代谢组学在家养动物遗传育种中的应用
2019-02-28周萌景军红毛瑞涵郭静王志鹏
周萌,景军红,毛瑞涵,郭静,王志鹏
代谢组学在家养动物遗传育种中的应用
周萌1,2,3,景军红1,2,3,毛瑞涵1,2,3,郭静1,2,3,王志鹏1,2,3
1. 农业农村部鸡遗传育种重点试验室,哈尔滨 150030 2. 黑龙江省普通高等学校动物遗传育种与繁殖重点试验室,哈尔滨 150030 3. 东北农业大学动物科学技术学院,哈尔滨 150030
代谢组学是使用分析化学技术对生物样品(如乳液、血浆、血清等)中的大量小分子代谢物进行全面鉴定和定量分析,已经广泛应用于生物医学、营养学、作物学和畜牧学研究中。最初,代谢组学主要应用于畜牧生产中的非遗传学研究。目前,随着生理基因组学、生理遗传学研究的增多,越来越多的研究者开始利用代谢组学的技术和方法开展动物遗传育种研究。本文综述了代谢组学检测技术与平台特点及代谢组学在动物遗传学与基因组中的应用,着重总结了动物代谢分子遗传参数估计、品系(品种)间代谢图谱差异、代谢组全基因组关联分析、筛选影响重要经济性状的生物标记等领域的研究进展,讨论了代谢组学研究还需亟待解决的问题。本文通过综述代谢组学在动物育种中的研究进展,旨在为进一步利用代谢组学技术开展动物重要经济性状的遗传基础研究提供参考。
代谢组学;家养动物;遗传学;动物育种
代谢组学(metabonomics)是对生物体内所有代谢分子进行定量分析,寻找代谢分子与生理、病理变化的相对关系的研究,是继基因组学和蛋白质组学之后新近发展起来的研究某一时刻细胞内所有代谢分子集合的一门学科[1,2]。与其他组学相比,代谢组学具有以下特点:基因和蛋白质表达的微小变化在代谢分子上得到的放大使代谢组学检测更加容易;代谢组学技术需要相对完整的代谢分子信息库,但其远没有全基因组测序及大量表达序列标签的数据库复杂;代谢分子种类远小于基因和蛋白质的数量,其物质的分子结构也更为简单[3]。
随着高通量测序技术和生物信息学的发展,代谢组学也已成为生物学研究的重要领域,已广泛应用在人类生物医学研究中,并取得了丰硕的成果,特别是在疾病的生物标志物研究、疾病机理研究等方面都展现出巨大的应用潜力与优势[4~8]。但是,在畜牧生产和研究中代谢组学的应用相对较少。根据已发表的相关文献,其应用主要集中在评估胴体品质和牛奶质量、预测饲料效率或剩余饲料摄入量(residual feed intake, RFI)、生殖生理学研究、营养生理学研究、疾病检测的生物标记开发和药物使用检测等非遗传学研究方面。而利用代谢组学开展动物遗传学研究长期被忽视,直到最近以精确、高通量为特点的动物表型组学以及生理基因组学、生理遗传学研究的开展,科研人员在解析动物重要经济性状遗传机制的研究中才更多地使用了代谢组学的技术和方法[9]。
动物基因组上的变异不一定直接对动物表型产生影响,而相关代谢分子的变化可能是该变异的最 终结果。换言之,基因组上的变异需通过相关代谢分子的转化而最终传递到表型。这可能是科研人员在将表型和遗传变异直接进行关联分析时,遗漏一些变异位点的原因之一,利用代谢组学数据则可以筛查出这些遗漏的位点[10]。随着代谢组学的快速发展,与其他组学数据的整合研究将有助于解析动物生命活动许多复杂的调控机制。
本文比较了代谢组学检测技术的优缺点,概述了利用代谢组学技术在主要家养动物体液中所检测到的代谢分子的分布情况,综述了关键代谢标志物在遗传育种领域中的应用,以期为进一步利用代谢组学技术开展动物重要经济性状的遗传基础研究提供参考。
1 代谢组学的分类及检测平台
1.1 代谢组学的分类
代谢组学的研究对象大都是相对分子质量1000以内的小分子物质,到目前为止,有114 100个代谢分子被收录到人类代谢组学数据库[11]。根据不同的理化属性可以将代谢组学所包含的物质主要分为氨基酸类(amino acid)、肽类(peptide)、碳水化合物类(carbohydrate)、能量类(energy)、脂类(lipid)、核苷酸(nucleotide)、维生素和辅助因子(cofactors and vitamins)及外源化合物类(xenobiotics);根据代谢分子的产生来源则可以分为内源性代谢分子和外源性代谢分子。根据代谢组学研究策略的不同,代谢组学研究可分为非靶标代谢组学和靶标代谢组学[9],这两种研究策略相辅相成,从发现到验证,从“无假设”到“假设驱动”,共同组成了代谢组学研究[12]。代谢组学包含物质及研究策略的分类特点见表1。
1.2 代谢组学检测平台
目前用于开展高通量代谢组学研究最常见的仪器平台主要包括核磁共振仪(nuclear magnetic resonance, NMR)、质谱仪(mass spectrometer, MS)、色谱-质谱联用仪(chromatograph-mass spectrometer, C-MS)。Goldansaz等[13]详细介绍了上述检测技术平台的特性(图1)及其应用。NMR是最早应用于代谢组学研究的高通量分析技术,其优势在于对样品检测前的预处理较为简单,且对样品无破坏性;其缺点在于检测灵敏度较低,只能检测μmol/L~mmol/L浓度的代谢分子。质谱技术具有高灵敏度和专属性等优势,能够检测nmol/L~pmol/L浓度的代谢分子。近年来,代谢组学研究普遍采用的方法是色谱-质谱联用技术,主要包括气相色谱-质谱(gas chromatography- mass spectrometry, GC-MS)和液相色谱-质谱(high performance liquid chromatography-mass spectrometry, LC-MS)联用技术。该技术的优势在于检测覆盖率达到前所未有的高度,可以检测到更多的小分子代谢产物,包括糖、糖醇、氨基酸、有机酸、脂肪酸和芳胺,以及包括大量次级代谢分子在内的数百种化学性质不同的化合物[14]。由于GC-MS需要对挥发性较低的代谢分子进行衍生化预处理,样品制备较为繁琐,甚至会引起样品的变化,因此与LC-MS相比,GC-MS没有被普遍使用。
2 代谢组学技术检测家养动物体液中关键代谢分子
在家养动物代谢组学研究中,主要是利用核磁技术和质谱技术测定血浆、血清、乳汁、尿液、粪便、瘤胃、肌肉、脂肪等样品的代谢分子。其中对血浆、血清、乳汁中的代谢组学研究最多,所检测到的代谢分子也最多。基于目前报道的检测结果,Goldansaz等[13]将在不同物种、不同样品中通过代谢组学技术所检测到的1070个代谢分子收录在LMDB数据库,该数据库中除了收集每个代谢分子的理化特性以外,重点收录了特定物种的特定样品内该代谢分子的标准化浓度等注释信息。通过检索该数据库,在不同动物的血浆中累计检测到408个代谢分子,在血清中累计检测到351个代谢分子,在乳汁中累计检测到422个代谢分子。上述检测到的代谢分子约70%来自牛和猪的组织或体液,在不同年份猪和牛各种组织或体液中所检测到代谢分子的分布情况见图2。随着敏感度更高的检测设备在动物代谢组学中的应用,动物各种组织或体液中的代谢分子将会被陆续检测出来。
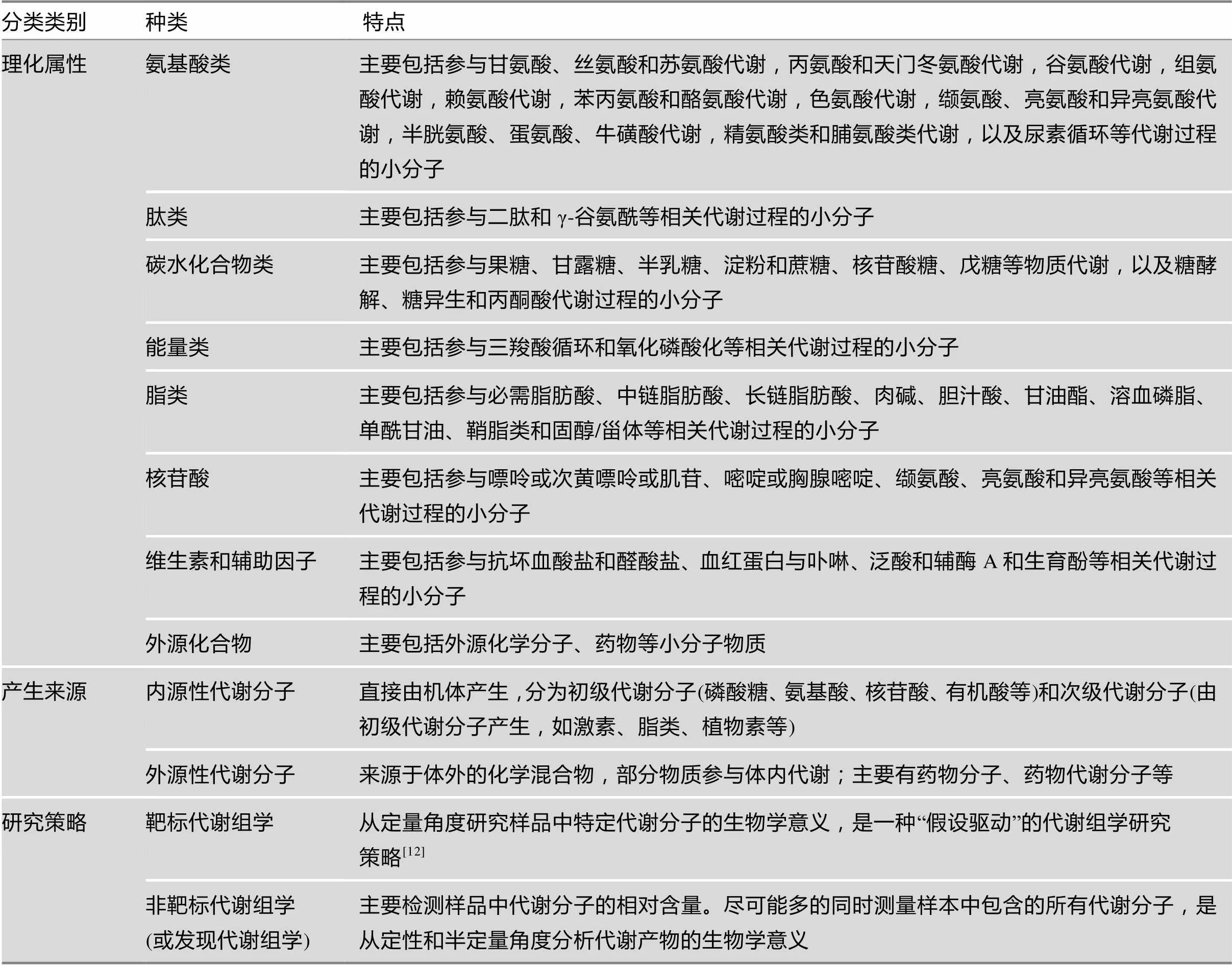
表1 代谢组学包含物质及研究策略的分类特点
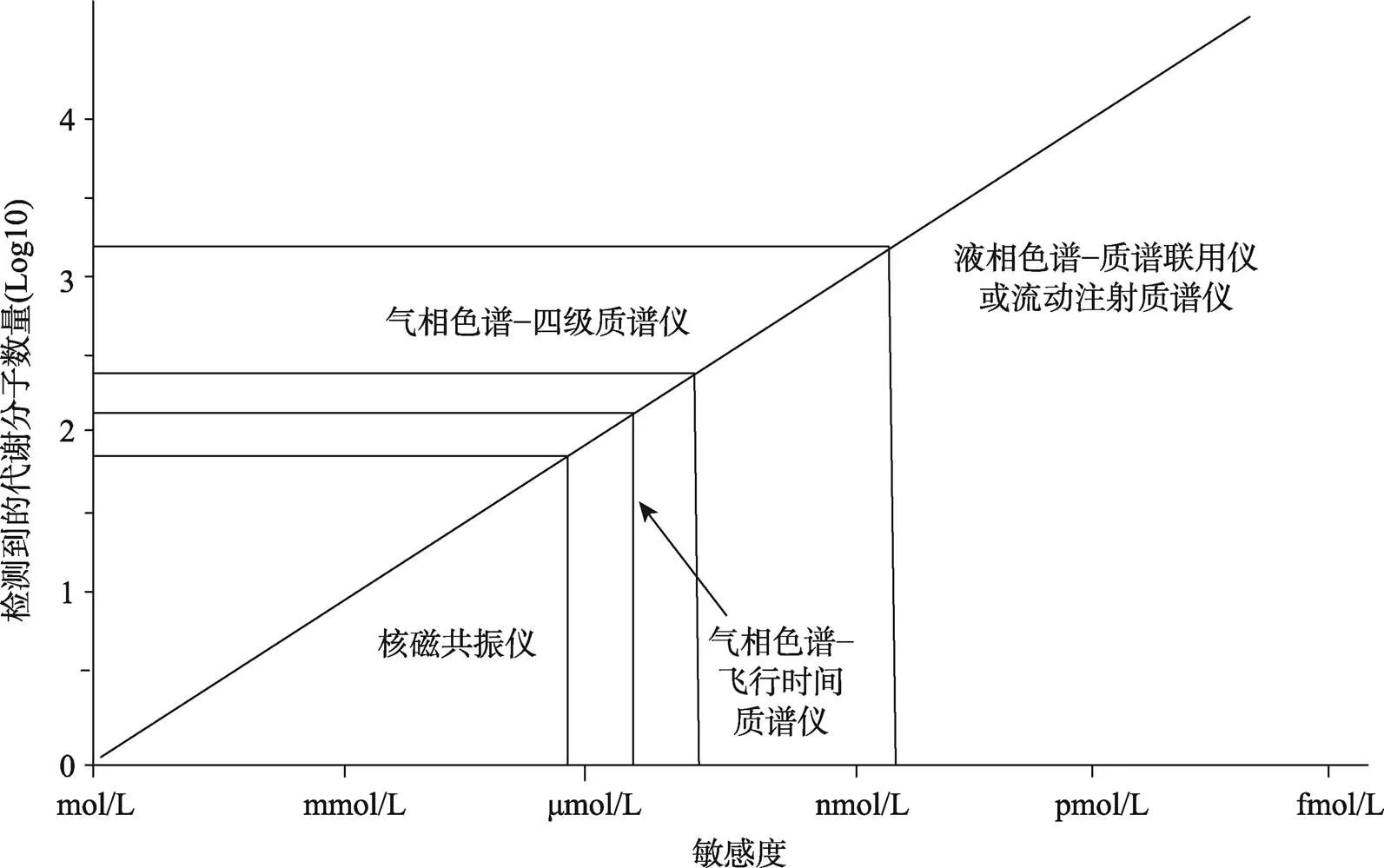
图1 不同代谢组学检测平台的敏感度
根据文献[13]修改绘制。
3 家养动物关键代谢标志物在遗传育种中的应用
如前所述,在家养动物上利用代谢组学的研究手段开展复杂性状遗传学机制的研究较少。目前所开展的相关研究主要集中在代谢分子遗传参数估计、筛选能够鉴别不同品种(系)的生物标记、代谢分子全基因组关联分析研究(genome-wide association studies with metaotypes, mGWAS)以及寻找代谢分子与重要经济性状的关系等方面。为了系统地解析乳品质或肉品质等相关性状的遗传学机制,目前对牛和猪的研究较多。相对而言,在其他家养动物上利用代谢组学开展遗传育种方面的研究较少,其研究多集中于营养生理学研究、疾病检测的生物标记开发和药物或食品添加剂使用效果检测等非遗传学研究。
3.1 动物代谢分子遗传参数估计
机体内的代谢水平会随时间而发生变化,更易受到环境和生活习惯的影响。因此,每个生物样品的代谢组学图谱仅仅是刻画了该个体在特定时间的代谢状态。在将代谢分子的表达量作为中间性状开展遗传学分析时,需要充分了解遗传因素解释这些代谢分子表型变异的份额,即需要对代谢分子表达水平开展遗传参数估计工作。如果期望将鉴定的代谢分子应用于动物遗传评估,估计这些代谢分子的遗传参数则更是一项必要的研究工作。在牛、猪和鸡等家养动物群体中,科研人员已经系统地估计了各类代谢分子的遗传参数。
对牛的代谢分子估计遗传参数的研究较多。Soyeurt等[15]检测了瑞士褐牛(Brown Swiss)、比利时蓝牛(Dual-Purpose Belgian Blue)、荷斯坦-弗里斯牛(Holstein Friesian)、泽西牛(Jersey)、蒙贝利亚牛(Montbeliarde)、诺曼地牛(Normande)等总计7700个牛乳样品,分析发现牛乳中不同长链脂肪酸代谢分子的遗传力在0.05~0.38之间,它们之间的遗传相关在−0.06~0.84之间。Stoop等[16]估计了1953只荷兰荷斯坦(Holstein)黑白花母牛尿素氮的遗传力,分析发现该代谢分子为低遗传力性状(2=0.14)。Oikonomou等[17]对初次泌乳奶牛的血清代谢分子开展了遗传参数估计,研究发现葡萄糖的遗传力为0.12~0.39,β-羟基丁酸酯为0.08~0.40,非酯化脂肪酸为0.08~ 0.35。Nogi等[18]对日本黑牛(Japanese Black cattle)进行了研究,单不饱和脂肪酸、饱和脂肪酸和多不饱和脂肪酸遗传力估计值分别为0.68、0.66和0.47。Buitenhuis等[19]对456只丹麦荷斯坦奶牛和436只丹麦泽西牛的牛乳开展了代谢分子的遗传力估计,其中乳清酸和β-羟基丁酸的遗传力均大于0.8。Wittenburg等[20]利用GC-MS测定了1295头奶牛乳中190种代谢分子的遗传力,发现这些代谢分子的广义遗传力为0~0.699,中位数为0.125。Gebreyesus等[21]对650头丹麦荷斯坦奶牛主要乳蛋白的遗传力进行估计,发现不同乳蛋白的遗传力为0.05~0.78。
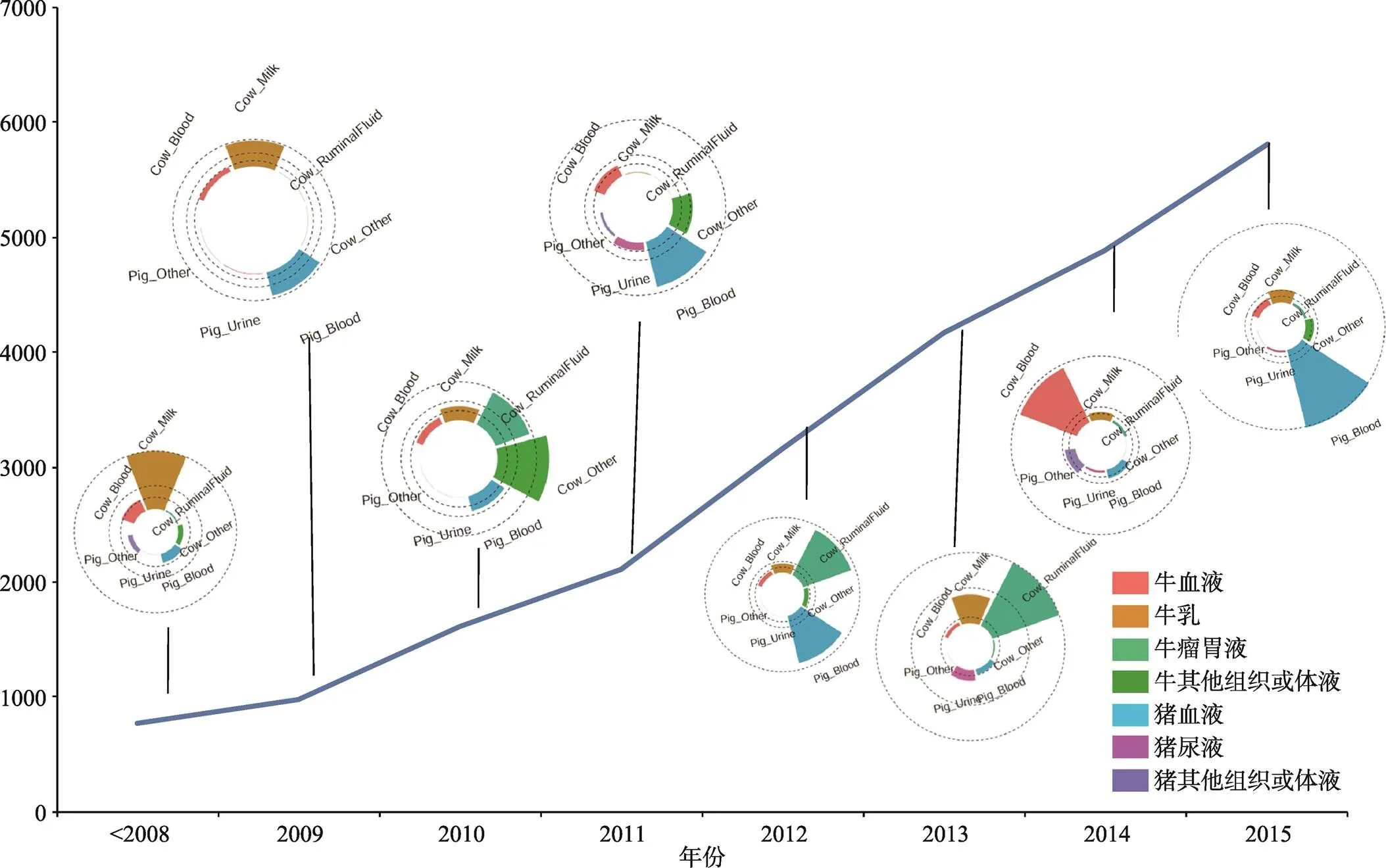
图2 在不同年份牛和猪中检测到的代谢分子数
数据来自于LMDB数据库。X轴表示年份,Y轴表示每年度在牛和猪不同组织或体液上检测到的代谢分子总数;每年度的圈图表示该年度在牛和猪不同组织或体液中检测到的代谢分子的分布情况,其中扇面颜色表示所检测的组织或体液类型,扇面的面积大小代表检测到的代谢分子数。
在对猪的研究中,重点估计了与肉质性状相关的各种长链脂肪酸的遗传力。Ntawubizi等[22]估计猪肉肌内脂肪酸的组成和参与多不饱和脂肪酸代谢的去饱和酶和延长酶活性指标的遗传参数,发现长链多不饱和脂肪酸的遗传力值通常在0.50以上。Ibáñez-Escriche等[23]对伊比利亚猪(Iberian pig)皮下脂肪组织的不同长链脂肪酸的遗传力进行估计,发现这些脂质代谢分子的遗传力范围为0.06~0.53。
另外,科研人员对特定几项鸡血液生化指标开展了遗传参数估计研究。Dong等[24]以肉鸡高、低脂双向选择品系为实验材料,估计了多项血液生化指标的遗传参数,研究发现总胆汁酸、肌酐、低密度脂蛋白胆固醇的遗传力为0.60~0.85。Zhang等[25]测定了332只广西黄鸡进食和禁食状况下的血液生化指标以及腹部脂肪性状,研究发现,在进食状态下,甘油三酯、总胆固醇等血液生化指标的遗传力较高,从0.26~0.60不等,在禁食状态下,这些血液生化指标的遗传力为0.22~0.59。
3.2 动物代谢分子在品种(系)中的鉴定
在对牛、猪和鸡等家养动物的代谢组学研究中,通过检测动物血清、组织、乳汁等研究材料,筛选出一些代谢分子作为区分品种(系)的生物标记物。目前,已有多篇文献报道了牛、猪、鸡的不同品种(系)间存在显著差异表达的代谢分子,这些代谢分子可以作为品种(系)鉴定的生物标记。例如:Karisa等[26]利用NMR技术检测了纯种安格斯牛(Augus)和杂交牛(58.3%安格斯、30.6%西门塔尔(Simmemal)和11.1%海福特牛(Hareford)等欧系肉牛)的血浆代谢图谱,发现肌酸、肉碱、马尿酸等代谢分子的表达在两个品种间差异显著。D'Alessandro等[27]分析发现可以用甘油-3-磷酸脱氢酶、甘油3-磷酸和甘油代谢分子来区分高脂的卡斯塔纳猪(Casertana)和瘦肉型大白猪(Large White);He等[28]用代谢组学方法比较肥胖型猪(宁乡品系)和瘦肉型杂交猪间的血清代谢图谱,发现宁乡品系猪血清的胰岛素、胰高血糖素、脂质、不饱和脂质、糖蛋白、肌肉肌醇、丙酮酸盐、苏氨酸、酪氨酸和肌酸均显著高于瘦肉型猪,而血清中葡萄糖和尿素则低于瘦肉型猪;Straadt等[29]通过NMR技术,检测了杜洛克/长白/约克夏(Duroc/ Landrace/Yorkshire)、伊比利亚/杜洛克(Iberian/Duroc)、伊比利亚/杜洛克/长白(Iberian/Duroc/Landrace)、曼加利萨/杜洛克(Mangalitza/Duroc)和曼加利萨/长白/约克夏(Mangalitza/Landrace/Yorkshire)5个杂交组合的猪背最长肌的代谢分子,发现氨基酸(丙氨酸、肌肽、异亮氨酸、甲硫氨酸、苯丙氨酸和缬氨酸)、乳酸盐、肌苷酸、肌苷、甘油和含胆碱的化合物等可以区分这些杂交猪;Bovo等[30]利用质谱技术检测了大白猪和杜洛克猪的血浆、血清,共检测到180种代谢分子,其中鞘磷脂在杜洛克猪中表达量较高,乙酰鸟氨酸在大白猪中表达量较高。Ji等[31]利用LC-MS检测来航鸡(Leghorn)、Fayoumi和商业肉鸡的脂肪组织,共检测到92种代谢分子,与商业肉鸡相比,Fayoumi和来航鸡脂肪组织中的肉碱、鸟苷、胞嘧啶、腺苷和磷酸戊糖含量显著增加,来航鸡脂肪组织中的3-磷酸甘油酸和磷酸烯醇丙酮酸含量较高;Baéza等[32]利用H-NMR技术,通过比较饲喂同样日粮、同一日龄的腹脂双向选择系公鸡的血浆,发现谷氨酰胺、组氨酸、甜菜碱等代谢分子的表达在两系间差异显著。
3.3 动物代谢分子的全基因组关联分析
基于基因组学研究技术,特别是全基因组SNP标记检测平台,将血清、血液、尿液等体液所检测到的代谢分子表达量作为表型值,开展了代谢分子全基因组关联分析,这是将代谢组学与基因组学耦合在一起的关键,也是目前代谢组学领域以遗传学研究为落脚点,鉴定代谢分子功能与遗传调控方面最主要的研究进展之一。从已有的文献报道可以发现,人类代谢组学mGWAS研究结果极为丰富,且已经筛选到多个与代谢分子表达显著相关的遗传标记或基因,通过进一步的分析发现这些代谢分子可以作为人类疾病的生物标记。家养动物mGWAS的报道相对较少,主要集中在牛和猪上,其目的是通过对某一类代谢分子的全基因组关联分析筛选出影响肉质或乳质的遗传标记或重要候选基因。例如,已筛选出DGAT1、FASN、SCD、ELOVL家族等重要候选基因与牛乳液和肌肉中各种不同链长的脂肪酸含量显著相关;筛选出FASD家族、ELOVL家族等重要候选基因与猪最长肌中脂肪酸含量显著相关。其他研究结果详见表2。在动物群体上开展mGWAS的样本量规模都比人类研究群体的规模小很多,这是由于:一方面,在饲养过程中可以有效的控制饲养环境,使动物所受到的环境因素趋于一致;另一方面,试验群体的遗传背景相似。
3.4 与动物重要经济性状相关的代谢分子鉴定
代谢组学所反映的小分子物质的产生和代谢能更直接、准确地反映生物体的生理状态和生理表型,是复杂表型的分子展现。相对于表型数据而言,代谢分子更容易进行标准化和批量化的分析检测,所检测得到的数据更为准确。筛选与重要经济性状或疾病相关的代谢分子是代谢组学研究的重要热点。在人类代谢组学研究方面,通过比较对照与处理组之间的代谢组学差异筛选出表征疾病的生物标记,或通过将代谢组学和其他组学整合分析筛选出影响复杂疾病发生、发展的重要代谢分子。
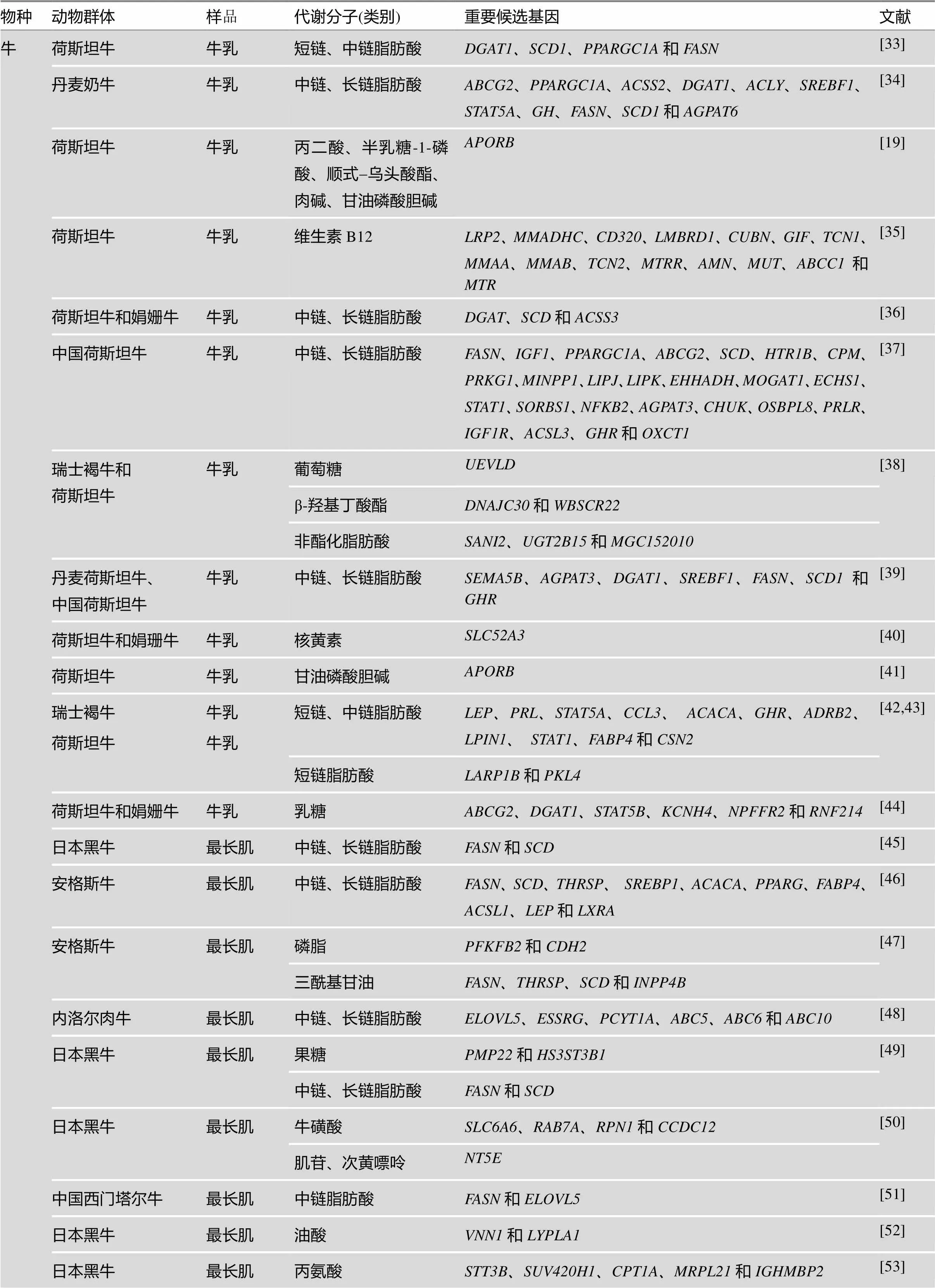
表2 牛、猪、鸡mGWAS研究结果汇总
续表
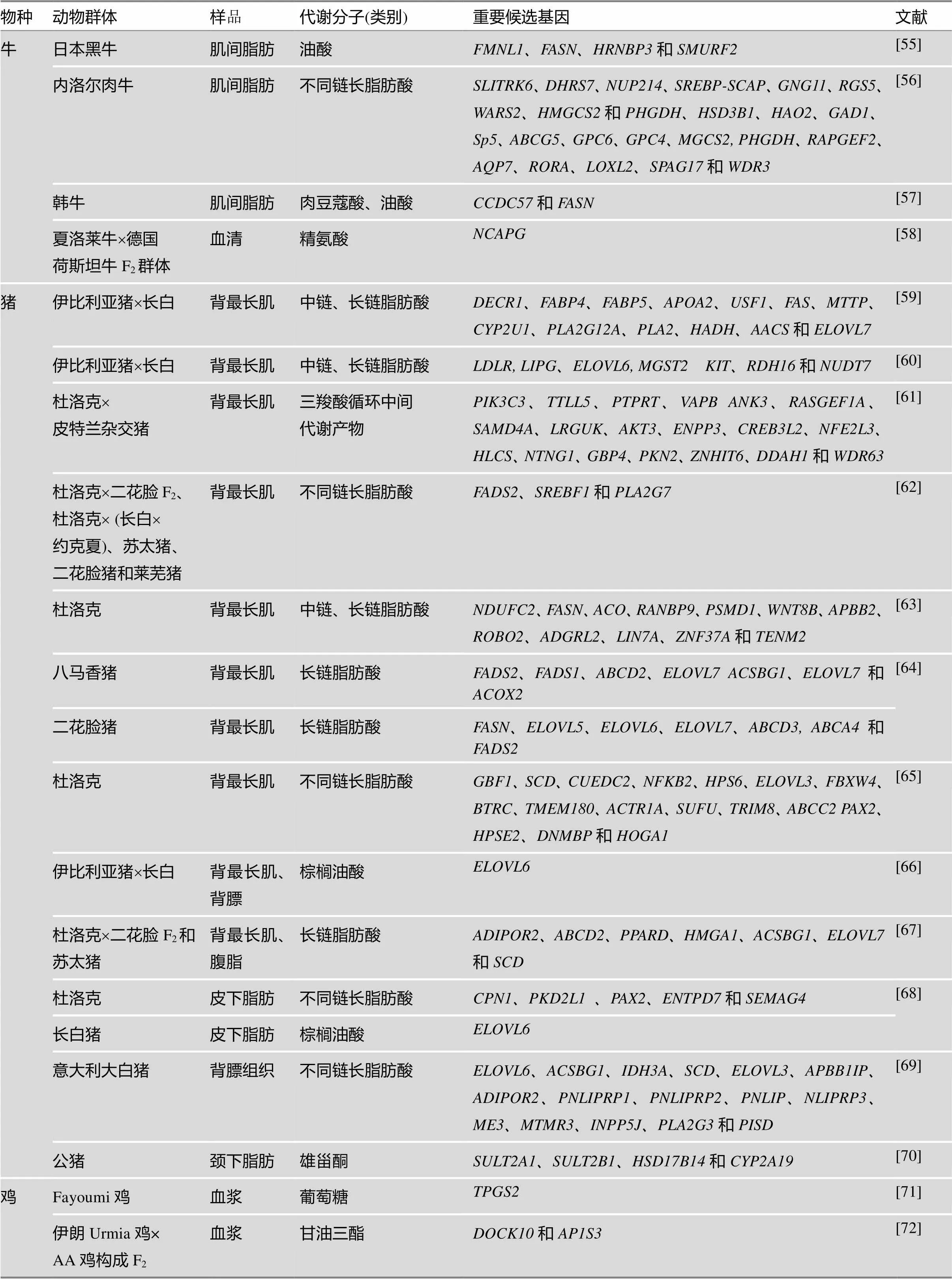
物种动物群体样品代谢分子(类别)重要候选基因文献 牛日本黑牛肌间脂肪油酸FMNL1、FASN、HRNBP3和SMURF2[55] 内洛尔肉牛肌间脂肪不同链长脂肪酸SLITRK6、DHRS7、NUP214、SREBP-SCAP、GNG11、RGS5、WARS2、HMGCS2和PHGDH、HSD3B1、HAO2、GAD1、Sp5、ABCG5、GPC6、GPC4、MGCS2, PHGDH、RAPGEF2、AQP7、RORA、LOXL2、SPAG17和WDR3[56] 韩牛肌间脂肪肉豆蔻酸、油酸CCDC57和FASN[57] 夏洛莱牛´德国荷斯坦牛F2群体血清精氨酸NCAPG[58] 猪伊比利亚猪´长白背最长肌中链、长链脂肪酸DECR1、FABP4、FABP5、APOA2、USF1、FAS、MTTP、CYP2U1、PLA2G12A、PLA2、HADH、AACS和ELOVL7[59] 伊比利亚猪´长白背最长肌中链、长链脂肪酸LDLR, LIPG、ELOVL6, MGST2 KIT、RDH16和NUDT7[60] 杜洛克´皮特兰杂交猪背最长肌三羧酸循环中间代谢产物PIK3C3、TTLL5、PTPRT、VAPB ANK3、RASGEF1A、SAMD4A、LRGUK、AKT3、ENPP3、CREB3L2、NFE2L3、HLCS、NTNG1、GBP4、PKN2、ZNHIT6、DDAH1和WDR63[61] 杜洛克´二花脸 F2、杜洛克´ (长白´约克夏)、苏太猪、二花脸猪和莱芜猪背最长肌不同链长脂肪酸FADS2、SREBF1和PLA2G7[62] 杜洛克背最长肌中链、长链脂肪酸NDUFC2、FASN、ACO、RANBP9、PSMD1、WNT8B、APBB2、ROBO2、ADGRL2、LIN7A、ZNF37A和TENM2[63] 八马香猪背最长肌长链脂肪酸FADS2、FADS1、ABCD2、ELOVL7 ACSBG1、ELOVL7和ACOX2[64] 二花脸猪背最长肌长链脂肪酸FASN、ELOVL5、ELOVL6、ELOVL7、ABCD3, ABCA4和FADS2 杜洛克背最长肌不同链长脂肪酸GBF1、SCD、CUEDC2、NFKB2、HPS6、ELOVL3、FBXW4、BTRC、TMEM180、ACTR1A、SUFU、TRIM8、ABCC2 PAX2、HPSE2、DNMBP和HOGA1[65] 伊比利亚猪´长白背最长肌、背膘棕榈油酸ELOVL6[66] 杜洛克´二花脸F2和苏太猪背最长肌、腹脂长链脂肪酸ADIPOR2、ABCD2、PPARD、HMGA1、ACSBG1、ELOVL7和SCD[67] 杜洛克皮下脂肪不同链长脂肪酸CPN1、PKD2L1 、PAX2、ENTPD7和SEMAG4[68] 长白猪皮下脂肪棕榈油酸ELOVL6 意大利大白猪背膘组织不同链长脂肪酸ELOVL6、ACSBG1、IDH3A、SCD、ELOVL3、APBB1IP、ADIPOR2、PNLIPRP1、PNLIPRP2、PNLIP、NLIPRP3、ME3、MTMR3、INPP5J、PLA2G3和PISD[69] 公猪颈下脂肪雄甾酮SULT2A1、SULT2B1、HSD17B14和CYP2A19[70] 鸡Fayoumi鸡血浆葡萄糖TPGS2[71] 伊朗Urmia鸡× AA鸡构成F2血浆甘油三酯DOCK10和AP1S3[72]
在动物群体中,目前主要是利用研究代谢分子与重要经济性状之间的相关性或差异分析来筛选生物标记物,育种者也期望能够在育种实践中利用这些筛选到的代谢分子作为生物标记物,并由此应用于标记辅助选择中,从而提高选择的准确性。目前,一些代谢分子已被筛选出来作为奶牛产奶性状或疾病的生物标记;尿碳酸酐酶-Ⅵ等代谢分子可以作为猪肾病诊断的生物标记;甘油磷脂等代谢分子可以作为鸡腹水综合征的生物标记。其他相关研究结果详见表3。
随着家养动物基因组数据的积累,研究者开始整合分析代谢组学与基因组、转录组、表观组、表型组的数据,尝试构建出“遗传标记或基因-代谢分子-表型”的关系网络,从而筛选出相关的生物标记,同时进一步解析相关性状的遗传机制。如Weikard等[75]整合肉牛基因组、代谢分子和生产性能数据,发现“基因、基因-精氨酸、肉碱-牛生长性状和体脂性状”的通路,从而推测精氨酸、肉碱可以作为生长性状和体脂性状的生物标记物;Tetens等[47]通过整合基因组、代谢组学数据,发现“基因-甘油磷酸胆碱-酮体耐受性”的关系;Ha等[38]通过比较产犊前3周、产犊后4周、产犊后13周3个时间点的代谢产物,结合GWAS和基因组富集分析,筛选出参与脂质和类固醇代谢的代谢分子与奶牛哺乳早期代谢适应性性状相关;Widmann等[58]和Weikard等[75]整合表型、代谢组学和基因组学数据推断基因上的突变位点,通过调控精氨酸代谢而影响NO通路,从而促进血管平滑肌收缩,最终影响牛的生长;Lundén等[74]发现了“基因-三甲胺-鱼腥味性状”的关系,Chu等[92]利用此关系在北京油鸡群体中剔除基因有害等位基因,从而培育出没有鱼腥味性状的北京油鸡。
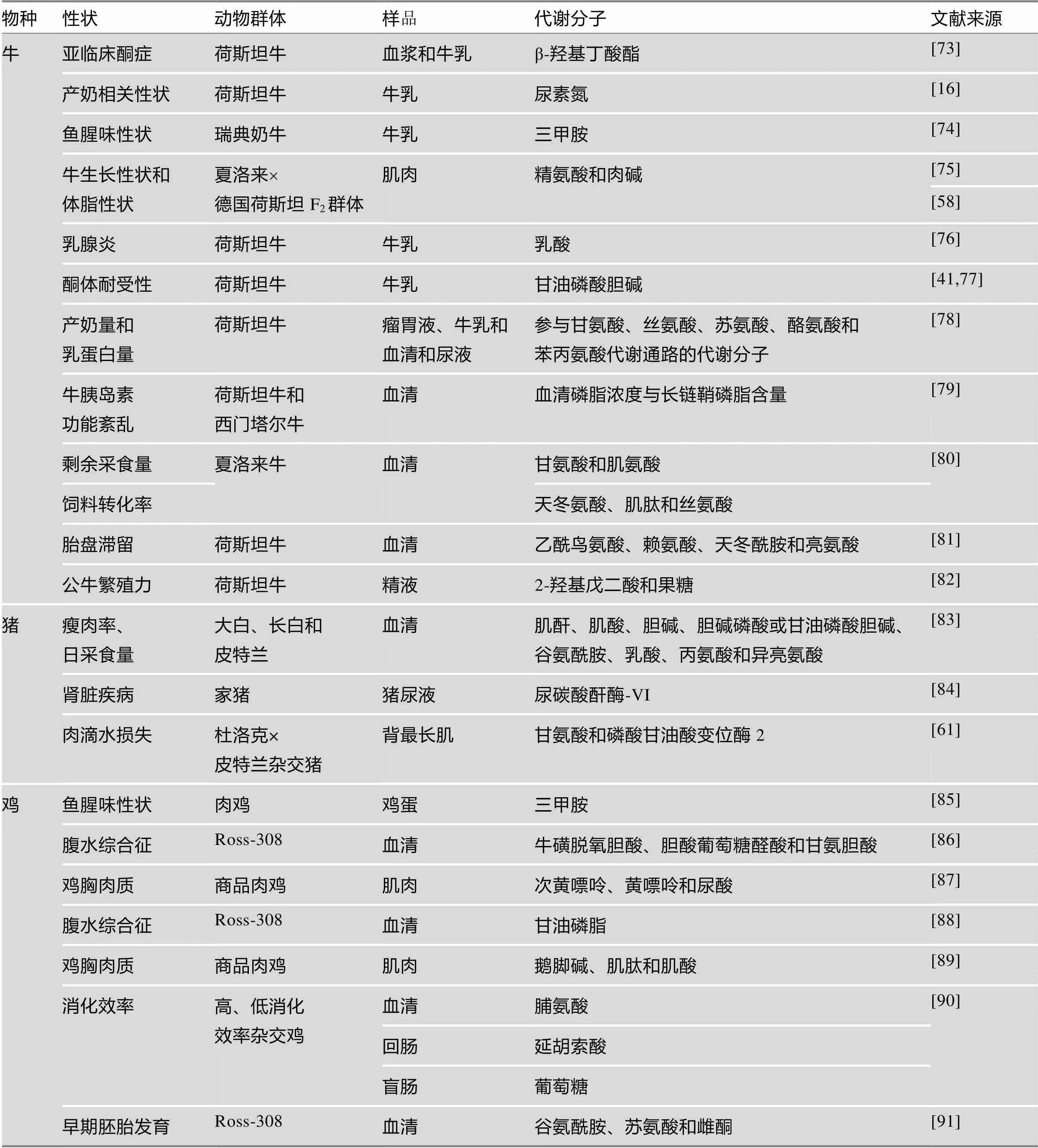
表3 牛、猪、鸡代谢分子与重要经济性状之间的关系
4 结语与展望
目前,在人和家养动物群体中,代谢组学已广泛应用于遗传学、功能基因组学、疾病预测、药物设计等领域[93]。同时,代谢组学的研究也为动物育种技术开辟了新思路,如利用特定生理状态的标记代谢分子对动物进行选育,或利用代谢组学信息理解动物潜在的遗传差异。随着代谢组学技术的日趋臻熟,该技术将会更为广泛地应用于牛、猪、鸡等家养动物重要经济性状的选育上,而且将会有助于阐明复杂的生物学机制问题。
当然,代谢组学研究还有很多亟待解决的问题。例如,生物体代谢产物的变化除受生理刺激和遗传因素影响,与环境因素也密切相关,所以在代谢组学样品收集和实验设计时应该考虑代谢对环境条件的敏感性;其次,目前已知的代谢组学技术平台在仪器的灵敏度、检测的覆盖度上仍存在一定的局限性,检测到的所有代谢分子可能也只代表了基因变异的一小部分,所以应该提高代谢组学检测技术的灵敏性、稳定性、广谱性和特异性;另外,代谢组学研究产生的海量高维数据,需要利用多元统计、生物信息等多种数据挖掘技术进行分析,代谢组学数据的分析是代谢组学研究的重要组成部分之一,但目前相关的研究方法还不够成熟,仍需要进一步的完善。
随着代谢组学数据的积累,将基因组、转录组、表观组、蛋白组与代谢组学、表型组整合分析将成为今后的研究热点,代谢组学信息的使用和发展也将有助于各种组学数据的整合分析。例如通过整合分析,筛选出影响重要经济性状的“基因-蛋白质-代谢分子-表型”网络或通路;通过将代谢组学和表观遗传组整合分析,发现某些代谢分子与染色质活跃区域的关系,从而更全面地解析机体内的代谢分子遗传学机制。
[1] Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data., 1999, 29(11): 1181–1189.
[2] Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge., 2010, 11(12): 855–866.
[3] Robinette SL, Holmes E, Nicholson JK, Dumas ME. Genetic determinants of metabolism in health and disease: from biochemical genetics to genome-wide associations., 2012, 4(4): 30.
[4] Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B, Grallert H, Xu T, Bader E, Huth C, Mittelstrass K, Döring A, Meisinger C, Gieger C, Prehn C, Roemisch-Margl W, Carstensen M, Xie L, Yamanaka-Okumura H, Xing G, Ceglarek U, Thiery J, Giani G, Lickert H, Lin X, Li Y, Boeing H, Joost HG, de Angelis MH, Rathmann W, Suhre K, Prokisch H, Peters A, Meitinger T, Roden M, Wichmann HE, Pischon T, Adamski J, Illig T. Novel biomarkers for pre-diabetes identified by metabolomics., 2012, 8: 615.
[5] Reinehr T, Wolters B, Knop C, Lass N, Hellmuth C, Harder U, Peissner W, Wahl S, Grallert H, Adamski J, Illig T, Prehn C, Yu Z, Wang-Sattler R, Koletzko B. Changes in the serum metabolite profile in obese children with weight loss., 2015, 54(2): 173–181.
[6] Kordalewska M, Markuszewski MJ. Metabolomics in cardiovascular diseases., 2015, 113: 121–136.
[7] Mirsaeidi M, Banoei MM, Winston BW, Schraufnagel DE. Metabolomics: applications and promise in mycobacterial disease., 2015, 12(9): 1278–1287.
[8] Shajahan-Haq AN, Cheema MS, Clarke R. Application of metabolomics in drug resistant breast cancer research., 2015, 5(1): 100–118.
[9] Fontanesi L. Metabolomics and livestock genomics: insights into a phenotyping frontier and its applications in animal breeding., 2016, 6(1): 73–79.
[10] Suhre K, Gieger C. Genetic variation in metabolic phenotypes: study designs and applications., 2012, 13(11): 759–769.
[11] Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. HMDB 4.0: the human metabolome database for 2018., 2018, 46(D1): D608–D617.
[12] Wang L, Bo T, Meng X. The booming “metabolomics” research techniques and methods., 2014, 34(01): 75–78.王磊, 薄涛, 蒙昔. 正在蓬勃兴起的“代谢组学”研究技术与方法. 生命的化学, 2014, 34(01): 75–78.
[13] Goldansaz SA, Guo AC, Sajed T, Steele MA, Plastow GS, Wishart DS. Livestock metabolomics and the livestock metabolome: A systematic review., 2017, 12(5): e0177675.
[14] Dharuri H, Demirkan A, van Klinken JB, Mook-Kanamori DO, van Duijn CM, 't Hoen PA, van Dijk KW. Genetics of the human metabolome, what is next?, 2014, 1842(10): 1923–1931.
[15] Soyeurt H, Gillon A, Vanderick S, Mayeres P, Bertozzi C, Gengler N. Estimation of heritability and genetic correlations for the major fatty acids in bovine milk., 2007, 90(9): 4435–4442.
[16] Stoop WM, Bovenhuis H, van Arendonk JA. Genetic parameters for milk urea nitrogen in relation to milk production traits., 2007, 90(4): 1981–1986.
[17] Oikonomou G, Valergakis GE, Arsenos G, Roubies N, Banos G. Genetic profile of body energy and blood metabolic traits across lactation in primiparous Holstein cows., 2008, 91(7): 2814–2822.
[18] Nogi T, Honda T, Mukai F, Okagaki T, Oyama K. Heritabilities and genetic correlations of fatty acid compositions in longissimus muscle lipid with carcass traits in Japanese Black cattle., 2011, 89(3): 615–621.
[19] Buitenhuis AJ, Sundekilde UK, Poulsen NA, Bertram HC, Larsen LB, Sørensen P. Estimation of genetic parameters and detection of quantitative trait loci for metabolites in Danish Holstein milk., 2013, 96(5): 3285– 3295.
[20] Wittenburg D, Melzer N, Willmitzer L, Lisec J, Kesting U, Reinsch N, Repsilber D. Milk metabolites and their genetic variability., 2013, 96(4): 2557–2569.
[21] Gebreyesus G, Lund MS, Janss L, Poulsen NA, Larsen LB, Bovenhuis H, Buitenhuis AJ. Short communication: Multi-trait estimation of genetic parameters for milk protein composition in the Danish Holstein., 2016, 99(4): 2863–2866.
[22] Ntawubizi M, Colman E, Janssens S, Raes K, Buys N, de Smet S. Genetic parameters for intramuscular fatty acid composition and metabolism in pigs., 2010, 88(4): 1286–1294.
[23] Ibáñez-Escriche N, Magallón E, Gonzalez E, Tejeda JF, Noguera JL. Genetic parameters and crossbreeding effects of fat deposition and fatty acid profiles in Iberian pig lines., 2016, 94(1): 28–37.
[24] Dong JQ, Zhang H, Jiang XF, Wang SZ, Du ZQ, Wang ZP, Leng L, Cao ZP, Li YM, Luan P, Li H. Comparison of serum biochemical parameters between two broiler chicken lines divergently selected for abdominal fat content., 2015, 93(7): 3278–3286.
[25] Zhang HL, Xu ZQ, Yang LL, Wang YX, Li YM, Dong JQ, Zhang XY, Jiang XY, Jiang XF, Li H, Zhang DX, Zhang H. Genetic parameters for the prediction of abdominal fat traits using blood biochemical indicators in broilers., 2018, 59(1): 28–33.
[26] Karisa BK, Thomson J, Wang Z, Li C, Montanholi YR, Miller SP, Moore SS, Plastow GS. Plasma metabolites associated with residual feed intake and other productivity performance traits in beef cattle., 2014, 165: 200–211.
[27] D'Alessandro A, Marrocco C, Zolla V, D’AndreaM, Zolla L. Meat quality of the longissimus lumborum muscle of Casertana and Large White pigs: metabolomics and proteomics intertwined., 2011, 75(2): 610–627.
[28] He Q, Ren P, Kong X, Wu Y, Wu G, Li P, Hao F, Tang H, Blachier F, Yin Y. Comparison of serum metabolite compositions between obese and lean growing pigs using an NMR-based metabonomic approach., 2012, 23(2): 133–139.
[29] Straadt IK, Aaslyng MD, Bertram HC. An NMR-based metabolomics study of pork from different crossbreeds and relation to sensory perception., 2014, 96(2): 719–728.
[30] Bovo S, Mazzoni G, Galimberti G, Calò DG, Fanelli F, Mezzullo M, Schiavo G, Manisi A, Trevisi P, Bosi P, Dall'Olio S, Pagotto U, Fontanesi L. Metabolomics evidences plasma and serum biomarkers differentiating two heavy pig breeds., 2016, 10(10): 1741–1748.
[31] Ji B, Middleton JL, Ernest B, Saxton AM, Lamont SJ, Campagna SR,Voy BH. Molecular and metabolic profiles suggest that increased lipid catabolism in adipose tissue contributes to leanness in domestic chickens., 2014, 46(9): 315–327.
[32] Baéza E, Jégou M, Gondret F, Lalande-Martin J, Tea I, Le Bihan-Duval E, Berri C, Collin A, Métayer-Coustard S, Louveau I, Lagarrigue S, Duclos MJ. Pertinent plasma indicators of the ability of chickens to synthesize and store lipids., 2015, 93(1): 107–116.
[33] Stoop WM, Schennink A, Visker MH, Mullaart E, van Arendonk JA, Bovenhuis H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids., 2009, 92(9): 4664–4675.
[34] Bouwman AC, Bovenhuis H, Visker MH, van Arendonk JA. Genome-wide association of milk fatty acids in Dutch dairy cattle., 2011, 12: 43.
[35] Rutten MJ, Bouwman AC, Sprong RC, van Arendonk JA, Visker MH. Genetic variation in vitamin B-12 content of bovine milk and its association with SNP along the bovine genome., 2013, 8(4): e62382.
[36] Buitenhuis B, Janss LL, Poulsen NA, Larsen LB, Larsen MK, Sørensen P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle., 2014, 15: 1112.
[37] Li C, Sun DX, Zhang SL, Wang S, Wu XP, Zhang Q, Liu L, Li YH, Qiao L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in chinese holstein., 2014, 9(5): e96186.
[38] Ha NT, Gross JJ, van Dorland A, Tetens J, Thaller G, Schlather M, Bruckmaier R, Simianer H. Gene-based mapping and pathway analysis of metabolic traits in dairy cows., 2015, 10(3): e0122325.
[39] Li X, Buitenhuis AJ, Lund MS, Li C, Sun D, Zhang Q, Poulsen NA, Su G. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations., 2015, 98(11): 8152–8163.
[40] Poulsen NA, Rybicka I, Larsen LB, Buitenhuis AJ, Larsen MK. Short communication: genetic variation of riboflavin content in bovine milk., 2015, 98(5): 3496– 3501.
[41] Tetens J, Heuer C, Heyer I, Klein MS, Gronwald W, Junge W, Oefner PJ, Thaller G, Krattenmacher N. Polymorphisms within the APOBR gene are highly associated with milk levels of prognostic ketosis biomarkers in dairy cows., 2015, 47(4): 129–137.
[42] Pegolo S, Cecchinato A, Mele M, Conte G, Schiavon S, Bittante G. Effects of candidate gene polymorphisms on the detailed fatty acids profile determined by gas chromatography in bovine milk., 2016, 99(6): 4558–4573.
[43] Duchemin SI, Bovenhuis H, Megens HJ, van Arendonk JAM, Visker MHPW. Fine-mapping of BTA17 using imputed sequences for associations with de novo synthesized fatty acids in bovine milk., 2017, 100(11): 9125–9135.
[44] Lopdell TJ, Tiplady K, Struchalin M, Johnson TJJ, Keehan M, Sherlock R, Couldrey C, Davis SR, Snell RG, Spelman RJ, Littlejohn MD. DNA and RNA-sequence based GWAS highlights membrane-transport genes as key modulators of milk lactose content., 2017, 18(1): 968.
[45] Ishii A, Yamaji K, Uemoto Y, Sasago N, Kobayashi E, Kobayashi N, Matsuhashi T, Maruyama S, Matsumoto H, Sasazaki S, Mannen H. Genome-wide association study for fatty acid composition in Japanese Black cattle., 2013, 84(10): 675–682.
[46] Saatchi M, Garrick DJ, Tait RG Jr, Mayes MS, Drewnoski M, Schoonmaker J, Diaz C, Beitz DC, Reecy JM. Genome-wide association and prediction of direct genomic breeding values for composition of fatty acids in Angus beef cattle.,2013, 14: 730.
[47] Buchanan JW, Reecy JM, Garrick DJ, Duan Q, Beitz DC, Koltes JE, Saatchi M, Koesterke L, Mateescu RG. Deriving gene networks from SNP associated with triacylglycerol and phospholipid fatty acid fractions from ribeyes of angus cattle., 2016, 7: 116.
[48] Lemos MV, Chiaia HL, Berton MP, Feitosa FL, Aboujaoud C, Camargo GM, Pereira AS, Albuquerque LG, Ferrinho AM, Mueller LF, Mazalli MR, Furlan JJ, Carvalheiro R, Gordo DM, Tonussi R, Espigolan R, Silva RM, de Oliveira HN, Duckett S, Aguilar I, Baldi F. Genome-wide association between single nucleotide polymorphisms with beef fatty acid profile in Nellore cattle using the single step procedure., 2016, 17: 213.
[49] Sasago N, Abe T, Sakuma H, Kojima T, Uemoto Y. Genome-wide association study for carcass traits, fatty acid composition, chemical composition, sugar, and the effects of related candidate genes in Japanese Black cattle., 2017, 88(1): 33–44.
[50] Uemoto Y, Ohtake T, Sasago N, Takeda M, Abe T, Sakuma H, Kojima T, Sasaki S. Effect of two non- synonymous ecto-5'-nucleotidase variants on the genetic architecture of inosine 5'-monophosphate (IMP) and its degradation products in Japanese Black beef., 2017, 18(1): 874.
[51] Zhu B, Niu H, Zhang W, Wang Z, Liang Y, Guan L, Guo P, Chen Y, Zhang L, Guo Y, Ni H, Gao X, Gao H, Xu L, Li J. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array., 2017, 18(1): 464.
[52] Kawaguchi F, Kigoshi H, Nakajima A, Matsumoto Y, Uemoto Y, Fukushima M, Yoshida E, Iwamoto E, Akiyama T, Kohama N, Kobayashi E, Honda T, Oyama K, Mannen H, Sasazaki S. Pool-based genome-wide association study identified novel candidate regions on BTA9 and 14 for oleic acid percentage in Japanese Black cattle., 2018, 89(8): 1060–1066.
[53] Sasago N, Takeda M, Ohtake T, Abe T, Sakuma H, Kojima T, Sasaki S, Uemoto Y. Genome-wide association studies identified variants for taurine concentration in Japanese Black beef., 2018, 89(8): 1051–1059.
[54] Chen L, Ekine-Dzivenu C, Vinsky M, Basarab J, Aalhus J, Dugan ME, Fitzsimmons C, Stothard P, Li C. Genome- wide association and genomic prediction of breeding values for fatty acid composition in subcutaneous adipose and longissimus lumborum muscle of beef cattle., 2015, 16: 135.
[55] Uemoto Y, Abe T, Tameoka N, Hasebe H, Inoue K, Nakajima H, Shoji N, Kobayashi M, Kobayashi E. Whole- genome association study for fatty acid composition of oleic acid in Japanese Black cattle., 2011, 42(2): 141–148.
[56] Cesar AS, Regitano LC, Mourão GB, Tullio RR, Lanna DP, Nassu RT, Mudado MA, Oliveira PS, do Nascimento ML, Chaves AS, Alencar MM, Sonstegard TS, Garrick DJ, Reecy JM, Coutinho LL. Genome-wide association study for intramuscular fat deposition and composition in Nellore cattle., 2014, 15: 39.
[57] Bhuiyan MSA, Kim YK, Kim HJ, Lee DH, Lee SH, Yoon HB, Lee SH. Genome-wide association study and prediction of genomic breeding values for fatty-acid composition in Korean Hanwoo cattle using a high-density single- nucleotide polymorphism array., 2018, 96(10): 4063–4075.
[58] Widmann P, Reverter A, Fortes MR, Weikard R, Suhre K, Hammon H, Albrecht E, Kuehn C. A systems biology approach using metabolomic data reveals genes and pathways interacting to modulate divergent growth in cattle., 2013, 14(1): 798.
[59] Ramayo-Caldas Y, Mercadé A, Castelló A, Yang B, Rodríguez C, Alves E, Díaz I, Ibáñez-Escriche N, Noguera JL, Pérez-Enciso M, Fernández AI, Folch JM. Genome- wide association study for intramuscular fatty acid composition in an Iberian × Landrace cross., 2012, 90(9): 2883–2893.
[60] Muñoz M, Rodríguez MC, Alves E, Folch JM, Ibañez- Escriche N, Silió L, Fernández AI. Genome-wide analysis of porcine backfat and intramuscular fat fatty acid composition using high-density genotyping and expression data., 2013, 14: 845.
[61] Welzenbach J, Neuhoff C, Heidt H, Cinar MU, Looft C, Schellander K, Tholen E, Große-Brinkhaus C. Integrative analysis of metabolomic, proteomic and genomic data to reveal functional pathways and candidate genes for drip loss in pigs., 2016, 17(9): 1426.
[62] Zhang W, Yang B, Zhang J, Cui L, Ma J, Chen C, Ai H, Xiao S, Ren J, Huang L. Genome-wide association studies for fatty acid metabolic traits in five divergent pig populations., 2016, 6: 24718.
[63] Sato S, Uemoto Y, Kikuchi T, Egawa S, Kohira K, Saito T, Sakuma H, Miyashita S, Arata S, Suzuki K. Genome-wide association studies reveal additional related loci for fatty acid composition in a Duroc pig multigenerational population., 2017, 88(10): 1482–1490.
[64] Zhang J, Zhang Y, Gong H, Cui L, Huang T, Ai H1, Ren J, Huang L, Yang B. Genetic mapping using 1.4M SNP array refined loci for fatty acid composition traits in Chinese Erhualian and Bamaxiang pigs., 2017, 134(6): 472–483.
[65] Viterbo VS, Lopez BIM, Kang H, Kim H, Song CW, Seo KS. Genome wide association study of fatty acid composition in Duroc swine., 2018, 31(8): 1127–1133.
[66] Corominas J, Ramayo-Caldas Y, Puig-Oliveras A, Pérez-Montarelo D, Noguera JL, Folch JM, Ballester M. Polymorphism in the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs., 2013, 8(1): e53687.
[67] Yang B, Zhang W, Zhang Z, Fan Y, Xie X, Ai H, Ma J, Xiao S, Huang L, Ren J. Genome-wide association analyses for fatty acid composition in porcine muscle and abdominal fat tissues., 2013, 8(6): e65554.
[68] van Son M, Enger EG, Grove H, Ros-Freixedes R, Kent MP, Lien S, Grindflek E. Genome-wide association study confirm major QTL for backfat fatty acid composition on SSC14 in Duroc pigs., 2017, 18(1): 369.
[69] Zappaterra M, Ros-Freixedes R, Estany J, Davoli R. Association study highlights the influence of ELOVL fatty acid elongase 6 gene region on backfat fatty acid composition in Large White pig breed., 2018, 12(12): 2443–2452.
[70] Duijvesteijn N, Knol EF, Bijma P. Boar taint in entire male pigs: a genomewide association study for direct and indirect genetic effects on androstenone., 2014, 92(10): 4319–4328.
[71] van Goor A, Ashwell CM, Persia ME, Rothschild MF, Schmidt CJ, Lamont SJ. Quantitative trait loci identified for blood chemistry components of an advanced intercross line of chickens under heat stress., 2016, 17: 287.
[72] Javanrouh-Aliabad A, Vaez Torshizi R, Masoudi AA, Ehsani A. Identification of candidate genes for blood metabolites in Iranian chickens using a genome-wide association study., 2018, 59(4): 381–388.
[73] Geishauser T, Leslie K, Tenhag J, Bashir A. Evaluation of eight cow-side ketone tests in milk for detection of subclinical ketosis in dairy cows., 2000, 83(2): 296–299.
[74] Lundén A, Marklund S, Gustafsson V, Andersson L. A nonsense mutation in the FMO3 gene underlies fishy off-flavor in cow’s milk., 2002, 12(12): 1885–1888.
[75] Weikard R, Altmaier E, Suhre K, Weinberger K, Hammon H, Albrecht E, Setoguchi K, Takasuga A, Küehn C. Metabolomic profiles indicate distinct physiological pathways affected by two loci with major divergent effect on Bos taurus growth and lipid deposition., 2010, 42A(2): 79–88.
[76] Melzer N, Wittenburg D, Hartwig S, Jakubowski S, Kesting U, Willmitzer L, Lisec J, Reinsch N, Repsilber D. Investigating associations between milk metabolite profiles and milk traits of Holstein cows., 2013, 96(3): 1521–1534.
[77] Klein MS, Buttchereit N, Miemczyk SP, Immervoll AK, Louis C, Wiedemann S, Junge W, Thaller G, Oefner PJ, Gronwald W. NMR metabolomic analysis of dairy cows reveals milk glycerophosphocholine to phosphocholine ratio as prognostic biomarker for risk of ketosis., 2012, 11(2): 1373–1381.
[78] Sun HZ, Wang DM, Wang B, Wang JK, Liu HY, Guan LL, Liu JX. Metabolomics of four biofluids from dairy cows: potential biomarkers for milk production and quality., 2015, 14(2): 1287–1298.
[79] Humer E, Khol-Parisini A, Metzler-Zebeli BU, Gruber L, Zebeli Q. Alterations of the lipid metabolome in dairy cows experiencing excessive lipolysis early postpartum., 2016, 11(7): e0158633.
[80] Meale SJ, Morgavi DP, Cassar-Malek I, Andueza D, Ortigues-Marty I, Robins RJ, Schiphorst AM, Laverroux S, Graulet B, Boudra H, Cantalapiedra-Hijar G. Exploration of biological markers of feed efficiency in young bulls., 2017, 65(45): 9817–9827.
[81] Dervishi E, Zhang G, Mandal R, Wishart DS, Ametaj BN. Targeted metabolomics: new insights into pathobiology of retained placenta in dairy cows and potential risk biomarkers., 2018, 12(5): 1050–1059.
[82] Velho ALC, Menezes E, Dinh T, Kaya A, Topper E, Moura AA, Memili E. Metabolomic markers of fertility in bull seminal plasma., 2018, 13(4): e0195279.
[83] Rohart F, Paris A, Laurent B, Canlet C, Molina J, Mercat MJ, Tribout T, Muller N, Iannuccelli N, Villa-Vialaneix N, Liaubet L, Milan D, San Cristobal M. Phenotypic prediction based on metabolomic data for growing pigs from three main European breeds., 2012, 90(13): 4729–4740.
[84] Nishita T, Yatsu J, Watanabe K, Ochiai H, Ichihara N, Orito K, Arishima K. Urinary carbonic anhydrase VI as a biomarker for kidney disease in pigs., 2014, 202(2): 378–380.
[85] BoltonW, CarterTC, Morley JR. The hen’s egg: genetics of taints in eggs from hens fed on rapeseed meal., 1976, 17(3): 313–320.
[86] Shen Y, Shi S, Tong H, Guo Y, Zou J. Metabolomics analysis reveals that bile acids and phospholipids contribute to variable responses to low-temperature-induced ascites syndrome., 2014, 10(6): 1557–1567.
[87] Abasht B, Mutryn MF, Michalek RD, Lee WR. Oxidative stress and metabolic perturbations in wooden breast disorder in chickens., 2016, 11(4): e0153750.
[88] Shi S, Shen Y, Zhang S, Zhao Z, Hou Z, Zhou H, Zou J, Guo Y.Combinatory evaluation of transcriptome and metabolome profiles of low temperature-induced resistant ascitessyndrome in Broiler Chickens., 2017, 7(1): 2389.
[89] Sundekilde UK, Rasmussen MK, Young JF, Bertram HC. High resolution magic angle spinning NMR spectroscopy reveals that pectoralis muscle dystrophy in chicken is associated with reduced muscle content of anserine and carnosine., 2017, 217: 151–154.
[90] Beauclercq S, Nadal-Desbarats L, Hennequet-Antier C, Gabriel I, Tesseraud S, Calenge F, Le Bihan-Duval E, Mignon-Grasteau S. Relationships between digestive efficiency and metabolomic profiles of serum and intestinal contents in chickens., 2018, 8: 6678.
[91] Peng ML, Li SN, He QQ, Zhao JL, Li LL, Ma HT. Based serum metabolomics analysis reveals simultaneous interconnecting changes during chicken embryonic development., 2018, 102(5): 1210– 1219.
[92] Chu Q, Zhang J, Zhu S, Zhang Y, Wang H, Geng A, Liu H. The detection and elimination of flavin-containing monooxygenase 3 gene T329S mutation in the Beijing You chicken., 2013, 92(12): 3109–3112.
[93] Li H, Jiang Y, He FC. Recent development of metabonomics and its applications in clinical research., 2008, 30(4): 389–399.
李灏, 姜颖, 贺福初. 代谢组学技术及其在临床研究中的应用. 遗传, 2008, 30(4): 389–399.
Applications of metabonomics in animal genetics and breeding
Meng Zhou1,2,3, Junhong Jing1,2,3, Ruihan Mao1,2,3, Jing Guo1,2,3, Zhipeng Wang1,2,3
Metabolomics uses advanced analytical chemistry techniques to comprehensively identify, quantify, and characterize a large number of small molecule metabolites in biological samples (e.g., milk, plasma, and serum). It is routinely used in biomedical, nutritional, crop and farm animal research. Metabolomic analyses in farm animals have been initiated in many non-genetic application fields. Recently, it is being increasingly used in animal breeding with the emergence of physiological genomics/genetics and refined phenotypic description. In this review, we describe the features of metabolomics platforms and approaches, and summarize the metabolomics applications in animal genetics and genomics with a focus on some key areas, such as the heritability estimates of metabolomic profiles, identification differences metabolites between lines or breeds, genome-wide association studies with metabotypes, biomarker discovery for economic traits. Moreover, we also discuss the potential applications based on current livestock metabolomics studies. The intent of this review is to provide a critical overview of the trends in the applications of metabolomics in animal breeding, aiming to provide a reference for further studies on the genetic background of the important traits of farm animals combined metabolomics with genomics.
metabonomics; domestic animals; genetics; animal breeding
2018-08-08;
2018-10-17
国家自然科学基金项目(编号:31101709),国家留学基金委项目(编号:201308230102),东北农业大学东农学者计划“学术骨干”项目(编号:15XG14)和农业部鸡遗传育种重点实验室开放课题(编号:CGB-201706)资助 [Supported by the Natural Science Foundation of China (No. 31101709), China Scholarship Council (No. 201308230102), Academic Backbone Project of Northeast Agricultural University (No. 15XG14) and the Open Projects of Key Laboratory of Chicken Genetics and Breeding Ministry of Agriculture and Rural Affairs (No. CGB-201706)]
周萌,硕士研究生,专业方向:动物分子数量遗传学。E-mail: 1341417113@qq.com
王志鹏,博士,副教授,研究方向:动物分子数量遗传学。E-mail: wangzhipeng@neau.edu.cn
10.16288/j.yczz.18-226
2019/1/14 13:15:25
URI: http://kns.cnki.net/kcms/detail/11.1913.R.20190114.1315.006.html
(责任编委: 李明洲)
