Characterization and Evaluation of OsLCT1 and OsNramp5 Mutants Generated Through CRISPR/Cas9-Mediated Mutagenesis for Breeding Low Cd Rice
2019-02-19LiuSongmeiJiangJieLiuYangMengJunXuShoulingTanYuanyuanLiYoufaShuQingyaoHuangJianzhong
Liu Songmei, Jiang Jie, Liu Yang, Meng Jun, Xu Shouling, Tan Yuanyuan, Li Youfa,Shu Qingyao, Huang Jianzhong
Characterization and Evaluation ofandMutants Generated Through CRISPR/Cas9-Mediated Mutagenesis for Breeding Low Cd Rice
Liu Songmei1, 2, Jiang Jie1, 2, Liu Yang1, 2, Meng Jun3, Xu Shouling1, Tan Yuanyuan1, Li Youfa4,Shu Qingyao1, 2, Huang Jianzhong1
(National Key Laboratory of Rice Biology / Institute of Nuclear Agricultural Sciences, Zhejiang University, Hangzhou 310058, China; Hubei Collaborative Innovation Center for Grain Industry, Jingzhou 434025, China; Zhejiang Provincial Key Laboratory of Agricultural Resources and Environment / College of Environmental and Natural Resource Sciences, Zhejiang University, Hangzhou 310058, China; Jiaxing Academy of Agricultural Sciences, Jiaxing 314016, China)
To explore how rice (L.) can be safely produced in Cd-polluted soil,andmutant lines were generated by CRISPR/Cas9-mediated mutagenesis. One ofmutant () and two ofmutants (and) were evaluated for grain Cd accumulation and agronomic performances. In paddy field soil containing approximately 0.9 mg/kg Cd,grains contained approximately 40% (0.17 mg/kg) of the Cd concentration of the wild type parental line, less than the China National Food Safety Standard (0.20 mg/kg). Bothmutants showed low grain Cd accumulation (< 0.06 mg/kg) in the paddy (approximately 0.9 mg/kg Cd) or in pots in soil spiked with 2 mg/kg Cd. However, onlyshowed normal growth and yield, whereas the growth ofwas severely impaired. The study showed thatcould be used to produce rice grains safe for human consumption in lightly contaminated paddy soils andused in soils contaminated by much higher levels of Cd.
cadmium; rice;;; genome-editing; heavy metal contamination; CRISPR; Cas9
Cadmium (Cd) is a non-essential heavy metal that is toxic to virtually all living organisms, including plants (Peng et al, 2018). Given its long biological half-life ranging from 10 to 30 years, Cd accumulated in the human body with age poses a serious health risk even at low-level chronic exposure (Clemens et al, 2013; Aziz et al, 2015). In the general population, Cd exposure is mainly through the food chain. Rice (L.), which feeds more than half of the world’s population, is the major source of Cd intake in regions, where it is the dietary staple (Meharg et al, 2013; Yao et al, 2015; Sun et al, 2017; FAO/WHO, 2018). Thus, from a public health perspective, there is a pressing demand to minimize Cd accumulation in rice grains.
Heavy metal contamination in agricultural soils and foods owing to anthropogenic activities is recognized as a global problem (Arao et al, 2010; Smolders and Mertens, 2013). Identification and termination of the contamination sources and phytoextraction of contaminants will reduce the concentration of heavy metals in contaminated soils in the long term. Growing non-food crops on heavily contaminated soils is also a sensible option. Agronomic practices such as reduction of metal phytoavailability, management of paddy water and application of fertilizer are effective in reducing grain Cd levels (Zhao et al, 2015). However, many of these measures are expensive and in some cases impractical (Arao and Ae, 2003).
In addition to field-based measures, breeding varieties that show low Cd accumulation in grains, acting on the entry point of Cd into the food chain, is a practical option to allow the use of Cd-contaminated paddy fields for rice production. Substantial genotypic variation in grain Cd accumulation exists in rice germplasm, which could be exploited to breed low-Cd- accumulated varieties (Zeng et al, 2008; Pinson et al, 2015). Breeding rice varieties that accumulate low Cd concentrations involves identification of quantitative trait loci for low Cd accumulation, and introgression and pyramiding of such loci into high-yielding and locally adapted cultivars through marker-assisted breeding (Zhang et al, 2011; Abe et al, 2017). Unfortunately, current breeding programs seldom target the low Cd accumulation trait, partly because of the high cost of Cd selection with regard to determination of Cd concentrations and the long complex process of selection. Inclusion of low Cd accumulation as an essential selection criterion in breeding may also hinder progress towards development of other desirable economic traits (Grant et al, 2008; Zhao et al, 2015).
In recent years, several important metal transporters involved in Cd accumulation have been identified in rice. OsHMA2 (Heavy Metal ATPase 2) is implicated to be a zinc and Cd transporter, facilitating root- to-shoot translocation of these metals (Takahashi et al, 2011; Satoh-Nagasawa et al, 2012). Loss of OsHMA2 function in insertion mutants results in reduced leaf and grain Cd concentrations as well as a dramatic decline in growth and yield (Yamaji et al, 2013). OsHMA3 is involved in the sequestration of Cd into the vacuole of root cells. Its over-expression significantly reduces Cd accumulation in the straw and grain, whereas its knockout elevates translocation of Cd from root to shoot (Miyadate et al, 2010; Ueno et al, 2010; Sasaki et al, 2014; Shao J F et al, 2017). Knockout of() may dramatically reduce grain Cd accumulation (by 43%–55%), but its function as a Cd transporter remains elusive (Shimo et al, 2011).
OsLCT1, a rice homolog of wheat low affinity cation transporter 1, is localized to the plasma membrane and its expression is strongly increased in nodes and leaf blades during grain ripening (Uraguchi et al, 2011). OsLCT1 is detected in the uppermost node, around large vascular bundles and in diffuse vascular bundles, and might be involved in mediating the translocation of remobilized Cd and certain other elements from the leaf blades to the grain (Uraguchi et al, 2011). Plants in whichis down-regulated accumulate approximately half of Cd in the grains as that of control plants, whereas the contents of other metals and plant growth are not negatively affected (Uraguchi et al, 2011). OsNramp5 (natural resistance- associated macrophage protein 5) is localized on the distal side of the plasma membrane in the exo- and endo-dermis of the root and is a major transporter of Cd into the root stele (Sasaki et al, 2012). Knockout/ knockdown ofcan drastically reduce the contents of manganese (Mn) and Cd in the shoot and grain, but reports of the effects on plant growth and yield are contradictory. Sasaki et al (2012) observed a significant negative effect on plant growth, however, Ishikawa et al (2012) and Tang et al (2017) reported that agronomic and yield-related traits are barely affected.
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system, CRISPR/Cas9, is an efficient and precise genome editing technique and has been used in a number of plant species, including rice (Ishizaki, 2016; Jung et al, 2017). In the present study, we generatedandmutants by CRISPR/Cas9-mediated mutagenesis and analyzed Cd accumulation and plant growth to explore and evaluate the possibility of generating low-Cd-accumulated rice genotypes with minimal reduction of plant growth and yield.
Materials and Methods
CRISPR/Cas9 vector construction and rice transformation
To generateandmutants, one target site was chosen forin exon 1 (Fig. 1-A) and two sites for, one in exon 7 and the other in exon 9 (Fig. 1-B). The sgRNAs for these target sites were designed by searching UniProt for the precise positions (http://www.uniprot.org/) and the CRISPR-P program was used to minimize off-target effects (Lei et al, 2014). DNA oligos were synthesized (TsingKe, Hangzhou, China) for construction of CRISPR/Cas9 vectors using the pHun4c12s plasmid harboring a CYP81A6-hpRNAi element (Lu et al, 2017), which was modified from pHun4c12 (Xu et al, 2014). Three plasmids, corresponding to the target sites, i.e. pH_lct1×1, pH_nramp5×7 and pH_nramp5×9, were constructed and transformed into(Xu et al, 2014).
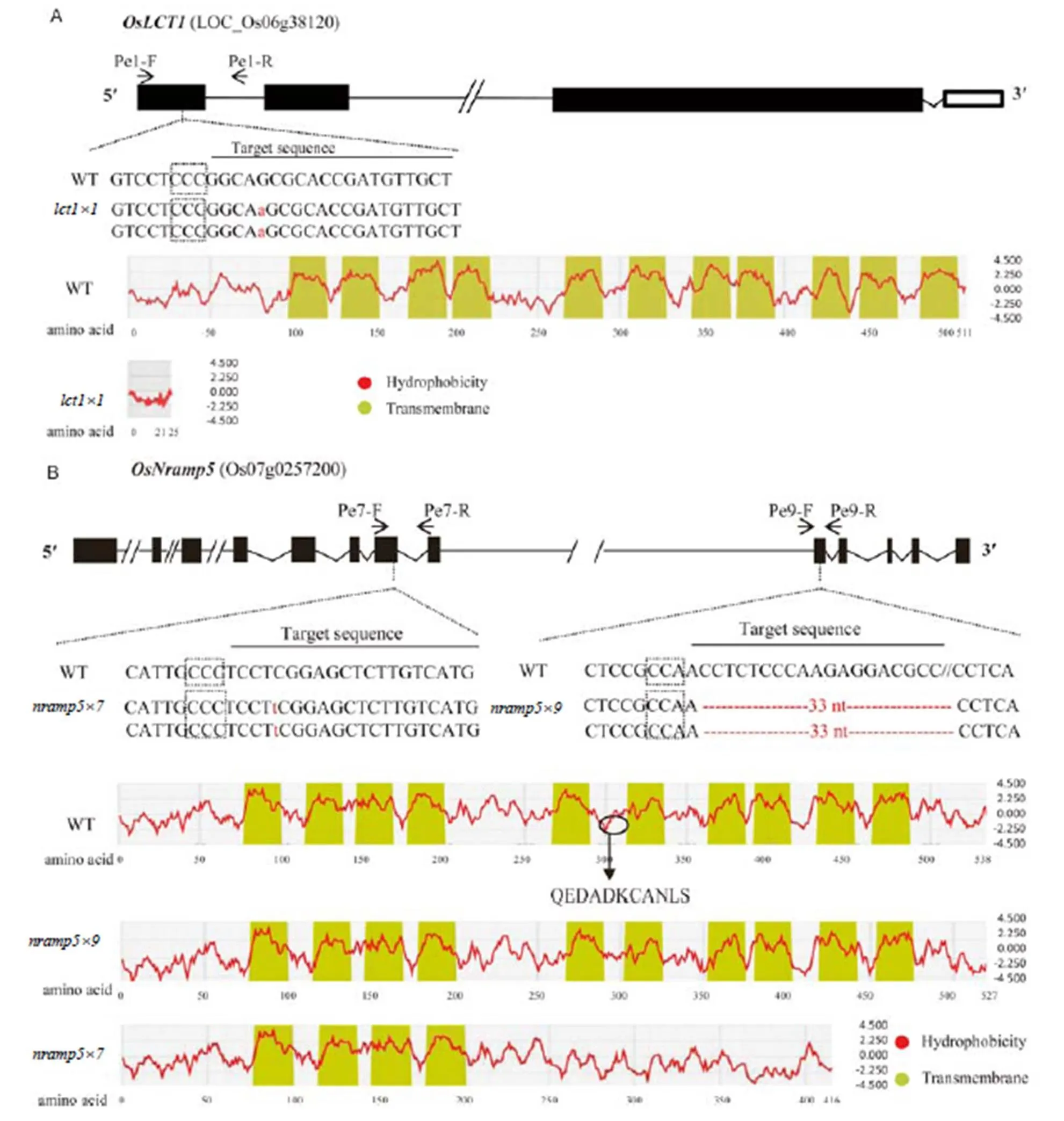
Fig. 1. Schematic diagram of the genes and sgRNA target sites for CRISPR/Cas9-mediated mutagenesis of(A) and(B), together with a hydrophobic plot of the wild type (WT) and mutant proteins.
Exons, introns, and untranslated regions are indicated by solid boxes, lines, and hollow boxes, respectively. The protospacer adjacent motif sequences (Meharg et al, 2013) prior to the target sequences are boxed with dot lines. The primers Pe1-F/-R, Pe7-F/-R and Pe9-F/-R used for genotyping the mutations and their positions are indicated by arrows. Mutations within or around the target sequences of the mutant lines,, andare shown in lowercase letters (for insertion) or ‘-’ (for deletion). The hydrophobicity (red curves) and the estimated transmembrane domains (filled boxes) of the predicted proteins were drawn using the SOSUI program (http://harrier.nagahama-i-bio.ac.jp/sosui/). For, the position of the deletion is shown in a circle in the WT hydrophobic plot and the deleted amino acid sequence is shown by uppercase letters.
Calli were induced from mature seeds of therice cultivar Xidao 1 and transformed with the three constructed CRISPR/Cas9 vectors by- mediated transformation in accordance with the method of Li et al (2016). Transgenic plantlets were regenerated from-resistant calli and acclimatized in a moist growth chamber (28 ºC with a 12 h photoperiod) for one week before being transplanted to experimental facilities.
Mutation detection in T0 plants
For detection of transgenes and mutations in regenerated T0plants, total genomic DNA was extracted from leaf tissues following a modified cetyltrimethylammonium bromide method (Zhang et al, 2014). The presence of the() gene was determined by PCR using the primers HygR_F (5′-AGAAGAAGATGTTGGCG ACCT-3′) and HygR_R (5′-GTCCTGCGGGTAAATA GCT-3′) (Li et al, 2014). Site-specific mutations were detected by PCR amplification using primer pairs flanking the designated target sites in eitheror, i.e. Pe1-F (5′-ATGACCGGCGAGGC AAGCAA-3′) and Pe1-R (5′-ATCATATCTTGAC CAAGTCG-3′) for(Fig. 1-A), and Pe7-F (5′-TGAGCATCGTGAAGCCGCCG-3′) and Pe7-R (5′-AATGCAAGAACAGATTGTGG-3′), and Pe9-F (5′-GACGGGTGCAGGTTCTTCCT-3′) and Pe9-R (5′-AGTAATAATGAATGAATGAC3′) for exons 7 and 9 of, respectively (Fig. 1-B). The following generalized PCR program was used: 5 min at 94.0 ºC, followed by 35 cycles of 30 s at 94.0 ºC, 30 s at 60.4 ºC (for Pe-1F/R and Pe-7F/R) or 55.2 ºC (for Pe-9F/R), and 30 s at 72.0 ºC, with a final extension of 7 min at 72.0 ºC.
The PCR products were first subjected to high resolution melting (HRM) analysis for mutations according to Li et al (2018), and putative mutants were sequenced (TsingKe, Hangzhou, China) and mutation sequences were decoded using the DSDecode program (http://skl.scau. edu.cn/dsdecode) (Liu et al, 2015). Mutations of a few selected plants were further confirmed by clone sequencing.
Development of transgene-free mutant lines
To obtain homozygous transgene-free mutants, T1seedlings were foliar sprayed with 1000 mg/L bentazon (approximately 100 mL/m2) at about the four-leaf stage in accordance with the method of Lu et al (2017). At least five surviving T1plants from each independent T0plant were selected for further analysis of the presence of T-DNA and site-specific mutations. Eventually, one transgene-free() and twomutant lines (and) were identified (Fig. 1). The T2plants and advanced-generation seeds were used for pot and field experiments.
Pot and field experiments
Pot experiments were conducted in the greenhouse at Zijingang Campus, Zhejiang University, Hangzhou, Zhejiang Province, China, in 2016 and 2017. Treatments with Cd (0, 2 or 10 mg/kg) were established by spiking CdCl2·5/2H2O solution to pre-dried soils taken from a paddy field of Zhejiang Zhijiang SeedTec Ltd, China. Following application of fertilizer (4.0 g urea, 2.4 g KH2PO4and 3.9 g KCl per 10 kg soil) as well as Cd spiking, the soils were mixed, watered and left for one month before transplanting seedlings. Rice seedlings were grown in plastic pots (diameter 30 cm), each containing 16 kg of air-dried (< 5 mm) soils prepared as above. After transplanting, the pots were placed in a greenhouse in which the temperature ranged from 21 ºC (night) to 33 ºC (day) and the rice plants were kept flooded with a 2–3 cm layer of water during the entire growing period. The control and each Cd treatment consisted of six pots, and each individual pot contained six plants, consisting either of three plants each ofand Xidao 1, or of two plants each of,, and Xidao 1.
Field experiments were performed in 2017 at two sites in Zhejiang Province, China, namely one site in Hangzhou without Cd contamination at the Experimental Farm of Zhejiang Zhijiang SeedTec. Ltd, and a site in a Cd-contaminated paddy field in Wenling (Tang et al, 2009), The mutant lines,andwere grown alongside the parental wild- type (WT) line Xidao 1, with 6 × 6 plants in each replicate. Routine field practices were applied in the experiment. At maturity, six plants were randomly sampled per line, and the plant height, panicle number, seed-setting rate and grain number per plant were recorded and analyzed using the SPSS 20.0 statistical software package.
Cd concentration measurement
To analyze the Cd concentration in the soils, soil samples were collected at 10 cm depth one month after CdCl2·5/2H2O spiking (for the pot experiment) or on the day of harvesting (for the field experiment). The soil samples were dried, ground into powder, sieved (< 0.154 mm), and used for mineral measurement. For analysis of the Cd concentration in rice grains, seeds were dried at 105 ºC for 30 min and kept at 60 ºC for 24 h. Brown rice grains were obtained by dehulling the seeds and were ground into powder and sieved (< 0.154 mm).
Digestion of soil samples was performed using a microwave digestion system (Mars6, USA) at 190 ºC for 50 min with 0.100 g sample, 5 mL HNO3, 1 mL HF and 1 mL HClO4. Brown rice grain samples were digested using the same method but at 160 ºC for 40 min with 0.200 g sample in 7 mL HNO3. The digested solution was concentrated at 140 ºC for 3 h until less than 1 mL of solution remained, and then diluted to 30 mL with ultrapure water (Hu et al, 2011; Meng et al, 2018). The Cd concentration was analyzed using an atomic absorption spectrometer AA800 (PerkinElmer, USA) in 2016, and by inductively coupled plasma- mass spectrometry ELAN DRC-e (PerkinElmer, USA) in 2017.
Results
Mutations of OsLCT1 and OsNramp5 in T0 plants and development of homozygous transgene-free mutant lines
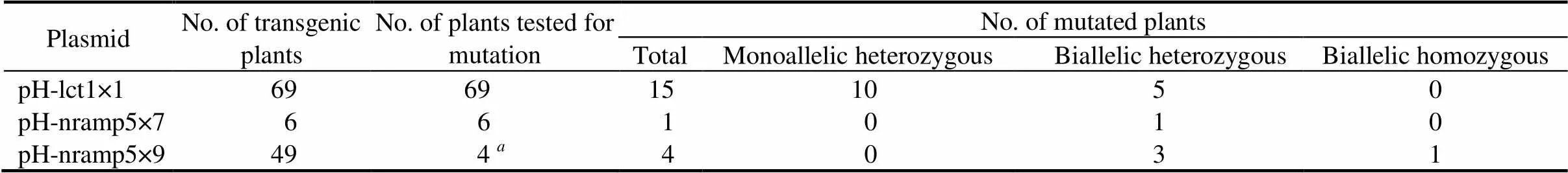
Table 1. Development and analysis of T0 rice plants for targeted mutagenesis of OsLCT1 and OsNramp5
Due to technical reasons, we failed to amplify the fragment encompassing the target site in exon 9 (Fig. 1-B) for the other plants.
A total of 124 HPT-positive T0plants were raised from transformation with the three CRISPR/Cas9 vectors (Table 1). Due to the difficulty in designing reliable primers for PCR amplification of the fragment encompassing the target site in exon 9 of, a number of PCRs failed and consequently only four plants were included for mutation analysis of the 49 T0plants transformed with pH-nramp59 (Table 1). Two-thirds of the mutated pH-lct1×1 T0plants were monoallelic heterozygous and the remaining mutants carried biallelic heterozygous mutations, i.e. two different mutations at the same target site (Table 1).
Seeds of the 20 mutated T0plants were harvested and grown as T1plant lines. After bentazon treatment at the seedling stage, about ten surviving plants of each T1line were sequenced for the target site. Consequently, 15 types of mutation were identified, ranging from a single nucleotide (nt) insertion/deletion to deletion of short fragments up to 49 nt (Fig. 2).
All T1plants of three selected lines, one each derived from the three CRISPR/Cas9 vectors, were further tested for both the presence of T-DNA and mutation. Seeds of these verified transgene-free, homozygous mutant plants, designated as the mutant lines,and, were harvested and used for further evaluation (Fig. 1).
Bothandharbored a 1-nt insertion, which generated a premature stop codon in the respective genes and resulted in significantly shortened encoded proteins, particularly for(Fig. 1). Indeed,was predicted to produce a protein of only 25 amino acids (a.a.) (Fig. 1-A). On the other hand, themutation was predicted to encode protein which was 122 a.a shorter than that of the WT and the amino acids following the mutation site were quite different to those of the WT, resulting in the loss of six predicted transmembrane domains (Fig. 1-B). In contrast, themutation (a 33-nt deletion) would result in the loss of an 11-a.a. fragment from a.a. positions 295 to 306, but not affect the number and property of transmembrane domains (Fig. 1-B).

Fig. 2. Mutations identified within and around the target sites ofandgenerated through CRISPR/Cas9-mediated genome editing in rice in T1generation.
The protospacer adjacent motif sequences are boxed. The 20-nt target sequences are underlined. Dashes indicate deletions and lowercase letters indicate inserted nucleotides.
Effects of OsLCT1 and osnramp5 mutations on Cd concentration in brown rice grains
In the 2016 pot experiment, the Cd concentration in the non-spiked control soil was 0.25 mg/kg, whereas the Cd concentrations in the soils spiked with 2 and 10 mg/kg Cd were 2.01 and 11.29 mg/kg, respectively. Under the 2 mg/kg Cd treatment, the Cd concentration in brown rice grains increased substantially to 2.13 and 1.29 mg/kg in Xidao 1 and, respectively, by contrast,mutants retained a low Cd concentration in the brown rice grains, i.e., 0.04–0.06 mg/kg (Fig. 3-A). Under the 10 mg/kg Cd treatment, Cd concentrations in brown rice grains of all the tested lines rose drastically to 6.02 mg/kg in Xidao 1, 0.44 mg/kg in theand 0.46 mg/kg in themutants.
In the 2017 pot experiment,andwere further tested with Xidao 1 as the control. The Cd concentration in non-spiked and spiked soils was similar to those observed in 2016, namely 0.24 mg/kg in control pots, and 2.01 and 10.70 mg/kg in the 2 and 10 mg/kg Cd treatments, respectively. Similar to the results of the 2016 experiment, significantly and markedly lower Cd concentrations were observed in brown rice grains of the twomutant lines compared with that of Xidao 1 (Fig. 3-B). For example, the Cd concentration increased by 23.4 and 80.4 times in brown rice grains of Xidao 1 grown in soil spiked with 2 and 10 mg/kg Cd compared with that of plants grown in non-spiked soil, whereas the respective increases in the two mutants were 2.6–8.3 and 56.7–64.5 times (Fig. 3-B). Brown rice grains of the two mutants consistently contained extremely low Cd levels (< 0.06 mg/kg) when grown in non-spiked soils as well as in Cd-spiked soils of the 2 mg/kg treatment (Fig. 3-B).
The effects ofandmutations on Cd concentration in brown rice grains were further assessed through two field experiments at Hangzhou and Wenling, Zhejiang Province, China, in 2017. The soils at the experimental sites contained Cd concentrations of 0.24 ± 0.03 mg/kg at Hangzhou and 0.90 ± 0.04 mg/kg at Wenling. Similar to the pot experiments, brown rice grains of Xidao 1 and the three mutant lines all contained extremely low Cd concentrations (< 0.04 mg/kg) when grown in non-contaminated paddy soils at Hangzhou (Fig. 3-C). At Wenling, the Cd concentration of bothmutants remained extremely low (0.02 mg/kg), whereas that of Xidao 1 andincreased significantly to 0.47 and 0.17 mg/kg, respectively (Fig. 3-C). Compared with Xidao 1, the grain Cd concentrations ofwere decreased by 45.9% and 63.0% at Hangzhou and Wenling, respectively (Fig. 3-C). The decrease in Cd concentration was even greater inandthan in, i.e., 67.2% and 64.4% at Hangzhou, and 96.6% and 96.6% at Wenling, respectively (Fig. 3-C).
Impacts of OsLCT1 and osnramp5 mutations on plant growth and yield
Agronomic performances of Xidao 1 and its mutants were investigated in the field experiment at Wenling, Zhejiang Province in 2017. No visible phenotypic differences were observed between plants of Xidao 1 and the mutant linesandat all growth stages when grown in paddy fields (Fig. 4-A). However, the growth ofplants was significantly impaired, resulting in plants significantly shorter (-9.43%) than those of Xidao 1 at the maturity stage (Fig. 4-A).
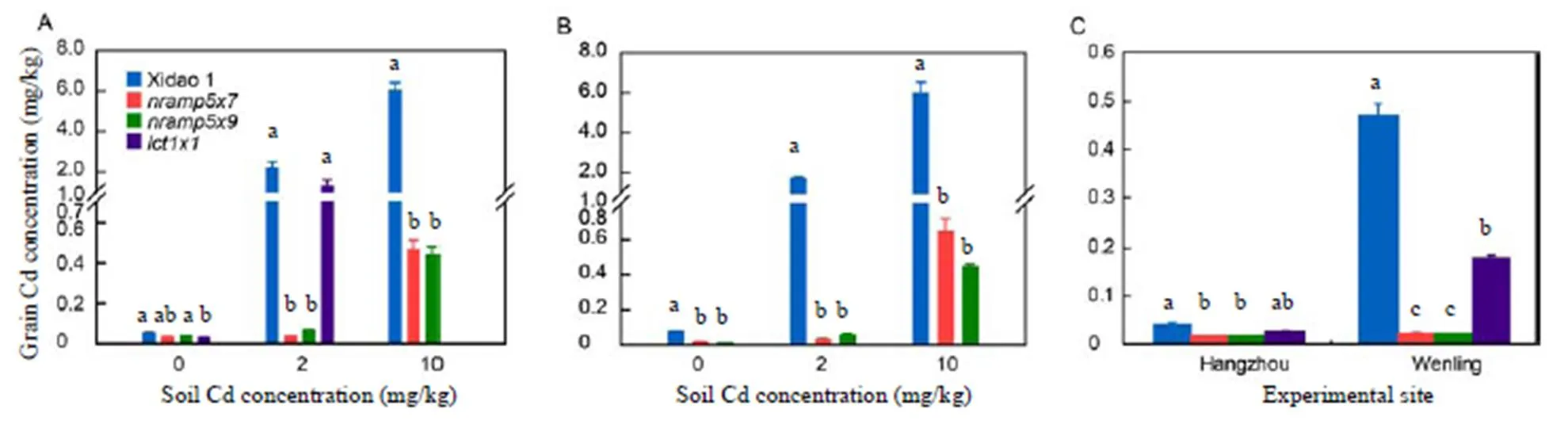
Fig. 3. Cadmium content in brown rice grains of the mutant lines,,and wild type Xidao 1.
A, Pot experiment in 2016. B, Pot experiment in 2017. C, Field experiment in 2017.
Plants were grown in soil spiked with 2 or 10 mg/kg Cd. Different letters above columns indicate significant difference atthe 0.05 level. The data presented are Mean ± SE (= 3).
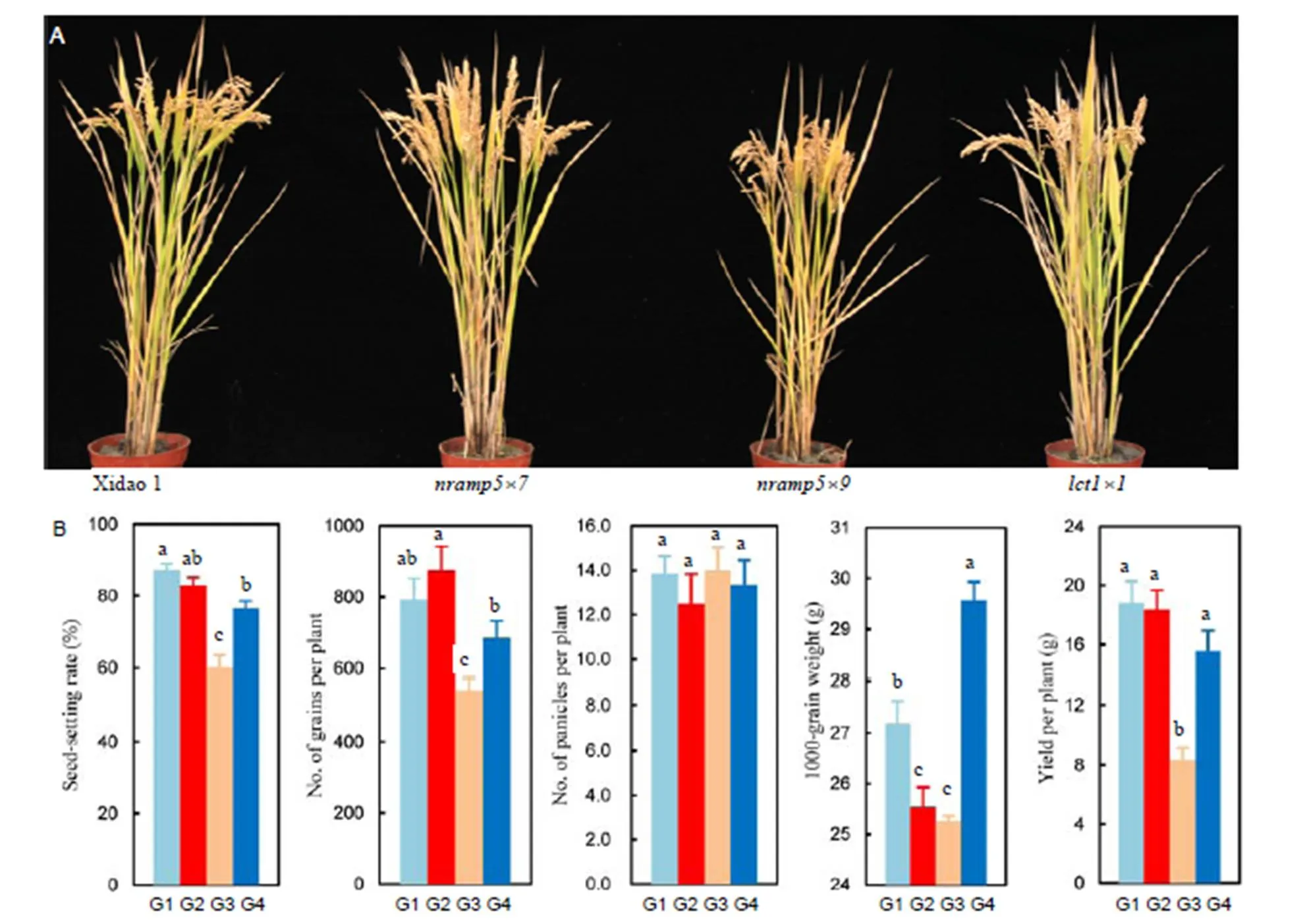
Fig. 4. Plant morphology (A) and yield-related traits (B) of Xidao 1 and itsandmutants grown in a paddy field at Hangzhou, China, in 2017.
G1, Xidao 1; G2,; G3,; G4,.
Data are presented as Mean ± SE (= 6). Different letters above columns indicate significant differences at the 0.05 level.
Compared with Xidao 1, no significant differences were observed in total grain number and panicle number per plant of, whereas seed-setting rate was significantly decreased (-13.7%) and 1000-grain weight increased (8.8%) with significantly increased seed length (data not shown). Consequently, the yield per plant ofwas 17.3% lower than that of Xidao 1. The mutant lineshowed similar seed-setting rate, panicle number per plant and number of grains per plant compared to Xidao 1, but 1000-grain weight was significantly lower (-6.1%). Consequently, the seed yield per plant ofwas not significantly different from that of Xidao 1 (Fig. 4-B). Except for panicle number per plant, in the mutant, the seed-setting rate, total grain number per plant and 1000-grain weight were all significantly reduced by 31%, 32% and 7%, respectively, compared with those of Xidao 1 (Fig. 4-B). Not surprisingly,showed extremely lower yield per plant (only 55.7%) than that of Xidao 1 (Fig. 4-B).
Discussion
The CRISPR/Cas9 system has been widely applied in plant functional genomics studies. Theoretically, it will empower plant breeders to develop crop cultivars more efficiently, thus promoting a new paradigm in plant breeding. However, its successful application in plant breeding has so far largely been limited to traits that can be achieved by conventional mutagenesis, e.g. fragrance and waxy grains in rice (Baysal et al, 2016; Shao G N et al, 2017; Zhang et al, 2017). Whether the technology can also be used for improvement of complex traits, which are the main breeding targets, awaits more exemplary studies (Wright et al, 2016; Jung et al, 2017). The present investigation aimed to provide such an instance on the manipulation of Cd accumulation in rice grains by targeted mutagenesis of Cd transporter genes.
Mutational effect of OsNramp5 and OsLCT1
In this study, theandmutants showed significantly decreased Cd concentrations in the grains (Fig. 3). However, the two mutations had significantly different impacts on plant growth. Although the 33-nt deletion would only result in a loss of 11 a.a. of the OsNramp5 protein (Fig. 1-B), the growth ofplants was severely stunted (Fig. 4-A) and grain yield was substantially reduced (Fig. 4-B). The 1-nt insertion in exon 7 ofwas expected to substantially truncate its encoded protein with the last six transmembrane domains removed (Fig. 1-B), but this had no negative effect on plant growth and yield (Fig. 4-B). These results of the impact ofmutation on rice growth and yield are overall consistent with previous reports. Sasaki et al (2012) reported that total disruption significantly affects plant growth and reduces yield, while Ishikawa et al (2012) and Tang et al (2017) reported that, based on observation of an ion beam- mutant and CRISPR/Cas9-generated mutant lines, respectively, mutations ofresult in significantly reduced Cd accumulation without significant effects on agronomic performances. Uraguchi et al (2011) reported that knockdown mutants ofcould reduce Cd accumulation in rice grains without affecting plant growth and the accumulation of other mineral elements. Themutant generated in this study is the first knockoutmutant. Similar tosuppression, the knockout mutation significantly reduced Cd accumulation ingrains (Fig. 3), however, with negative impact on seed-setting rate but increased 1000-grain weight, the overall growth ofremained similar to that of Xidao 1 (Fig. 4-B).
Most genes that control important agronomic traits usually play multiple roles in plant growth and defense. Hence, mutation of these genes often affects the performance of other traits, causing the pleiotropic effects. The inconsistency among the present results for the two mutations and those reported previously strongly suggests thatandperform multiple functions in rice growth. Their roles might be (partially) complemented by other genes in certain genotypes when they are mutated, e.g. in Huazhan (Tang et al, 2017). It is also possible that certain mutations, e.g., themutation in the present study, disrupted its function for Cd transport but did not affect other, yet unknown, functions associated with plant growth. Furthermore, somaclonal variations are not uncommon and could result in inferior performance of progenies from transgenic plants (Shu et al, 2002). Therefore, additional studies are also needed to assess whether the reduced agronomic performance is due toandmutations or is caused by somaclonal variations.
Usefulness of nrmap5×7 and lct1×1 for rice production in Cd-polluted paddy fields
In China, the Cd concentration in soils of most Cd-polluted paddy fields is less than 1.5 mg/kg (Zhao et al, 2015). The pot and field experiments eloquently demonstrated that the mutant linecould produce grains with extremely low Cd concentrations even when grown in heavily polluted soils (Fig. 3). Thus, it shows promise to maintain a grain Cd concentration below the limit of the China National Food Safety Standard (i.e., 0.2 mg/kg) when grown in Cd-contaminated paddy soils with Cd concentrations of 2.0 mg/kg or higher. However, the utility of themutantneeds further evaluation. The mutant would be safe to grow in paddy soils with Cd contamination up to 1.0 mg/kg based on the results of the paddy field experiment, but the pot experiments also indicated thatis much more sensitive than themutants to the increase in soil Cd concentration.
Given that the field experiments were extremely limited in area at both locations, further evaluation of the mutants’ agronomic performance is needed before large-scale production. If yield is eventually proven to be negatively impacted, then a cost-effect evaluation should be performed for comparison of the relative merits of other intervention measures, such as liming or application of passivation agents (Zhao et al, 2015; Zhu et al, 2016). On the other side, further breeding may also ameliorate the negative impact observed in the mutants, either resulting from somaclonal variation (i.e., elimination of background mutations) or from pleiotropic effect ofandmutations (i.e. introducing beneficial genes). If the yield is barely affected, the mutant lines could not only directly be used in rice production in areas with Cd contamination, but also be served as low-Cd accumulated germplasm for future rice breeding programs.
In conclusion, this study demonstrated that it is feasible to generate yield-competitive, low-Cd- accumulated rice lines, e.g. by targeted mutagenesis of, but a sufficient number of mutant plants should be generated and evaluated.
Acknowledgements
We thank workers in the farms of Zhejiang Zhijiang SeedTec Ltd and Wenling for taking care of the paddy fields. We also appreciate the assistance in Cd measurement of Mr. Wu Zhongchang and Mr. Xu Jiming from the College of Life Sciences, Zhejiang University, Hangzhou, China. This work was supported by the Zhejiang Provincial S & T Project on Breeding of Agricultural (Food) Crops (Grant No. 2016C02050-2).
Abe T, Ito M, Takahashi R, Honma T, Sekiya N, Shirao K, Kuramata M, Murakami M, Ishikawa S. 2017. Breeding of a practical rice line ‘TJTT8’ for phytoextraction of cadmium contamination in paddy fields., 63(4): 388–395.
Arao T, Ae N. 2003. Genotypic variations in cadmium levels of rice grain., 49(4): 473–479.
Arao T, Ishikawa S, Murakami M, Abe K, Maejima Y, Makino T. 2010. Heavy metal contamination of agricultural soil and countermeasures in Japan., 8(3): 247–257.
Aziz R, Rafiq M T, Li T Q, Liu D, He Z L, Stoffella P J, Sun K W, Yang X E. 2015. Uptake of cadmium by rice grown on contaminated soils and its bioavailability/toxicity in human cell lines (Caco-2/HL-7702)., 63(13): 3599–3608.
Baysal C, Bortesi L, Zhu C, Farré G, Schillberg S, Christou P. 2016. CRISPR/Cas9 activity in the ricegene does not induce off-target effects in the closely related paralog., 36: 108.
Clemens S, Aarts M G M, Thomine S, Verbruggen N. 2013. Plant science: The key to preventing slow cadmium poisoning., 18(2): 92–99.
FAO/WHO. 2018. Evaluation of certain food additives and contaminants (seventy-third report of the joint FAO/WHO expert committee on food additives). WHO Technical Report Series.
Grant C A, Clarke J M, Duguid S, Chaney R L. 2008. Selection and breeding of plant cultivars to minimize cadmium accumulation., 390: 301–310.
Hu L F, McBride M B, Cheng H, Wu J J, Shi J C, Xu J M, Wu L S. 2011. Root-induced changes to cadmium speciation in the rhizosphere of two rice (L.) genotypes., 111(3): 356–361.
Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa N K, Nakanishi H. 2012. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice., 109: 19166–19171.
Ishizaki T. 2016. CRISPR/Cas9 in rice can induce new mutations in later generations, leading to chimerism and unpredicted segregation of the targeted mutation., 36: 165.
Jung C, Capistrano-Gossmann G, Braatz J, Sashidhar N, Melzer S. 2017. Recent developments in genome editing and applications in plant breeding., 137(1): 1–9.
Lei Y, Lu L, Liu H Y, Li S, Xing F, Chen L L. 2014. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR- system in plants., 7(9): 1494–1496.
Li S, Liu S M, Liu Y H, Lu H P, Tan Y Y, Huang J Z, Wei P C, Shu Q Y. 2018. HRM-facilitated rapid identification and genotyping of mutations induced by CRISPR/Cas9 mutagenesis in rice., 18: 184–191.
Li W L, Xu B B, Song Q J, Liu X M, Xu J M, Brookes P C. 2014. The identification of ‘hotspots’ of heavy metal pollution in soil-rice systems at a regional scale in eastern China., 472: 407–420.
Li W X, Wu S L, Liu Y H, Jin G L, Zhao H J, Fan L J, Shu Q Y. 2016. Genome-wide profiling of genetic variation in- transformed rice plants., 17(12): 992–996.
Liu W Z, Xie X R, Ma X L, Li J, Chen J H, Liu Y G. 2015. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations., 8: 1431–1433.
Lu H P, Liu S M, Xu S L, Chen W Y, Zhou X, Tan Y Y, Huang J Z, Shu Q Y. 2017. CRISPR-S: An active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants., 15(11): 1371–1373.
Meharg A A, Norton G, Deacon C, Williams P, Adomako E E, Price A, Zhu Y G, Li G, Zhao F J, McGrath S, Villada A, Sommella A, de Silva P M C S, Brammer H, Dasgupta T, Islam M R. 2013. Variation in rice cadmium related to human exposure., 47(11): 5613–5618.
Meng J, Zhong L B, Wang L, Liu X M, Tang C X, Chen H L, Xu J M. 2018. Contrasting effects of alkaline amendments on the bioavailability and uptake of Cd in rice plants in a Cd- contaminated acid paddy soil., 25(9): 8827–8835.
Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Satoh-Nagasawa N, Watanabe A, Fujimura T, Akagi H. 2010. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles., 189(1): 190–199.
Peng X X, Bai N N, Wang H H. 2018. Isolation and expression profiles of cadmium stress-responsive rice WRKY15 transcription factor gene., 32(2): 103–110. (in Chinese with English abstract)
Pinson S R M, Tarpley L, Yan W G, Yeater K, Lahner B, Yakubova E, Huang X Y, Zhang M, Guerinot M L, Salt D E. 2015. Worldwide genetic diversity for mineral element concentrations in rice grain., 55(1): 294–311.
Sasaki A, Yamaji N, Yokosho K, Ma J F. 2012. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice., 24(5): 2155–2167.
Sasaki A, Yamaji N, Ma J F. 2014. Overexpression ofenhances Cd tolerance and expression of Zn transporter genes in rice., 65(20): 6013–6021.
Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, Takahashi H, Watanabe A, Akagi H. 2012. Mutations in rice () heavy metal ATPase 2 () restrict the translocation of zinc and cadmium., 53(1): 213–224.
Shao G N, Xie L H, Jiao G A, Wei X J, Sheng Z H, Tang S Q, Hu P S. 2017. CRISPR/CAS9-mediated editing of the fragrant genein rice., 31: 216–222. (in Chinese with English abstract)
Shao J F, Fujii-Kashino M, Yamaji N, Fukuoka S, Shen R F, Ma J F. 2017. Isolation and characterization of a rice line with high Cd accumulation for potential use in phytoremediation., 410: 357–368.
Shimo H, Ishimaru Y, An G, Yamakawa T, Nakanishi H, Nishizawa N K. 2011. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice., 62(15): 5727–5734.
Shu Q Y, Cui H R, Ye G Y, Wu D X, Xia Y W, Gao M W, Altosaar I. 2002. Agronomic and morphological characterization of-transformed Bt rice plants., 127(3): 345–352.
Smolders E, Mertens J. 2013. Cadmium.: Alloway B J. Heavy Metals in Soils: Trace Metals and Metalloids in Soils and Their Bioavailability. Dordrecht: Springer Netherlands: 283–311.
Sun Y L, Liu H M, Xu Q G. 2017. Effects of cadmium stress on rice seed germination characteristics., 31(4): 425–431. (in Chinese with English abstract)
Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa N K. 2011. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice., 62(14): 4843–4850.
Tang L, Mao B G, Li Y K, Lv Q M, Zhang L P, Chen C Y, He H J, Wang W P, Zeng X F, Shao Y, Pan Y L, Hu Y Y, Peng Y, Fu X Q, Li H Q, Xia S T, Zhao B R. 2017. Knockout ofusing the CRISPR/Cas9 system produces low Cd-accumulatingrice without compromising yield., 7(1): 14438.
Tang X J, Shen C F, Shi D Z, Cheema S A, Khan M I, Zhang C K, Chen Y X. 2009. Heavy metal and persistent organic compound contamination in soil from Wenling: An emerging e-waste recycling city in Taizhou area, China., 173: 653–600.
Ueno D, Yamaji N, Kono I, Huang C F, Ando T, Yano M, Ma J F. 2010. Gene limiting cadmium accumulation in rice., 107: 16500–16505.
Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, Nagamura Y, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T. 2011. Low- affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains., 108: 20959–20964.
Uraguchi S, Kamiya T, Clemens S, Fujiwara T. 2014. Characterization of OsLCT1, a cadmium transporter fromrice ()., 151(3): 339–347.
Wright A V, Nuñez J K, Doudna J A. 2016. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering., 164: 29–44.
Xu R F, Li H, Qin R Y, Wang L, Li L, Wei P C, Yang J B. 2014. Gene targeting using the-mediated CRISPR-Cas system in rice., 7(1): 5.
Yamaji N, Xia J X, Mitani-Ueno N, Yokosho K, Ma J F. 2013. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2., 162(2): 927–939.
Yao W Y, Sun L, Zhou H, Yang F, Mao D H, Wang J R, Chen L H, Zhang G Y, Dai J P, Xiao G Y, Chen C Y. 2015. Additive, dominant parental effects control the inheritance of grain cadmium accumulation in hybrid rice., 35(1): 39.
Zeng F R, Mao Y, Cheng W D, Wu F B, Zhang G P. 2008. Genotypic and environmental variation in chromium, cadmium and lead concentrations in rice., 153(2): 309–314.
Zhang H L, Huang J Z, Chen X Y, Tan Y Y, Shu Q Y. 2014. Competitive amplification of differentially melting amplicons facilitates efficient genotyping of photoperiod- and temperature- sensitive genic male sterility in rice., 34(4): 1765–1776.
Zhang J S, Zhang H, Botella J R, Zhu J K. 2017. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of thegene in elite rice varieties., 60(5): 369–375.
Zhang X Q, Zhang G P, Guo L B, Wang H Z, Zeng D L, Dong G J, Qian Q, Xue D W. 2011. Identification of quantitative trait loci for Cd and Zn concentrations of brown rice grown in Cd-polluted soils., 180(2): 173–179.
Zhao F J, Ma Y B, Zhu Y G, Tang Z, McGrath S P. 2015. Soil contamination in China: Current status and mitigation strategies., 49(2): 750–759.
Zhu H H, Chen C, Xu C, Zhu Q H, Huang D Y. 2016. Effects of soil acidification and liming on the phytoavailability of cadmium in paddy soils of central subtropical China., 219: 99–106.
(Managing Editor: Li Guan)
Copyright © 2019, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2019.01.002
27 September 2018;
26 November 2018
Huang Jianzhong (jzhuang@zju.edu.cn)
杂志排行
Rice Science的其它文章
- Improvements of TKC Technology Accelerate Isolation of Transgene-Free CRISPR/Cas9-Edited Rice Plants
- Production of Two Elite Glutinous Rice Varieties by Editing Wx Gene
- Development and Application of CRISPR/Cas System in Rice
- Rapid Creation of New Photoperiod-/Thermo-Sensitive Genic Male-Sterile Rice Materials by CRISPR/Cas9 System
- CRISPR/Cas9-Mediated Adenine Base Editing in Rice Genome
- Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice
