Bile-derived circulating extracellular miR-30d-5p and miR-92a-3p as potential biomarkers for cholangiocarcinoma
2020-03-03HyeSookHnMiJinKimJoungHoHnJieunYunHeeKyungKimYewonYngKiBeKimSeonMeePrk
Hye Sook Hn , , Mi Jin Kim , Joung-Ho Hn , , , Jieun Yun , Hee Kyung Kim , , Yewon Yng , Ki Be Kim , Seon Mee Prk ,
a Department of Internal Medicine, Chungbuk National University Hospital, Cheongju 28644, Korea
b Departments of Internal Medicine, Chungbuk National University College of Medicine, Cheongju 28644, Korea
c Department of Pharmaceutical Engineering, College of Science Engineering, Cheongju University, Cheongju 28644, Korea
Keywords: Bile Biomarkers Cholangiocarcinoma MicroRNA
ABSTRACT Background: Cholangiocarcinoma (CCA) is from cholangiocytes, and therefore bile is a potentially rich source of biomarkers for CCA. The aim of the study was to identify and validate microRNAs (miRNAs) in bile samples that are differentially expressed between benign biliary disease (BBD) and CCA. Methods: Bile samples from 106 patients with obstructive biliary disease were allocated consecutively to a discovery set (10 patients with BBD and 11 with CCA) and then a validation set (48 patients with BBD and 37 with CCA). An miRNA microarray platform was used to screen 1209 miRNAs in the discovery set. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) was used to validate the profiling results in the discovery and validation sets. In addition, the levels of carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA) were determined from patient serum samples. Results: Microarray profiling showed that miR-30d-5p and miR-92a-3p were significantly upregulated in bile from the CCA group compared with those from the BBD group. qRT-PCR results indicated that the expression levels of miR-30d-5p and of miR-92a-3p were significantly upregulated in the CCA group compared to the BBD group, validating the miRNA microarray results. Pathway analysis suggested that putative target genes of miR-30d-5p and of miR-92a-3p were involved in CCA-associated signalling path- ways, such as Hippo, Wnt, p53, MAPK, and EGFR. Receiver operating curve analysis revealed that the areas under the curve for bile miR-30d-5p, miR-92a-3p, serum CA19-9, and CEA were 0.730, 0.652, 0.675, and 0.603, respectively, and bile miR-30d-5p showed the best diagnostic performance with a sensitivity of 81.1% and a specificity of 60.5%. Conclusions: The levels of extracellular miR-30d-5p and miR-92a-3p in bile were significantly higher in patients with CCA than those in patients with BBD. Bile-derived circulating extracellular miR-30d-5p and miR-92a-3p are potential biomarkers for discriminating CCA from BBD.
Introduction
Cholangiocytes, the epithelial cells of the bile duct, comprise a small proportion of the total cellular component of the liver, but play an essential role in bile secretion and transport of bil- iary and blood components [1] . Cholangiocarcinomas (CCAs) arise from cholangiocytes and as intrahepatic, perihilar, or distal CCAs based on tumor localization [2 , 3] . Although patients with early CCA can be cured by surgical tumor resection, a large majority of pa- tients are initially diagnosed with unresectable, locally advanced or metastatic disease, and experience a relapse after initial surgical resection. Consequently, the prognosis of CCA patients is dismal, and these patients have a median overall survival of 6 -12 months and a five-year survival rate of only approximately 10% [2 , 3] .
CCA is a frequent cause of bile duct stricture or obstruction, leading to retention of bile, and jaundice is the most common symptom in patients with CCA. Patients with suspected bile duct stricture or obstruction are usually assessed by diagnostic imaging techniques including ultrasonography, computed tomography, and magnetic resonance cholangiopancreatography. However, these imaging techniques have limited sensitivity for the diagnosis of CCA [2 , 3] . Moreover, cytopathological evaluation of intraductal bile aspiration cytology, biliary brush cytology, fine-needle aspiration biopsy, or intraductal forceps biopsy, which are confirmative diagnostic tests for CCA, also have poor cell yields due to the desmoplastic microenvironment of CCA and therefore has low sensitivity for the diagnosis of CCA [2] . Serum levels of carbohy- drate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA) are usually elevated in patients with CCA; however, neither 0has sufficient sensitivity nor specificity to be used for differential diagnosis of benign biliary disease (BBD) or early detection of CCA [2 , 3] . Therefore, novel and reliable biomarkers are urgently needed for discriminating CCA from BBD.
MicroRNAs (miRNAs) are a class of highly conserved, endoge- nously expressed, small non-coding RNA molecules of 18-25 nucleotides in length that repress protein translation by binding to target mRNAs [4] . Functionally, miRNAs can act as either onco- genes or tumor suppressors, and many miRNAs have already been employed as diagnostic, prognostic, or predictive biomarkers in multiple cancer types [5] . One of the most remarkable aspects of miRNAs is their high degree of stability in the extracellular environment as enclosure of miRNAs in small vesicles such as apoptotic bodies, microvesicles, or exosomes, which protects them from degrading enzymes [6 , 7] . Most studies on circulating extracellular miRNAs measured miRNAs in peripheral blood, such as serum or plasma. However, recent studies have reported the potential usefulness of miRNAs as tumor biomarkers in body fluids other than blood [6 , 7] . For CCA, an accurate tumor-derived miRNA profile is more likely to exist in bile than in blood, as potential biomarkers may be directly secreted into bile by adjacent CCAs.
Therefore, in the current study, we investigated bile samples for the presence of extracellular miRNAs that are differentially ex- pressed between BBD and CCA. Following the identification of such miRNAs in bile, their potential as biomarkers for discriminating CCA from BBD was assessed and compared with that of the com- monly used serum tumor markers, CA19-9 and CEA.
Methods
Patients and bile collection
Bile and serum samples from 106 patients with obstructive bil- iary disease between January 2011 and September 2014 were al- located to a discovery set ( n = 21; 10 patients with BBD and 11 with CCA) and then a validation set ( n = 85; 48 patients with BBD and 37 with CCA) in chronological order. The workflow diagram of the study design is shown in Fig. 1 . BBD was diagnosed based on the clinical characteristics and the absence of malignant cells in the brush cytology specimen or bile duct biopsy. CCA was con- firmed by cytopathological diagnosis based on the presence of ade- nocarcinoma cells after brushing, biopsy, or surgical resection. Bile samples from patients with CCA were obtained from those show- ing no evidence of other malignancies.
Bile samples were collected at the time of diagnostic or ther- apeutic bile drainage via endoscopic retrograde cholangiopancre- atography (ERCP) or percutaneous transhepatic biliary drainage. Aspiration of bile was performed after cannulation of the biliary tree before injection of contrast medium. Bile samples were ini- tially stored at 4-8 °C, and then rapidly centrifuged at 1792 g for 10 min. The supernatant was aliquoted into microcentrifuge tubes and stored at -80 °C until use.
The study protocol was reviewed and approved by the Institu- tional Review Board of Chungbuk National University Hospital (IRB approval number: 2011-12-099) and written informed consent was obtained from all study participants.
RNA extraction and miRNA microarray analysis
Total RNA from 400 μL of each bile supernatant was extracted using Trizol LS Reagent (Molecular Research Center, Cincinnati, OH, USA), according to the manufacturer’s instructions, quantified by absorbance at 260 nm, and evaluated using a Bioanalyzer 2100 (Ag- ilent Technologies, San Carlos, CA, USA) to confirm integrity. RNA peaks in each bile sample were detected at 25-200 nucleotides (nt).
Candidate miRNAs were identified by miRNA microarray ex- pression profiling of the discovery set using the Agilent Human miRNA Microarray Release 16.0 platform, which contains 1209 miRNA probes. GeneSpring GX 12 software (Agilent Technolo- gies) and the DAVID Bioinformatics Resource (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA) were used for integrative data analysis and vi- sualization.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
The miRNAs differentially expressed in the microarray profiling of the discovery set were validated by qRT-PCR in the discovery and validation sample sets. Total RNA was extracted as described above. To quantify miRNA expression derived from the miRNA mi- croarray, 3.7 μL of total RNA was reverse transcribed using the Rotor-Gene Q platform and the miScript PCR Starter Kit (QIAGEN Korea Ltd., Seoul, South Korea) according to the manufacturer’s protocol. miRNA expression was normalized to the total RNA con- centration measured using Quant-iT RiboGreen RNA Reagent and Kit (Invitrogen, Grand Island, NY, USA). Quant-iT RiboGreen RNA Reagent is an ultrasensitive fluorescent nucleic acid stain for quan- titating RNA [8] , and the RiboGreen RNA quantification kit allowed accurate RNA concentration measurement [9 , 10] . Chemically syn- thesized RNA oligonucleotides (Cosmo Genetech Co. Ltd., Seoul, South Korea) corresponding to the target miRNAs were used to generate standard curves ranging from 30 to 3 ×104copies. All samples were purified for triplicate runs of qRT-PCR.
miRNA target gene prediction and pathway analysis
Target genes of selected miRNAs validated by qRT-PCR were predicted using the SpidermiR module in R (R Foundation for Statistical Computing, Vienna, Austria) and TargetScan in DIANA- miRPath version 3.0 ( http://www.microrna.gr/miRPathv3 ). Pathway analysis of target genes was performed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Significant enrich- ment results were obtained with P values < 0.05.
Immunoassay for serum CA19-9 and CEA
The expression levels of serum CA19-9 and CEA were measured using an Architect i40 0 0SR chemiluminescent enzyme immunoas- say (Abbott Laboratories, Abbott Park, IL, USA).
Statistical analysis
The unpaired t -test was used to assess differences in liver chemistry profiles between the BBD and CCA groups, and the Mann-Whitney U test was used to assess differences in the expres- sion levels of bile miRNAs, serum CA19-9, and CEA. We plotted receiver operating characteristic (ROC) curves and calculated the area under the curve (AUC) of the significantly differentially ex- pressed bile miRNAs, serum CA19-9, and CEA to predict the perfor- mance in discriminating between CCA and BBD. The optimal cutoff points for candidate markers were determined based on the high- est combined sensitivity and specificity for detection in the ROC curve analysis. Risk scores were assigned to all patients according to a linear combination of the expression levels of the bile miRNAs, serum CA19-9, and CEA, weighted according to the regression co- efficient. Stepwise Cox regression and stratification analyses were also conducted. All statistical analyses were performed using IBM SPSS Statistics software, version 21.0 (IBM Corp., Armonk, NY, USA), Prism software, version 6.0 (GraphPad), or R software, version 3.4.1 (R Foundation for Statistical Computing). All P values were two- sided and values < 0.05 were considered statistically significant.
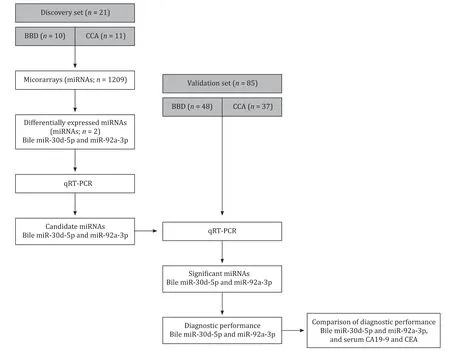
Fig. 1. Overview of the study design. BBD: benign biliary disease; CCA: cholangiocarcinoma; qRT-PCR: quantitative real-time reverse transcription polymerase chain reaction; CA19-9: carbohydrate antigen 19-9; CEA: carcinoembryonic antigen.
Results
Patient characteristics
Table 1 shows the baseline characteristics of the study partic- ipants in the discovery and validation sets. The majority of pa- tients were diagnosed with choledocholithiasis in the BBD group and with distal CCA in the CCA group. The median follow-up pe- riod was 80.7 months (range, 48.3 -92.2 months), and no cancers including CCA were identified in the BBD group during this pe- riod. Comparing the BBD and CCA patients in the validation set, total bilirubin, gamma-glutamyl transpeptidase, and alkaline phos- phatase, which are indicators of cholestasis-specific liver enzyme, and CA19-9 were all significantly higher in the CCA patient serum samples. The level of serum CEA was also higher in the CCA group, but this difference was not significant.
Selection of candidate bile miRNAs by miRNA microarray
To identify CCA-specific bile miRNAs, 1209 miRNA expression profiles of CCA (n = 11) and BBD (n = 10) bile samples in the discov- ery set were compared using an Agilent miRNA microarray plat- form. Microarray analysis revealed 43 miRNA variations with a fold change ≥1.5 that could discriminate between the BBD and CCA groups (F ig. 2 and Table 2) . Of the 43 miRNAs, only miR-30d-5p and miR-92a-3p showed significantly higher expression with a fold change ≥1.5 and P < 0.05 in bile samples from the CCA group than in those from the BBD group ( Table 2) . These two candidate miR- NAs were selected for further analysis.
Validation of miRNA microarray results using qRT-PCR
To confirm the results of the miRNA microarray profiling, the expression levels of miR-30d-5p and miR-92a-3p in the discov- ery set of bile samples were examined by qRT-PCR. The results showed that the expression levels of the two miRNAs were sig- nificantly upregulated in the CCA group compared with the BBD group ( P < 0.05, data not shown). Next, we assessed the expression levels of miR-30d-5p and miR-92a-3p by qRT-PCR in the validation set of bile samples (BBD, n = 48; CCA, n = 37). The median total RNA concentration in the 85 bile samples of the validation set was 59.7 ng/μL (range 12.0 -640.1 ng/μL), and the expression levels of miR-30d-5p and of miR-92a-3p were significantly upregulated in the CCA group compared with those in the BBD group ( P < 0.001 and P = 0.017, respectively; Fig. 3 ).
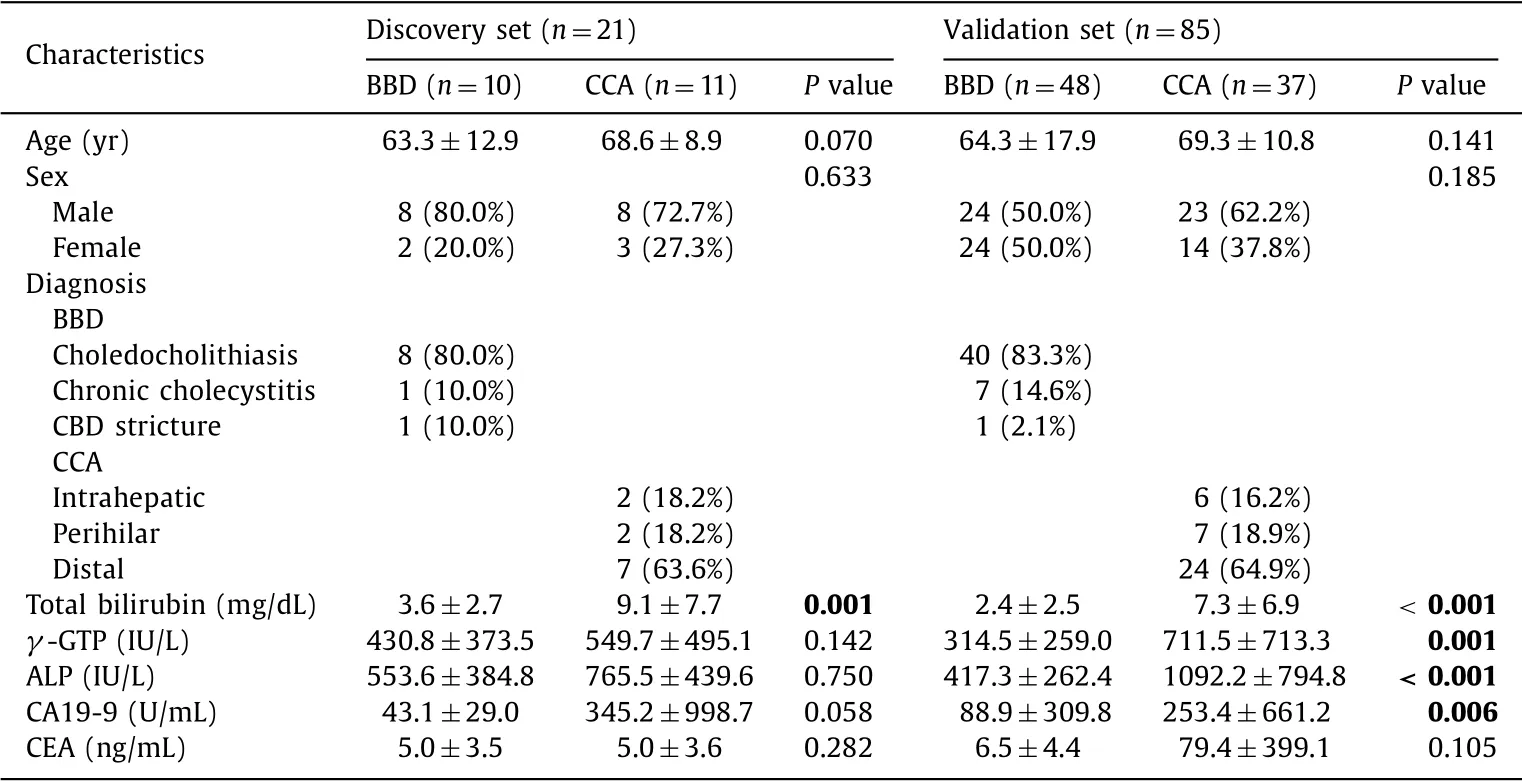
Table 1 Patient characteristics.

Fig. 2. Hierarchical clustering of 43 differentially expressed miRNAs with a fold change ≥1.5 in the BBD bile samples versus the CCA bile samples. BBD: benign biliary disease; CCA: cholangiocarcinoma.

Table 2 Microarray expression profile in the discovery set.
Target genes and pathway analysis of selected miRNAs
Using TargetScan, the number of target genes of miR-30d-5p and miR-92a-3p was predicted to be 1243 and 571 genes, re- spectively, of which 23 and 49 genes, respectively, were validated ( Table 3 ). KEGG pathway analysis revealed that these target genes were involved in many CCA-associated signaling pathways, sug- gesting their possible roles in CCA pathogenesis. Target genes of miR-30d-5p were involved in the Hippo, Wnt, and p53 signaling pathways ( Fig. 4 A), and miR-92a-3p in the MAPK, Hippo, and EGFR signaling pathways ( Fig. 4 B).
Performance of bile miR-30d-5p and miR-92a-3p, and of serum CA19-9 and CEA, as biomarkers for discriminating CCA from BBD
The performance of bile miR-30d-5p and miR-92a-3p, and of serum CA19-9 and CEA, as individual biomarkers for discriminat- ing CCA from BBD, was evaluated in the validation set ( Table 4 ). In the ROC analysis, the levels of bile miR-30d-5p showed an AUC of 0.730, with a sensitivity of 81.1% and a specificity of 60.5% ( Fig. 5 A). The levels of bile miR-92a-3p showed an AUC of 0.652, with a sen- sitivity of 65.7% and a specificity of 66.7% ( Fig. 5 B). The AUC for serum CA19-9 was 0.675, with a sensitivity of 70.3% and a speci- ficity of 64.6% ( Fig. 5 C). The AUC for serum CEA was 0.603, with a sensitivity of 64.9% and a specificity of 60.4% ( Fig. 5 D). Next, the performance of combinations of the miRNAs with the most com- monly used serum marker CA19-9 was tested. The AUC for the combination of two miRNAs and serum CA19-9 was 0.689, with a sensitivity of 70.3% and a specificity of 68.7% ( Fig. 5 E). The AUC for a combination of bile miR-30d-5p plus serum CA19-9 was 0.728, with a sensitivity of 81.1% and a specificity of 58.3% ( Fig. 5 F).
Discussion
This study showed that the expression levels of miR-30d-5p and miR-92a-3p in bile samples were specifically upregulated in patients with CCA compared with those in patients with BBD ( Fig. 6 ). This miRNA expression in bile was confirmed in a larger independent sample set using qRT-PCR, thereby validating the ini- tial microarray-based analysis. The present study also compared the expression level of bile miRNAs with that of the commonly used serum tumor markers in patients with CCA—serum CA19-9 and CEA. Of the four, bile miR-30d-5p showed the best perfor- mance for discriminating between CCA and BBD, with a sensitivity of 81.1%, a specificity of 60.5%, and an AUC of 0.730. Additionally, a combination of the two bile miRNAs and serum CA19-9 did not have better discriminating performance than the performance of miR-30d-5p alone for CCA.
Bile samples can be obtained when ERCP or percutaneous tran- shepatic biliary drainage is being performed for diagnosis or treat- ment of suspected biliary tract disease. Concentrations of potential biomarkers may be increased in bile because of impaired bile flow as a result of biliary tract strictures or obstruction. Furthermore, potential tumor-derived biomarkers may be directly secreted into bile by adjacent CCAs. Therefore, bile sampling could enhance diag- nostic performance relative to sampling circulating serum biomark- ers [11 , 12] . It has also been reported that human bile contains a large number of extracellular vesicles and miRNAs [13 , 14] . A re- cent meta-analysis reported that miRNAs in bile have a relatively higher diagnostic accuracy as biomarkers for CCA than miRNAs in serum or urine [15] . Several studies have reported the diagnostic performance of bile miRNA as a biomarker for biliary tract can- cer ( Table 5 ). Shigehara et al. demonstrated that the level of 10 of 667 bile miRNAs studied (miR-9, miR-145*, miR-105, miR-147b, let-7f-2*, let-7i*, miR-302c*, miR-199a-3p, miR-222*, and miR-942) were significantly higher in the biliary tract cancer group than in the BBD group (patients with choledocholithiasis) [13] . Moreover, the combination of miR-9 and miR-145 * is a suitable diagnostic biomarker for biliary tract cancer. Li et al. defined a novel bil- iary miRNA panel (derived from miRNA-laden microvesicles) for CCA diagnosis, combining five miRNAs (miR-16, miR-1274b, miR- 191, miR-486-3p, and miR-484) that were able to differentiate CCA from primary sclerosing cholangitis (PSC) or other bile duct ob- structions [14] . A recent study conducted by Voigtländer et al. re- vealed that the expression levels of miR-412, miR-640, miR-1537, and miR-3189 in bile were different between patients with PSC and PSC/CCA [16] .

Fig. 3. Comparison of ( A ) miR-30d-5p and ( B ) miR-92a-3p expression levels determined by qRT-PCR in the validation set bile samples from benign biliary disease (BBD) and cholangiocarcinoma (CCA). Statistical significance was determined by the Mann-Whitney U test. qRT-PCR: quantitative real-time reverse transcription polymerase chain reaction.

Fig. 4. KEGG pathway analysis of target genes of ( A ) miR-30d-5p and ( B ) miR-92a-3p.
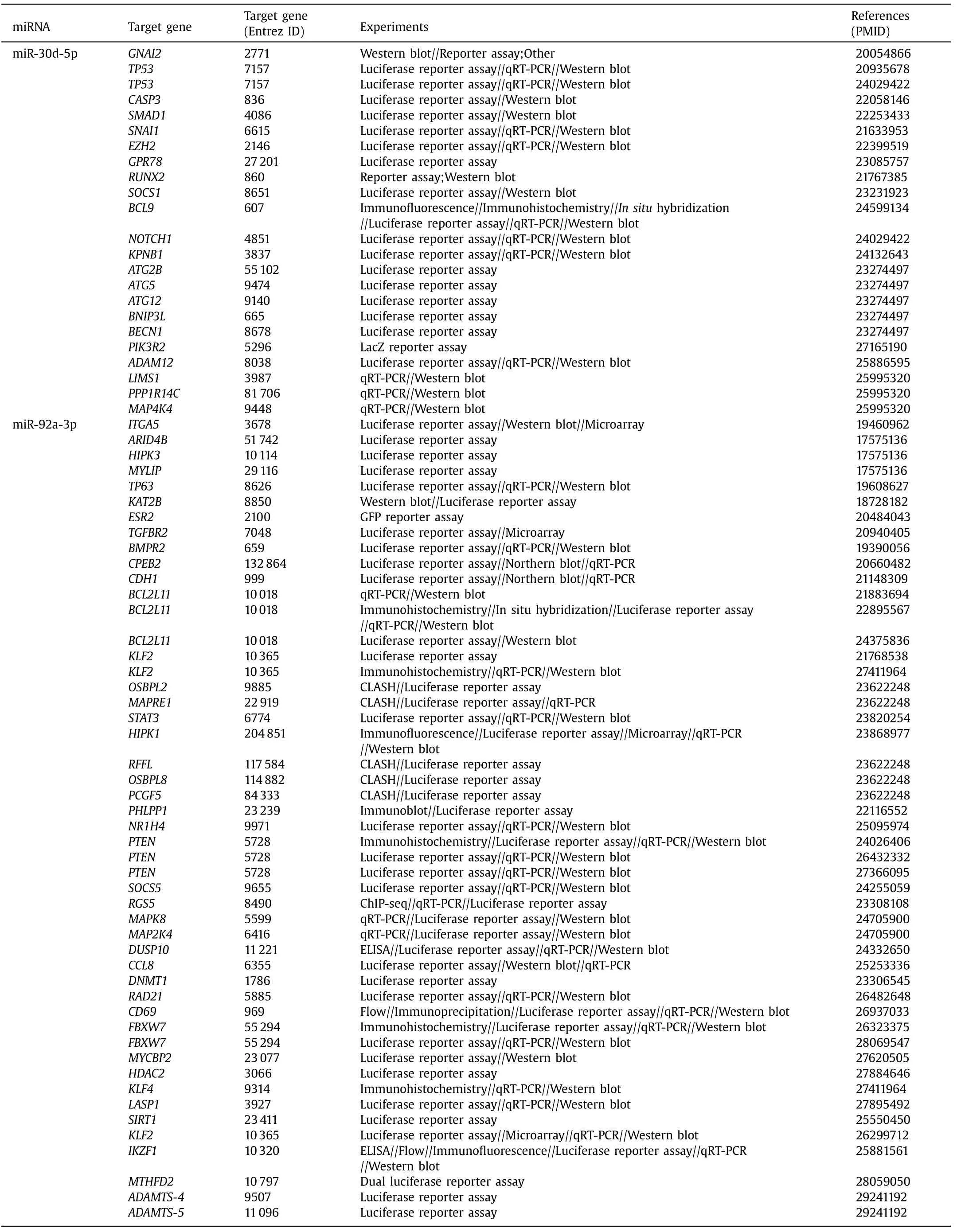
Table 3 Validated miR-30d-5p and miR-92a-3p targeted genes.
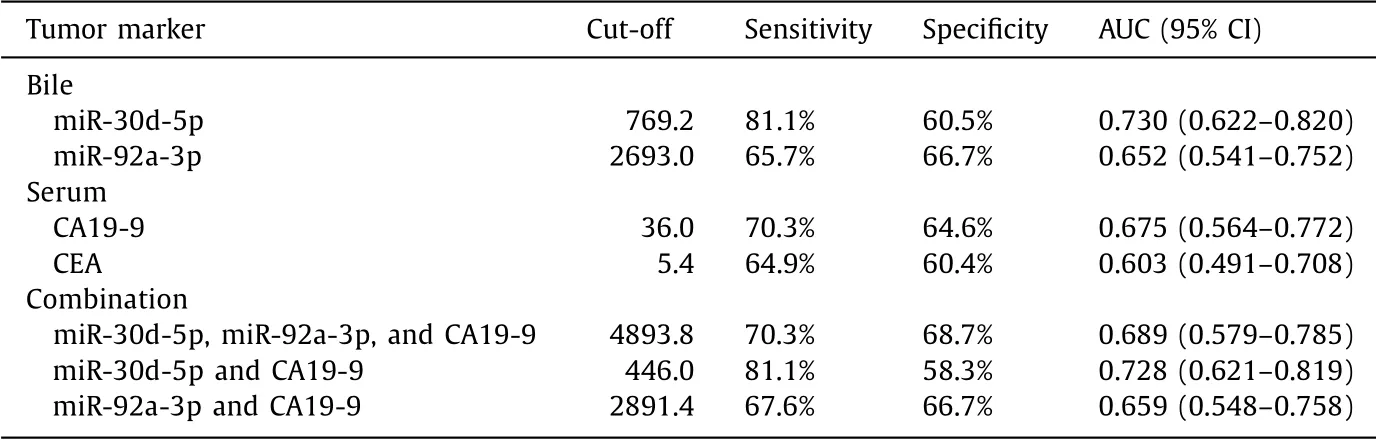
Table 4 Parameters determining the performance of bile miR-30d-5p and miR-92a-3p, and serum CA19-9 and CEA, as biomarkers for discriminating cholangiocarcinoma from benign biliary disease.
Table 5 Summary of published studies examining miRNAs in bile samples of cholangiocarcinoma.
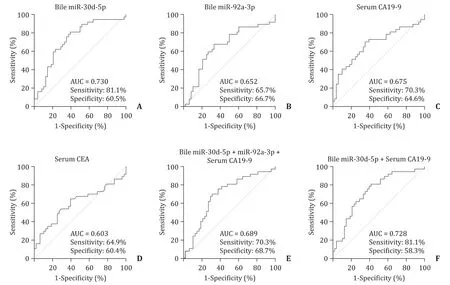
Fig. 5. Receiver operating characteristic curve analyses for discriminating cholangiocarcinoma from benign biliary disease. Each panel shows the area under the curve (AUC), sensitivity, and specificity values for ( A ) bile miR-30d-5p, ( B ) bile miR-92a-3p, ( C ) serum carbohydrate antigen 19-9 (CA19-9), ( D ) serum carcinoembryonic antigen (CEA), ( E ) bile miR-30d-5p + bile miR-92a-3p + serum CA19-9, and ( F ) bile miR-30d-5p + serum CA19-9.
Fig. 6. Bile-derived circulating extracellular miR-30d-5p and MiR-92a-3p are poten- tial biomarkers for discriminating cholangiocarcinoma from bening biliary disease.
In the aforementioned studies, distinct expression patterns of bile miRNAs were observed to differentiate CCA from BBD, but the miRNAs found to be significantly expressed in CCA are inconsis- tent [13-16] . An intuitive explanation for this difference is a lack of standardization of bile collection, storage, processing, normal- ization of miRNA values, and different study populations. As the method of bile processing varied in previous studies, this would have contributed to variations in the components of the bile sam- ples studied. Whereas Shigehara et al. used whole bile as a source for miRNA profiling [13] , Li et al. established that RNA originat- ing from floating cells is rapidly degraded and therefore focused only on the extracellular vesicles [14] . However, recent publica- tions show that a significant majority of extracellular miRNAs are actually not present in extracellular vesicles [17 , 18] . Therefore, to capture the more abundant vesicle-free miRNA fraction in bile and avoid RNA degradation, we used floating cell-free bile samples, preparing subcellular fractions with only one centrifugation at low speed, to get a complete representation of the small RNA spec- trum available for biomarker purposes. An additional difficulty is the normalization of miRNA values in biologic fluids. Most miRNA studies use U6, the endogenous control RNA in qRT-PCR, to nor- malize miRNA expression; however, there is accumulating evidence that U6 is not appropriate for this purpose [19] . Normalization to endogenous invariant controls would be an optimal choice, but data to guide the choice of these in bile samples are lacking. There- fore, in the present study, we used the nucleic acid dye RiboGreen to quantify cell-free RNA levels in bile. RiboGreen-based quantifi- cation of total cell-free RNA in a body fluid should make it easier to normalize the expression of each cell-free RNA against that of a housekeeping gene [9 , 10] . Another explanation for the reported variation in miRNAs in CCA is that there are likely strong geograph- ical differences in development of CCA caused by region-specific risk factors such as consumption of undercooked fish, liver fluke infestation, PSC, hepatitis B and C viruses, and malformations of the biliary tract [2 , 3] . In addition, most studies used bile samples from patients with BBD (i.e., PSC, choledocholithiasis, sphincter of Oddi dysfunction, chronic pancreatitis, cholangitis) derived from large heterogeneous groups as controls, each with a distinct bio- logical diversity [13 , 14 , 16] . Therefore, it is difficult to directly de- cide by comparison which of the target miRNAs found in previous studies is superior.
The present study showed that miR-30d-5p and miR-92a-3p were present in bile at higher levels in the CCA group than in the BBD group. Upregulation of certain miRNAs in cancer suggests that they have oncogenic characteristics. KEGG pathway analysis suggested that the predicted and validated target genes of miR- 30d-5p and miR-92a-3p were involved in various pathways, such as the Hippo, Wnt, p53, MAPK, and EGFR signaling pathways. Re- cently, miR-30d was found to be amplified in more than 30% of human epithelial tumors of various types [20] . The upregulation of miR-30d found in hepatocellular carcinoma, medulloblastoma, non-small cell lung cancer (in malignant effusion), breast can- cer, and prostate cancer could play a role in promoting the ad- vanced progression of patients with these malignancies. In con- trast to its oncogenic role, decreased expression of miR-30d has been observed in colon cancer, non-small cell lung cancer (in tis- sues), and renal carcinoma, and is associated with highly malig- nant phenotypes of these cancer cells [21-23] . He et al. investi- gated the biological function of miR-30d in gallbladder carcinoma and found that miR-30d could serve as a tumor suppressor by in- hibiting the proliferation and invasion of gallbladder cells by tar- geting lactate dehydrogenase-A (LDHA) inhibition [24] . LDHA cat- alyzes the conversion of pyruvate into lactate and may be involved in the acidic tumor microenvironment by regulating aerobic glycol- ysis (Warburg effect) [24] . MiR-92a, which belongs to the miR-17- 92 cluster, has also been reported in various cancers and exhibits a different role in carcinogenesis. It has been reported in several studies that miR-92a is an oncogene, particularly in colorectal can- cer, and has become a useful biomarker for early detection [25] . Furthermore, miR-92a plays a critical role in the tumor progres- sion of liver, lung and cervical cancers [26-28] . In a study of the molecular mechanisms underlying miR-92a upregulation in CCA, Zhu et al. showed that, compared with benign human biliary ep- ithelial cells, the miR-17-92 cluster is highly expressed in human CCA cells and phosphatase and tensin homolog (PTEN) is a direct target of the miR-17-92 cluster [29] . Therefore, inhibition of PTEN by members of the miR-17-92 cluster may represent an important mechanism for CCA growth. However, miR-92a also has a tumor- suppressive role in gastric, ovarian, and breast cancers [30-32] . The apparently discordant finding that one miRNA can function either as an oncogene or a tumor suppressor most likely reflects the con- tribution made by the miRNA to distinct biological processes as it regulates different target genes depending on the cancer type or even different microenvironments within the same cancer type.
The present study has some limitations. First, this study is from a single institution with a limited number of participating patients. In particular, the number of bile samples for miRNA microarray in the discovery set is small and additional validation is required once again with a larger number of bile samples from other institutions to confirm our findings. Second, we cannot evaluate the expression of miRNAs in CCA by comparison with the expression in normal bile as the invasive technique of biliary drainage cannot be per- formed on healthy volunteers. Nonetheless, to our knowledge, this is the first study to evaluate bile miRNAs in an Asian population as a biomarker for discriminating CCA from BBD and to compare the diagnostic value of bile miRNAs with serum CEA and CA19-9, which are commonly used in clinical practice.
In conclusion, the levels of circulating extracellular miR-30d-5p and miR-92a-3p in bile were significantly higher in patients with CCA than in patients with BBD. Therefore, bile-derived miR-30d-5p and miR-92a-3p are potential biomarkers for discriminating CCA from BBD. These miRNAs now warrant further pathophysiological investigation and underlying mechanisms in patients with CCA.
Acknowledgments
All bile samples and data in this study were provided by Chungbuk National University Hospital, a member of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare, and Family Affairs.
CRediT authorship contribution statement
Hye Sook Han:Formal analysis, Funding acquisition, Method- ology, Writing - original draft.Mi Jin Kim:Data curation, Formal analysis, Writing - original draft.Joung-Ho Han:Conceptualization, Funding acquisition, Supervision, Writing - review & editing.Jieun Yun:Investigation, Methodology.Hee Kyung Kim:Methodology.Yaewon Yang:Methodology.Ki Bae Kim:Methodology.Seon Mee Park:Data curation, Funding acquisition.
Funding
This research was supported by grants from a Basic Science Re- search Program through the National Research Foundation of Ko- rea (NRF), funded by the Ministry of Education, Science and Tech- nology ( 2017R1A5A2015541 and 2019R1A2C1007401 ). This study was also supported by a grant from the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Ko- rea ( 1120330 ) and a National Research Foundation of Korea grant funded by the Korea government ( NRF-2013R 1A 1A 2063994 ).
Ethical approval
This study was approved by the Institutional Review Board of Chungbuk National University Hospital (IRB approval number: 2011-12-099).
Competing interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the sub- ject of this article.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Impact of EBV infection and immune function assay for lymphoproliferative disorder in pediatric patients after liver transplantation: A single-center experience
- Hepatobiliary&Pancreatic Diseases International
- Intraoperative management and early post-operative outcomes of patients with coronary artery disease who underwent orthotopic liver transplantation
- Acute onset of autoimmune hepatitis in children and adolescents
- Liver stiffness as a predictor of hepatocellular carcinoma behavior in patients with hepatitis C related liver cirrhosis ✩
- Treatment and prognosis of hepatic epithelioid hemangioendothelioma based on SEER data analysis from 1973 to 2014
