquchi M, Suzuki T, Kurata N. 2006. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I., 119(2): 217–225.
Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2007. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice., 19(8): 2583–2594.
Ohashi E, Tsurimoto T. 2017. Functions of multiple clamp and clamp-loader complexes in eukaryotic DNA replication.,1042: 135–162.
Pan Q, Shai O, Lee L J, Frey B J, Blencowe B J. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing., 40(12): 1413–1415.
Peretz G, Arie L G, Bakhrat A, Abdu U. 2009. Thegene is required for homologous recombination repair during meiosis., 126: 677–686.
Shinohara M, Sakai K, Ogawa T, Shinohara A. 2003. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast., 164(3): 855–865.
Szakonyi D, Duque P. 2018. Alternative splicing as a regulator of early plant development., 9: 1174.
Udell C M, Lee S K, Davey S. 1998.andencode mammalian homologues of the fission yeastrad1cell cycle checkpoint control gene., 26(17): 3971–3976.
Uri A, Martha K, Veronika B I, Anna B, Trudi S. 2007. An essential role forin somatic and meiotic DNA damage responses., 120(6): 1042–1049.
Vasileva A, Hopkins K M, Wang X Y, Weissbach M M, Friedman R A, Wolgemuth D J, Lieberman H B. 2013. The DNA damage checkpoint protein RAD9A is essential for male meiosis in the mouse.,126(17): 3927–3938.
Venclovas C, Thelen M P. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes., 28(13): 2481–2493.
Wang C L, Wang Y, Cheng Z J, Zhao Z G, Chen J, Sheng P K, Yu Y, Ma W W, Duan E C, Wu F Q, Liu L L, Qin R Z, Zhang X, Guo X P, Wang J L, Jiang L, Wan J M. 2016. The role of OsMSH4 in male and female gamete development in rice meiosis., 67(5): 1447–1459.
Wang M, Wang K J, Tang D, Wei C X, Li M, Shen Y, Chi Z C, Gu M H, Cheng Z K. 2010. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice., 22(2): 417–430.
Wang Y, Copenhaver G P. 2018. Meiotic recombination: Mixing it up in plants., 69: 577–609.
Xu M, Bai L, Gong Y, Xie W, Hang H Y, Jiang T. 2009. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex., 284(31): 20457–20461.
Yan W, Chen Z F, Lu J W, Xu C J, Xie G, Li Y Q, Deng X W, He H, Tang X Y. 2017. Simultaneous identification of multiple causal mutations in rice., 7: 2055.
Yuan W Y, Li X W, Chang Y X, Wen R Y, Chen G X, Zhang Q F, Wu C Y. 2009. Mutation of the rice generesults in lack of bivalent formation in meiosis., 59(2): 303–315.
Zhang C B, Liu Y H, Hu Z S, An L L, He Y K, Hang H Y. 2011. Targeted deletion of mouseleads to deficient cellular DNA damage responses., 2(5): 410–422.
Zhang D B, Luo X, Zhu L. 2011. Cytological analysis and genetic control of rice anther development., 38(9): 379–390.
Zhou B B, Elledge S J. 2000. The DNA damage response: Putting checkpoints in perspective.,408: 433–439.
Zou L, Cortez D, Elledge S J. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin., 16(2): 198–208.
17 August 2019;
21 October 2019
Tang Xiaoyan (txy@frontier-ag.com); Wu Jianxin (wjxin@m.scnu.edu.cn); Chen Zhufeng(czf@frontier-ag.com)
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2020.05.005
(Managing Editor: Li Guan)
杂志排行
Rice Science的其它文章
- Genetic Variation Dissection of Rice Blast Resistance Using an Indica Population
- OsPS6 Plays Important Role in Anther Development and Microspore Formation
- Deciphering Rice Lesion Mimic Mutants to Understand Molecular Network Governing Plant Immunity and Growth
- RNA-Seq Study Reveals AP2-Domain-Containing Signalling Regulators Involved in Initial Imbibition of Seed Germination in Rice
- RNAi-Mediated Silencing of ITPK Gene Reduces Phytic Acid Content, Alters Transcripts of Phytic Acid Biosynthetic Genes, and Modulates Mineral Distribution in Rice Seeds
- Accumulation of Polyphenolic Compounds and Osmolytes under Dehydration Stress and Their Implication in Redox Regulation in Four Indigenous Aromatic Rice Cultivars
