吡啶取代苯酚配体构建的三个配合物的晶体结构
2019-07-10王寒晖朱敦如
余 柱 柏 健 王寒晖 曹 颖 朱敦如*,,2
(1南京工业大学化工学院,材料化学工程国家重点实验室,南京 211816)(2南京大学配位化学国家重点实验室,南京 210023)
0 Introduction
Thephenol-based “end-off”compartmental ligands usually contain twoadjacent,but different,donor sets[1-2].These ligands are able to coordinate two metal ions simultaneously,and the resulting binuclear complexes can be used to mimic dimetallic biosites in many proteins and native enzymes[3-9].During the past two decades,a large number of binuclear complexes with bridging phenolato group have been synthesized,and further investigations have revealed that some complexesareimportantmodel compoundsfor metalloenzymes such as phosphatase,urease,superoxide dismutase,catalase,tyrosinase,nuclease[10-20].In the synthesis of the dinucleating phenol-based compartmental ligands,the compound HL(Scheme 1),containing formyl,amino and pyridine functions,is usually used as a key precursor.We reported its crystal structure in 2012[21]and a binuclear manganese(Ⅱ)complex [Mn2L2](ClO4)2in 2007[22]. Recently, two mononuclear copper(Ⅱ)complexes,[Cu(HL)(CH3CN)(ClO4)]ClO4and cis-[Cu(HL)(NCS)2)]·0.5C4H8O2[23],and a mixed-bridged linear trinuclear copper(Ⅱ)complex with the HL derivative have been reported by us[24].As a continuation of our investigation of complexes based on HL,herein we reported the syntheses,crystal structures and spectral characterization of three novel complexes with HL,[Ca2L2(NO3)2](1),[Cd(HL)(NO3)2](2)and[Cd2L2(NO3)2]·H2O(3).Notably,the mononuclear cadmium complex 2 can be transformed to the binuclear cadmium complex 3 under basic conditions.

Scheme 1 Structure of phenol-based ligand HL
1 Experimental
1.1 Materials and measurements
All reagents and solvents were purchased from commercial sources and used as received,unless noted otherwise.The ligand HL was synthesized via previously reported methods with slight modification[25].Elemental analyses(C,H,N)were carried out with a Thermo Finnigan Flash 1112A elemental analyzer.The infrared spectra (IR)of the complexes were recorded on a Nicolet Avatar 380 FTIR instrument with KBr pellets in a range of 4 000~400 cm-1.Electrospray ionization mass spectrum (ESI-MS)was recorded with an LCQ ADVANTAGE MAX mass spectrometer,with MeOH on the mobile phase;the flow rate of the mobile phase was 0.2 cm3·min-1.The spray voltage,the capillary voltage,and the capillary temperature were 4 kV,40 V,and 260 ℃,respectively.
1.2 Syntheses of complexes 1~3
[Ca2L2(NO3)2](1):To a methanol solution(2 mL)of HL(69.5 mg,0.2 mmol)was added dropwise a solution of Ca(NO3)2·4H2O(47.2 mg,0.2 mmol)in 0.5 mL distilled water.The mixture was stirred for 4 hours at room temperature,and then a pale yellow precipitate was formed.After a filtration,the precipitate was washed with distilled water and THF,then dried thoroughly under vacuum to give a light yellow powder of 1 (76.8 mg,Yield:85.6%).The suitable single crystals for X-ray crystal structure determination were obtained by slow evaporation of the methanol solution of 1.Elemental analysis Calcd.for C42H40Ca2N8O10(%):C,56.24;H,4.49;N,12.49.Found(%):C,56.13;H,4.35;N,12.39.IR (KBr,cm-1):2 945,2 858,1 647,1 600,1 545,1 328,1 008,755.ESI-MS:m/z=834.58,386.58,348.58.
[Cd(HL)(NO3)2](2):The procedure was the same as that for 1 except using Cd(NO3)2·4H2O(61.7 mg;0.2 mmol)to replace Ca(NO3)2·4H2O.Yield:82.3%.Elemental analysis Calcd.for C21H21CdN5O8(%):C,43.20;H,3.63;N,12.00.Found(%):C,43.11;H,3.52;N,11.89.IR(KBr,cm-1):3 445,2 980,2 868,1 655,1 605,1 550,1 433,998,747.ESI-MS:m/z=983.78,521.17,348.57.
[Cd2L2(NO3)2]·H2O(3):A solution of NaOH(4 mg,0.1 mmol)in 5 mL methanol was added dropwise to a solution of 2(58.4 mg,0.1 mmol)in 10 mL methanol with stirring.Then the reaction mixture was heated for 1 h at 50℃.After the resulting suspension was filtered to remove any insoluble impurities,slow evaporation of the filtrate gave light yellow single crystals of 3(37.9 mg,Yield:71.6%).Elemental analysis Calcd.for C42H40Cd2N8O11(%):C,47.70;H,3.81;N,10.59.Found(%):C,47.83;H,3.94;N,10.41.IR(KBr,cm-1):3 568,2 953,2 871,1 645,1 606,1 552,1 427,1 003,757.
1.3 Crystal structure determination
The well-shaped single crystals of 1~3 were selected for X-ray diffraction study.The unit cell parameters and intensity data were collected on a Bruker SMARTAPEXⅡCCDdiffractometer using a graphitemonochromated Mo Kα (λ=0.071 073 nm)radiation.The structures were solved by direct methods and refined on F2by full matrix least squares procedures using SHELXTL-2018 software.All non-hydrogen atoms were anisotropically refined.All H atoms were fixed in calculated positions and refined isotropically.In 2,atoms O5 and O5A of one NO3-anion were found to be highly disordered with an occupancy factor of 0.70 and 0.30,respectively.In 3,lattice water O1w and O1wA were found to be highly disordered with an occupancy factor of 0.205 and 0.295,respectively,and the hydrogen atoms on the water were not calculated.The crystallographic data of 1~3 are summarized in Table 1 and the selected bond lengths and angles are given in Table 2.
CCDC:1465302,1;1465303,2;1473801,3.

Table 1 Crystal data and structure refinement for 1~3

Continued Table 1

Table 2 Selected bond distances(nm)and bond angles(°)for 1~3
2 Results and discussion
2.1 Crystal structure of 1
Single crystal X-ray structure analysis shows that 1 crystalizes in the monoclinic system with space group P21/c.A projection of the structure of 1 with the atomic labeling system is illustrated in Fig.1a.The asymmetric unit is composed of two Ca(Ⅱ) ions,two L-ligands and two NO3-anions.The two Ca(Ⅱ)ions are doubly bridged by two phenolatoμ2-O-ions to form a nearly parallelogram [Ca2O2]unit with a Ca1…Ca2 distance of 0.376 41(11)nm.The Ca1-O1(0.238 1 nm)and Ca2-O6(0.239 1 nm)distances are approximately equal,so do the Ca1-O6 (0.233 8 nm)and Ca2-O1(0.234 6 nm)distances(Table 2).Both Ca1 and Ca2 ions are eight-coordinated by three N atoms and one phenolato O atom from one L-,one phenolato O atom and an aldehyde O atom from another L-ligand,and two O atoms from one NO3-anion to show a distorted dodecahedron configuration[CaN3O5](Fig.1b,1c).This configuration can be further demonstrated by the SHAPE software with the continuous symmetry measures(CSM)[26]of Ca1 and Ca2 being calculated as 1.61 and 1.51,respectively.The L-ligand adopts a pentadentate coordination mode via two pyridyl N atoms and one N atom of tertiary amine,one phenolato O atom and one aldehyde O atom.These features are quite similar to those observed in two reported binuclear complexes[Co2L2](ClO4)2and[Mn2L2](BF4)2[27].The phenol ring with O1 makes dihedral angles of 33.0°and 12.9°with the pyridyl ring with N1 and N3,respectively.The dihedral angle between N1-pyridyl and N3-pyridyl rings is 21.4°.The phenol ring containing O6 makes dihedral angles of 29.2°and 29.1°with the pyridyl ring containing N5 and N7,respectively.The dihedral angle between N5-pyridyl and N7-pyridyl rings is 9.1°.All these values are smaller than those found in free HL ligand[21].The crystal structure of 1 is further stabilized by eight kinds of weak intermolecular C-H…Ohydrogen bonds interactions(Table 3 and Fig.2a).
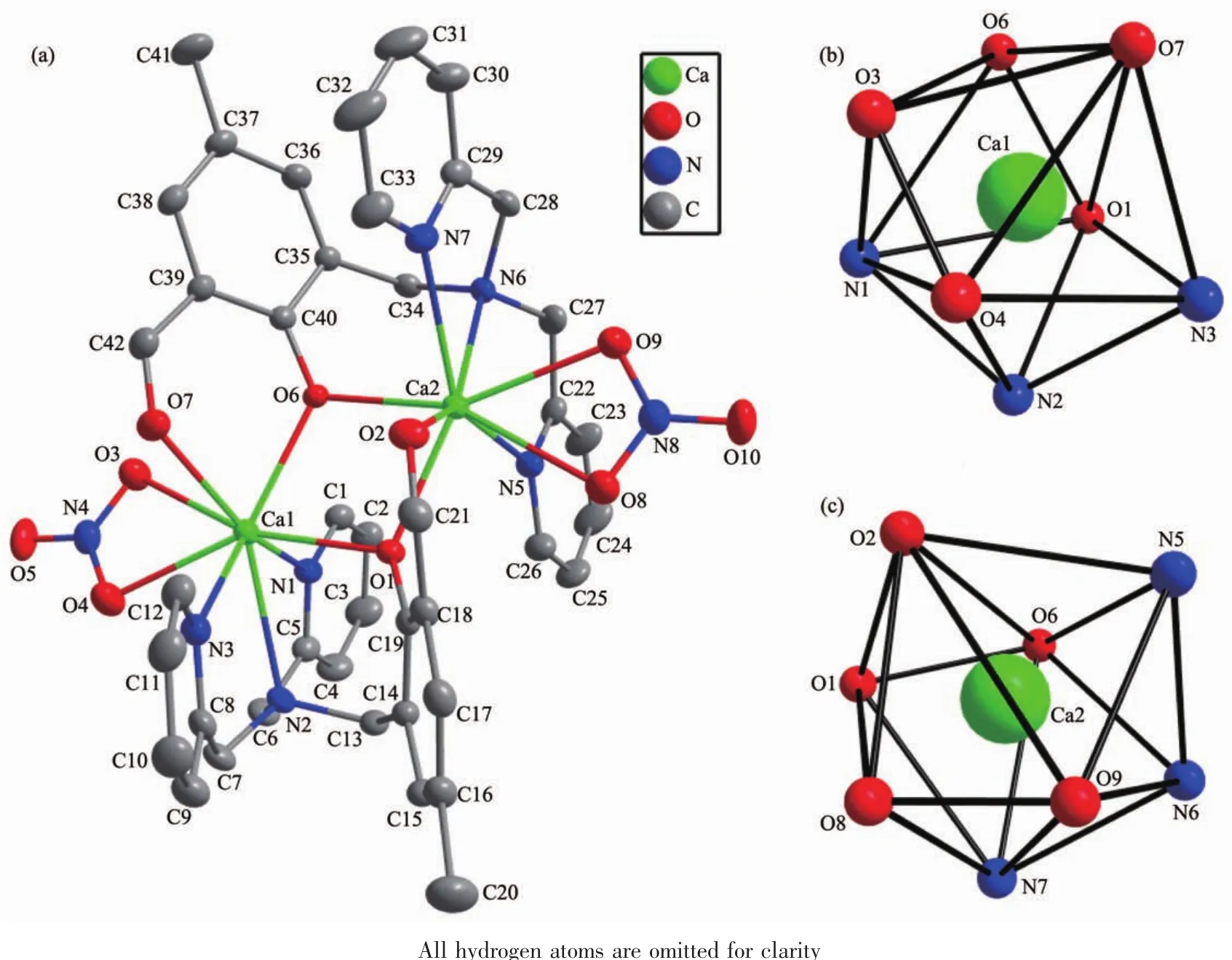
Fig.1 Projection of structure of 1 with 20%thermal ellipsoids probability(a);Distorted dodecahedron configuration of Ca1(b)and Ca2(c)

Table 3 Hydrogen bonding interactions in 1~3

Fig.2 Hydrogen bonding interactions in 1(a),2(b)and 3(c)
2.2 Crystal structure of 2

Fig.3 (a)Projection of structure of 2 with 20%thermal ellipsoids probability;(b)Distorted dodecahedron configuration of Cd1
The structure projection of 2 is presented in Fig.3a together with the atomic labeling system.The complex crystallizes in the triclinic system with space group P1.The asymmetric unit includes one Cd(Ⅱ) ion,one HL ligand and two NO3-anions.The Cd(Ⅱ)cation in 2 is eight-coordinated by three N atoms and one phenol O atom from HL ligand and four O atoms from two NO3-anions to exhibit a distorted dodecahedron configuration[CdN3O5](Fig.3b),whereas the aldehyde group does not take part in coordination.The configuration of the Cd1 ion is further determined by the SHAPE software with a CSM of 3.01.Different from that observed in 1,the phenol group in 2 remains protonated,and can be best regarded as being semicoordinated to the Cd1 ion with a Cd1-O1 distance of 0.265 7(2)nm.This bond length is quite longer than those found in the related literatures[28-29].The Cd1-N bond lengths are from 0.229 5(2)to 0.245 5(2)nm,which are in the normal ranges observed for the related Cd complex[30].The HL ligand adopts a tetradentate coordination mode through two pyridyl N atoms,one N atom of tertiary amine and one phenol O atom,which is very similar to those found in the related mononuclear complexes[23,31].The phenol ring makes dihedral angles of 38.5°and 30.8°with the pyridyl ring with N1 and N3,respectively.The dihedral angle between two pyridyl rings is 8.4°.All the numbers are smaller than those observed in free HL ligand[21].There are one kind of strong intramolecular O1-H1…O2 hydrogen bond and five kinds of weak intermolecular C-H…O hydrogen bonds interactions in 2(Table 3 and Fig.2b).Notably,each molecule of 2 connects the neighboring ones by two intermolecular C-H…O hydrogen bonds (C10-H10A…O8viiand C17-H17A…O8viii)to form a 2D layer parallel to the ab plane.These layers are further linked by theπ…π stacking interactions(π(N1-py)… πvi(N1-pyvi),Cg1 …Cg1vi0.361 6 nm,Symmetry codes:vi1-x,1-y,1-z)between two pyridyl rings to produce a 3D supramolecular network(Fig.4).

Fig.4 Three dimensional supramolecular network of 2
2.3 Crystal structure of 3
Single crystal X-ray structure analysis reveals that 3 crystalizes in the monoclinic system with space group C2/c.A structure projection of 3 with the atomic labeling system is shown in Fig.5a.The asymmetric unit consistsof one Cd(Ⅱ) ion,one L-ligand,one NO3-anion and a half lattice H2O molecule.The two Cd(Ⅱ)ions are doubly bridged by two phenolato μ2-O-ions to form a perfect parallelogram[Cd2O2]unit with a Cd1…Cd1idistance of 0.366 3(4)nm.The Cd1-O1ibond length(0.238 4 nm)is longer than Cd1-O1 one(0.223 9 nm)(Table 2).The Cd1 ion is sevencoordinated by three N atoms and one phenolato O atom from one L-,one phenolato O atom and an aldehyde O atom from another L-ligand,and one O atom from one NO3-anion to show a distorted monocapped trigonal prism configuration[CdN3O4](Fig.5b).Two triangular bases are composed of O1,O1i,O3 and O2,N3i,N1i,respectively,with a dihedral angle of 8.7°,while the single cap(N2i)is located on a side of the trigonal prism.The Cd1 ion′s configuration is further verified by the SHAPE software with a CSM of 0.85.The coordination mode of L-ligand is the same as that in 1.However,the two NO3-anions in 3 are monodentate mode and sit on the same side of the[Cd2O2]plane,which is quite different from those observed in 1.The Cd1-N and Cd1-O bond lengths are in the normal ranges[30].The phenol ring makes dihedral angles of 46.1°and 25.4°with the pyridyl ring with N1 and N3,respectively,which are smaller than those observed in free HL ligand[21].However,the dihedral angle between two pyridyl rings is 69.1°,which is close to that found in free HL ligand[21].There are four kinds of weak intermolecular C-H…O hydrogen bonds and one type of C-H…πinteraction in the structure of 3(Table 3 and Fig.2c).These interactions are very helpful to stabilize the crystal packing.

Fig.5 (a)Projection of structure of 3 with 20%thermal ellipsoids probability;(b)Distorted monocapped trigonal prism configuration of Cd1
2.4 Spectral characterization
In the IR spectra of complexes 1~3,a strong band at 1 647 cm-1(1),1 655 cm-1(2)and 1 645 cm-1(3)can be attributed to the C=O stretching vibration of the aldehyde group.Compared to 1 and 3,the larger value in 2 indicates that the aldehyde group does not take part in coordination.A band at 1 600 cm-1(1),1 605 cm-1(2)and 1 606 cm-1(3)can be assigned to the coordinated pyridyl ring vibrations[23].Two weak absorption peaks at 2 945 and 2 858 cm-1for 1,2 980 and 2 868 cm-1for 2,2 953 and 2 871 cm-1for 3,are assigned to the C-H stretching vibration.In addition,a broad band at 3 445 cm-1in 2 can be attributable to the O-H stretching vibration of phenol,while a broad band at 3 568 cm-1in 3 can be due to the O-H stretching vibration of the lattice water molecule,suggesting the existence of O-H…O hydrogen bonding interactions in 2 and 3.These features are in consistent with the X-ray crystallography results.
The structures of 1 and 2 in methanol solution were also investigated by electrospray ionization mass spectrometry(ESI-MS).Fig.S1a(Supporting Information)shows the positive ESI mass spectrum of 1 and the peak at m/z=348.58 is H2L+ion.Two peaks at m/z=386.58 and m/z=834.58 are[CaL]+and[Ca2L2(NO3)]+ion,respectively.The positive ESI mass spectrum of 2 is shown in Fig.S1b and a peak at m/z=348.57 is H2L+ion.The two peaks at m/z=521.17 and m/z=983.78 are[Cd(HL)(NO3)]+and[Cd2(L)2(NO3)]+ion,respectively.All these features are in agreement with the results of X-ray analysis.
3 Conclusions
Three new complexes with 2-(N,N-di(2-pyridylmethyl)aminomethyl)-6-aldehydo-4-methylphenol(HL),[Ca2L2(NO3)2](1),[Cd(HL)(NO3)2](2)and[Cd2L2(NO3)2]·H2O(3),have been prepared and characterized by IR,elemental analysis,ESI-MS spectra and X-ray crystal structure analysis.Both 1 and 3 are binuclear complexes doubly bridged by two phenolatoμ2-O-ions to form a[M2O2]parallelogram,while 2 is a mononuclear complex.Complex 2 can be transformed to 3 under basic conditions,showing that the deprotonated phenol group is beneficial for binding two metal ions to form a binuclear complex.
Supportinginformation isavailableat http://www.wjhxxb.cn
