N-((喹啉-8-基)亚甲基)水杨酰肼 Sn(Ⅳ)/Ag(Ⅰ)配合物的合成、晶体结构和DNA相互作用
2019-07-10王贝贝蔡红新梅雪澳张秀秀吴伟娜
王贝贝 蔡红新 梅雪澳 张秀秀 吴伟娜*, 王 元 陈 忠
(1河南理工大学化学化工学院,河南省煤炭绿色转化重点实验室,焦作 454000)(2河南理工大学物理与电子信息工程学院,焦作 454000)(3江西科技师范大学材料与机电学院,南昌 330013)
Schiff bases are an important class of ligands in coordination chemistry and have been found extensive application in many different fields[1-4].The Schiff base derivatives of acylhydrazone have attracted more attention due to their variable bonding modes towards metal ions and wide range of biological properties,such as antioxidant,anti-inflammatory,antibacterial and antitumor activities[5].Asone of the most important heterocycles,quinoline is frequently employed for the construction of transition metal complexes with significant pharmacological activities[6-7].Our group has reported the structuresof several Zn(Ⅱ)/Cd(Ⅱ) complexes with quinoline-containing acylhydrazone[8].Nevertheless,the investigation on Ag(Ⅰ) and Sn(Ⅳ) complexes with such type of ligands are relatively scarce.In fact,Ag(Ⅰ)-based complexes have been known for centuries,especially with respect to medical properties[9-10],and Sn(Ⅳ) compounds are widely used in catalysis,pesticides, polyvinyl chloride stabilizer, preservative,sterilization,anticancer drug and so on[11-13].Herein,we report the crystal structures and DNA binding properties of Sn(Ⅳ) and Ag(Ⅰ)complexes with N-((quinolin-8-yl)methylene)salicylhydrazide(HL,Scheme 1).
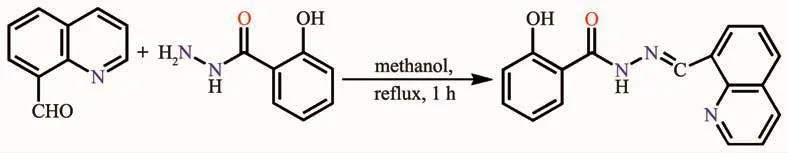
Scheme 1 Synthetic route of HL
1 Experimental
1.1 Materials and measurements
Solvents and starting materials for synthesis were purchased commercially and used as received.Elemental analysis was carried out on an Elemental Vario ELanalyzer.The IRspectra(ν=4 000~400 cm-1)were determined by the KBr pressed disc method on a Bruker V70 FT-IR spectrophotometer.1H NMR spectra of HL was acquired with Bruker AV400 NMR instrument in DMSO-d6solution with TMS as internal standard.The UV spectra were recorded on a Purkinje General TU-1800 spectrophotometer.The interactions between three compounds and EB-DNA are measured using literature method[14]via emission spectra on a Varian CARY Eclipse spectrophotometer with the pass width of emission and excitation being 5 nm.
1.2 Preparations of the ligand HL,complexes 1 and 2
As shown in Scheme 1,the ligand HL was prepared by condensation of quinoline-8-carbaldehyde(1.57 g,10 mmol)and 2-hydroxylbenzoyl hydrazine(1.52 g,10 mmol)in methanol solution(100 mL)under refluxing for 1 h.The white solid was filtered and washed three times by methanol.Yield:2.01 g(69%).Elemental analysis Calcd.for C17H13N3O2(%):C 70.09,H 4.50,N 14.42;Found(%):C 70.26,H 4.29,N 14.55.FT-IR(cm-1):ν(N-H)3 259,ν(O=C)1 651,ν(C=N)1 604,ν(quinoline C=N)1 575.1H NMR(400 MHz,DMSO-d6):δ12.13(1H,s,NH-C=O),12.01(1H,s,OH),9.73(1H,s,CH=N),8.96~8.98(1H)/8.36~8.44(2H)/7.94~8.09(2H)/7.59~7.72(2H)/7.40~7.42(1H)/6.91~6.95(2H)for Ar-H.
The complexes1 and 2 were generated by reaction of the ligand HL(5 mmol)with equimolar of Ph2SnCl2and AgNO3in methanol solution(10 mL),respectively.Crystals suitable for X-ray diffraction analysis were obtained by evaporating the corresponding reaction solutions at room temperature.
1:Yellow blocks.Anal.Calcd.for C29.25H23ClN3O2.25Sn(%):C 57.97,H 3.79,N 6.92;Found(%):C 57.76,H 3.82,N 6.75.FT-IR(cm-1):ν(N=C-O)1 624,ν(C=N)1 589,ν(C=N quinoline)1 575.
2:Colorless blocks.Anal.Calcd.for C18H17N4O6Ag(%):C 43.83,H 3.47,N 11.36;Found(%):C 43.62,H 3.77,N 11.16.FT-IR(cm-1):ν(N-H)3 446,ν(N=C-O)1 615,ν(C=N)1 586,ν(C=N quinoline)1 528,ν(NO3)1 478 and 1 302.
1.3 X-ray crystallography
The X-ray diffraction measurement for HL,complexes 1 and 2 were performed on a Bruker SMART APEXⅡCCD diffractometer equipped with a graphite monochromatized Mo Kα radiation (λ=0.071 073 nm)by usingφ-ω scan mode.Semi-empirical absorption correction was applied to the intensity data using the SADABS program[15].The structures were solved by direct methods and refined by full matrix least-square on F2using the SHELXTL-97 program[16].All non-hydrogen atoms were refined anisotropically.All the other H atoms were positioned geometrically and refined using a riding model.Details of the crystal parameters,data collection and refinements for HL,complexes 1 and 2 are summarized in Table 1.
CCDC:1883407,HL;1892778,1;1883405,2.
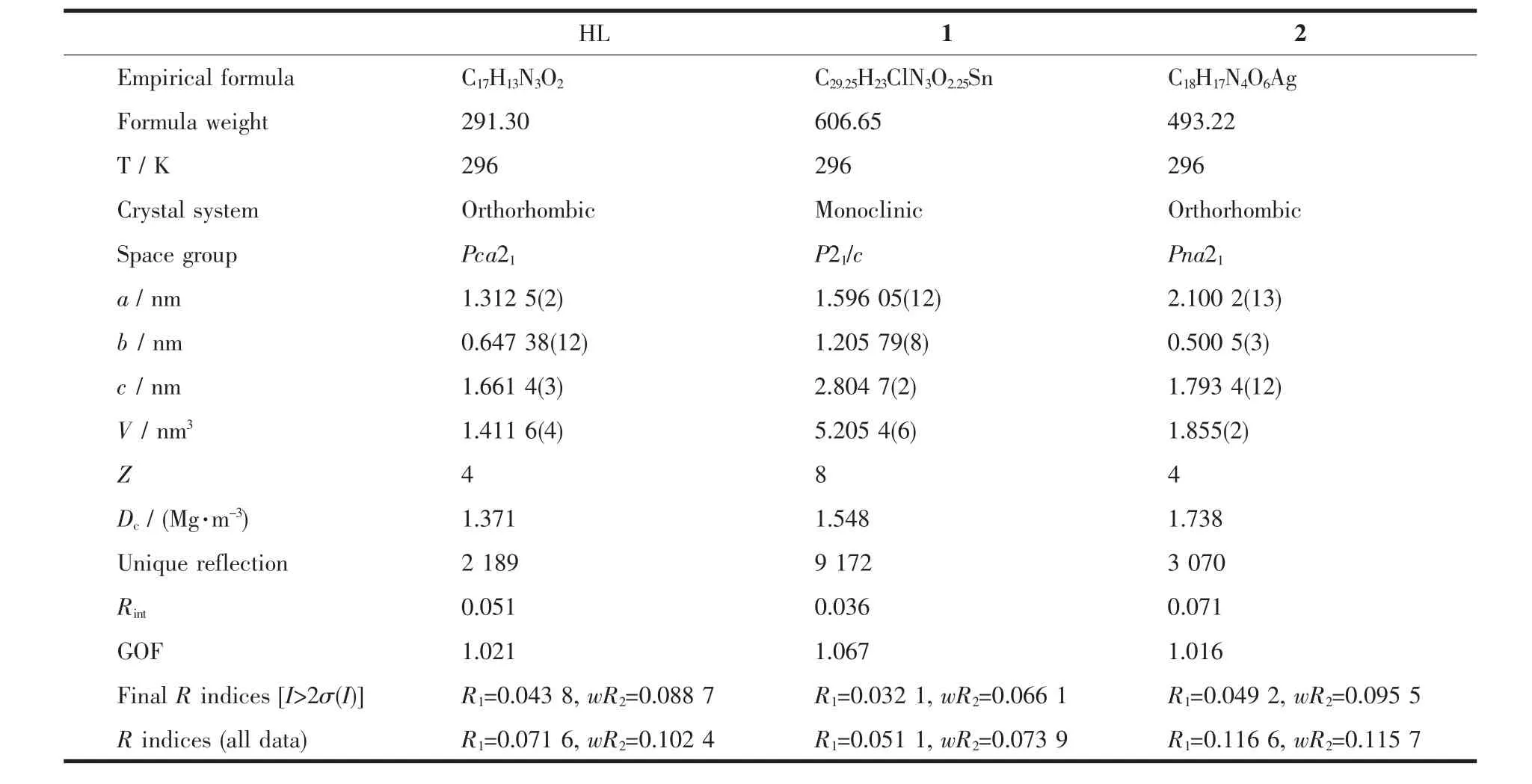
Table 1 Selected crystallographic data for HL,complexes 1 and 2
2 Results and discussion
2.1 Crystal structures description
A diamond drawing of HL,complexes 1 and 2 are shown in Fig.1a~c.Selected bond distances and angles are listed in Table 2.The bond length of carbonyl group in HL is 0.122 8(4)nm,which is similar as that in complex 2(0.124 9(15)nm),while obviously shorter than that in complex 1(0.128 1(4)and 0.128 9(4)nm).This fact shows that HL is anionic and neutral in complexes 1 and 2,respectively.
As shown in Fig.1a,the asymmetric unit of complex 1 contains two independent complexes molecules and a half of lattice water molecule.Each six-coordinated Sn(Ⅳ) ion is surrounded by two nitrogen atoms coming from quinoline and imine groups,one chloride ion,one oxygen atom coming from carbonyl group and two carbon atoms coming from two phenyl groups,exhibiting a distorted octahedral coordination geometry.There exist intramolecular OH…N hydrogen bonds(O2-H2…N3,O4-H4…N6 and O5-H5A…Cl1,with D…A distance being 0.257 8,0.259 1 and 0.341 0 nm,D-H…A angle being 145°,144°and 147°,respectively)in the structure.
In complex 2,the Ag(Ⅰ) ion is surrounded by one tridentate hydrazone ligand with ON2donor set and one bidentate nitrate anions,possessing a distorted square pyramid coordination geometry (τ=0.338).Furthermore,in the solid structure of 2,the complex molecules are linked by lattice methanol molecules into chains along c axis via various of intermolecular O-H…O hydrogen bonds(O2-H2A…O6,with D…A distance being 0.272 5(18)nm,D-H…A angle being 122.2°;O6-H6A…O4i,with D…A distance being 0.301 1(17)nm,D-H…A angle being 161.1°;O6-H6A…O5i,with D…A distance being 0.296 4(18)nm,D-H…A angle being 132.4°,Symmetry codes:i1.5-x,0.5+y,-0.5+z).The intramolecular O-H…O hydrogen bonds between amine N atoms and hydroxyl O atoms(N3-H3A…O2,with D…A distance being 0.259 8(15)nm,D-H…A angle being 130.7°)are also present.

Fig.1 Diamond drawing of HL(a),and complexes 1(b)and 2(c)with 10%thermal ellipsoids;Chain-like structures in complex 2 formed by hydrogen bonds shown in dashed line(d)

Table 2 Selected bond lengths(nm)and angles(°)in HL,complexes 1 and 2

Continued Table 2
2.2 IR spectra
The FT-IR spectral region for both complexes are more or less similar due to the similar coordination modes of the ligands.Theν(C=O)of the free ligand was 1 651 cm-1,and it disappeared in the spectra of complexes.Meanwhile,new N=C-O stretching vibration absorption was observed at 1 624 and 1 615 cm-1in complexes 1 and 2,respectively,revealing that the C=O in O=C-N moiety has enolizated and the oxygen atom coordinates to the metal ion in both complexes[8].The ν(C=N)bands of the imine group,quinoline ring in the hydrazone ligand shifted to lower frequency values in the complexes,indicating that the N atoms of such two units take part in the coordination[17-18].In addition,the general pattern of the IR spectroscopy for complex 2 supports the normal coordination of bidentate nitrate group[19].It is in accordance with the crystal structure study.
2.3 UV spectra

Fig.2 UV spectra of the ligand HL,complexes 1 and 2 in CH3OH solution at room temperature
The UV spectra of HL,complexes 1 and 2 were measured in CH3OH solution(c=20 μmol·L-1)at room temperature (Fig.2).The spectra of HL featured two bands located at 229 nm(ε=50 168 L·mol-1·cm-1)and 336 nm (ε=34 974 L·mol-1·cm-1),which could be assigned to characteristic π-π*transition centered on quinolone ring and the imine moiety[20],respectively.Both bands exhibited significant hyperchromicity in the spectra of both complexes(ε=93 738 and 57 310 L·mol-1·cm-1for complex 1;ε=81 974 and 53 125 L·mol-1·cm-1for complex 2),indicating that the ligand HL takes part in the coordination[21].
2.4 EB-DNA binding study by fluorescence spectra
It is well known that EB can intercalate nonspecifically into DNA,which causes it to fluoresce strongly.Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and a decrease in the fluorescence intensity[22-23].The effects of the ligand and complexes on the fluorescence spectra of EB-DNA system are presented in Fig.3.The fluorescence intensities of EB bound to ct-DNA at about 600 nm showed remarkable decreasing trends with the increasing concentration of the tested compounds,indicating that some EB molecules are released into solution after the exchange with the compounds.The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation:I0/I=1+Ksqr[22],where I0and I represent the fluorescence intensities in the absence and presence of quencher,respectively;Ksqis the linear Stern-Volmer quenching constant;r is the ratio of the concentration of quencher and DNA.In the quenching plots of I0/I versus r,Ksqvalues are given by the slopes.The Ksqvalues were 0.632 1,0.790 3 and 0.791 9 for the ligand HL,complexes 1 and 2,respectively.The results indicate that interactions of the complexes with DNA are stronger than that of the ligand HL,because the complexes have higher rigidity to bind the base pairs along DNA,thus increasing their binding abilities[14].

Fig.3 Emission spectra of EB-DNA system in the presence of HL(a)and complexes 1(b)and 2(c),respectively
