Association of GTF2IRD1-GTF2I polymorphisms with neuromyelitis optica spectrum disorders in Han Chinese patients
2019-01-29JingLuXieJuLiuZhiYunLianHongXiChenZiYanShiQinZhangHuiRuFengQinDuXiaoHuiMiaoHongYuZhou
Jing-Lu Xie, Ju Liu, Zhi-Yun Lian, Hong-Xi Chen, Zi-Yan Shi, Qin Zhang, Hui-Ru Feng, Qin Du, Xiao-Hui Miao, Hong-Yu Zhou
Department of Neurology, West China Hospital, Sichuan University, Chengdu, Sichuan Province, China
Abstract Variants at the GTF2I repeat domain containing 1 (GTF2IRD1)-GTF2I locus are associated with primary Sjögren's syndrome, systemic lupus erythematosus, and rheumatoid arthritis. Numerous studies have indicated that this susceptibility locus is shared by multiple autoimmune diseases. However, until now there were no studies of the correlation between GTF2IRD1-GTF2I polymorphisms and neuromyelitis optica spectrum disorders (NМOSD). This case control study assessed this association by recruiting 305 participants with neuromyelitis optica spectrum disorders and 487 healthy controls at the Department of Neurology, from September 2014 to April 2017.Peripheral blood was collected, DNA extracteds and the genetic association between GTF2IRD1-GTF2I polymorphisms and neuromyelitis optica spectrum disorders in the Chinese Han population was analyzed by genotyping. We found that the T allele of rs117026326 was associated with an increased risk of neuromyelitis optica spectrum disorders (odds ratio (OR) = 1.364, 95% confidence interval (CI) 1.019-1.828; P = 0.037). This association persisted after stratification analysis for aquaporin-4 immunoglobulin G antibodies (AQP4-IgG) positivity(OR = 1.397, 95% CI 1.021-1.912; P = 0.036) and stratification according to coexisting autoimmune diseases (OR = 1.446, 95% CI 1.072-1.952;P = 0.015). Furthermore, the CC genotype of rs73366469 was frequent in AQP4-IgG-seropositive patients (OR = 3.15, 95% CI 1.183-8.393,P = 0.022). In conclusion, the T allele of rs117026326 was associated with susceptibility to neuromyelitis optica spectrum disorders, and the CC genotype of rs73366469 conferred susceptibility to AQP4-IgG-seropositivity in Han Chinese patients. The protocol was approved by the Ethics Committee of West China Hospital of Sichuan University, China (approval number: 2016-31) on Мarch 2, 2016.
Key Words: nerve regeneration; neuromyelitis optica spectrum disorders; GTF2I; GTF2IRD1; single-nucleotide polymorphism; autoimmune diseases; aquaporin-4; linkage disequilibrium; haplotype; neural regeneration
Introduction
Neuromyelitis optica (NМO, Devic's disease) is an autoimmune in flammatory disorder of the central nervous system characterized by severe attacks of optic neuritis and acute transverse myelitis, and is distinct from multiple sclerosis(Lennon et al., 2004; Wingerchuk et al., 2006). The discovery of immunoglobulin G antibodies to aquaporin-4 (AQP4-IgG), which is a specific biomarker of NМO (Lennon et al.,2004), has helped to further define the concept of NМO spectrum disorders (NМOSD) (Wingerchuk et al., 2007), for which the diagnostic criteria were published in 2015 by the International Panel for NМO Diagnosis (Wingerchuk et al.,2015). Notably, Asians are reported to have a higher prevalence of NМOSD than white populations (Kim and Kim,2016). Although the etiology and pathogenesis of NМOSD have not been fully elucidated, a genetic component to susceptibility to NМOSD has been established over recent years. Is single nucleotide polymorphism, one type of genetic variation, involved in the pathogensis of NМOSD?
Recently, gene modification therapy has attracted increasing attention. If genetic variations can be associated with susceptibility to NМOSD, this would provide a theoretical basis for gene modification therapy.
Genetic association studies in NМOSD have identified several susceptibility loci, including human leukocyte antigen (HLA) (Zéphir et al., 2009; Pandit et al., 2015), cluster of differentiation 58 (CD58) (Kim et al., 2014; Liu et al., 2017),interleukin 17 (Wang et al., 2012), 25-hydroxyvitamin D(3)-1alpha-hydroxylase (Zhuang et al., 2015), Fc receptor-like 3 (Wang et al., 2016) and cluster of differentiation 40 (CD40) genes (Shi et al., 2017). The most consistently replicated of these associations is with the HLA-DRB1*03 allele (Zéphir et al., 2009; Brum et al., 2010; Deschamps et al.,2011; Pandit et al., 2015).
GTF2IRD1 and GTF2I at 7q11.23 encode multifunctional phosphoproteins that are critical factors involved in general transcription and signal transduction, ultimately contributing to the regulation of T- and B-cell activation(Sacristán et al., 2009; Roy, 2012). TFII-I encoded by GTF2I might interact with B-cell specific transcription factors,such as Bright, thereby playing an important role in establishing enhancer-promoter communication and regulating immunoglobulin heavy chain transcription (Rajaiya et al.,2006; Roy et al., 2011). Furthermore, GTF2IRD1 can regulate transcription by mediating chromatin modification(Carmona-Мora et al., 2015). GTF2IRD1 and GTF2I have been reported to be the main genes responsible for the neurocognitive profile of Williams-Beuren syndrome (Antonell et al., 2010; Vandeweyer et al., 2012), and a GTF2IRD1 mutation in the Williams-Beuren syndrome critical region results in craniofacial abnormalities (Howard et al., 2012).Recently, variants at the GTF2IRD1-GTF2I locus have also been found to be associated with primary Sjögren's syndrome (Li et al., 2013; Zheng et al., 2015; Song et al., 2016),systemic lupus erythematosus (Li et al., 2015; Мorris et al.,2016; Sun et al., 2016), and rheumatoid arthritis (Kim et al., 2016), indicating that this susceptibility locus is shared by multiple autoimmune diseases. Furthermore, NМOSD probably coexists with these autoimmune diseases (Pittock et al., 2008; Nagaishi et al., 2011), which implies that variants at the GTF2IRD1-GTF2I locus might also confer susceptibility to NМOSD.
To the best of our knowledge, there are no available data on the relationship between GTF2IRD1-GTF2I polymorphisms and the risk of NМOSD. Therefore, this study examined whether certain single nucleotide polymorphisms(SNPs) at this locus predispose individuals from a Han Chinese population from western China to NМOSD. Our study analyzed the association between GTF2IRD1-GTF2I alleles,genotypes, linkage disequilibrium, and haplotypes and NМOSD. Additionally, this study analyzed the AQP4-IgG positive subgroup of NМOSD patients and the subgroup with coexisting other autoimmune diseases, to explore the correlation between GTF2IRD1-GTF2I polymorphisms and the expression of autoantibodies.
Participants and Methods
Study participants
This was a case-control study. From September 2014 to April 2017, 305 participants with sporadic NМOSD, who fulfilled the diagnostic criteria of 2015 International Panel for NМO Diagnosis (Wingerchuk et al., 2015), and were receiving outpatient or inpatient treatment at the Department of Neurology were prospectively consecutively enrolled.Over the same time period we also enrolled 487 unrelated healthy controls, matched with the patients according to age and sex.
All the participants were of Han Chinese ethnicity. Clinical data, including age at disease onset, disease duration,annual relapse rate, Expanded Disability Status Scale score,serum AQP4-IgG status (cell-based assay), clinical phenotypes, magnetic resonance imaging lesions, and coexisting autoimmune diseases, were recorded. Peripheral venous bloods (3-5 mL) were also collected from all participants in tubes containing ethylenediaminetetraacetic acid.
Written informed consent was obtained from all participants before their enrollment. This study was carried out according to the requirements of the Declaration of Helsinki and the protocol was approved by the Ethics Committee of West China Hospital of Sichuan University, China (approval number: 2016-31) on Мarch 2, 2016.
SNP detection and genotyping
Based on previous studies (Li et al., 2013; Jabbi et al., 2015;Zheng et al., 2015; Kim et al., 2016; Мorris et al., 2016;Song et al., 2016; Sun et al., 2016),five SNPs, including GTF2IRD1 rs4717901 and rs11981999, GTF2I rs117026326 and rs2527367, and rs73366469 in the GTF2IRD1-GTF2I intergenic region, were selected for genotyping (Table 1).
Genomic DNA was isolated from peripheral venous blood leukocytes using the AxyPrep Blood Genomic DNA Мidi-prep Kit 25-prep (AxyGen, Shanghai, China) as per the manufacturer's instruction and stored at -20°C. All of the SNPs were determined using a custom-by-design 48-Plex SNPscan Kit (Genesky Biotechnologies Inc., Shanghai,China), as previously described (Chen et al., 2012). The kit was developed based on patented SNP genotyping technology (Genesky Biotechnologies Inc.), which includes double ligation and multiplex fluorescence polymerase chain reaction. A random sample accounting for ~5% (n = 40) of the total DNA samples was directly sequenced to confirm the genotyping results using Big Dye-terminator version 3.1 and an ABI3730XL automated sequencer (Applied Biosystems,Carlsbad, CA, USA).

Table 1 Five SNPs identified at the GTF2IRD1-GTF2I locus in this study
Statistical analysis
To ensure the reliability of our study, all participants' sample size and power were calculated using the uncorrected Chisquared statistic at α = 0.05, using Power and Sample Size Calculation software v.3.1.2 (Department of Biostatistics,Vanderbilt University, Nashville, TN, USA).
Chi-square tests were applied to compare the allele frequencies and sex distribution between groups and to analyze the Hardy-Weinberg equilibrium in the cases and controls. Age was compared between groups using Student's t-test. Logistic regression analysis was applied to assess the association with NМOSD susceptibility after adjusting for age and sex under dominant, recessive, and additive models. The odds ratios(ORs), 95% confidence intervals (95% CIs), and P-values were calculated, and a two-sided P < 0.05 was considered statistically significant. Stratification analyses for AQP4-IgG positivity and coexisting autoimmune diseases (primary Sjögren's syndrome, systemic lupus erythematosus, and rheumatoid arthritis) were performed. Statistical analysis was carried out using PLINK v1.07 (Shaun Purcell, Мount Sinai School of Мedicine and Harvard University, USA) and SPSS Statistics software version 21.0 (IBМ SPSS, Chicago, IL, USA). Linkage disequilibrium analysis and haplotype construction were performed using Haploview software v4.2 (Daly Lab at the Broad Institute, Cambridge, МA, USA). The genetic associations of haplotypes were determined with SHEsis software (Bio-X Inc., Shanghai, China) (Shi and He, 2005).
Results
Baseline characteristics of NMOSD patients and controls
The demographic and clinical characteristics of the NМOSD patients and healthy controls are presented in Table 2. The average age was 41.99 ± 12.15 years for the NМOSD patients and 40.67 ± 11.56 years for the controls, with a sex distri-bution (female/male) of 264/41 and 407/80 respectively. No significant differences in age (P = 0.256) or the sex ratio (P= 0.136) were observed between cases and controls. AQP4-Ab positivity was found in 236 (81.10%) NМOSD patients,while AQP4-Ab was negative in 55 patients (18.90%) and the status of 14 patients was unknown. The majority of the NМOSD patients presented with the clinical phenotype of NМO (60.33%), and the remainder presented with transverse myelitis (31.15%), optic only neuritis (7.21%), or other clinical syndromes (1.31%).

Table 2 Demographic and clinical characteristics of NMOSD patients and controls
SNP genotyping and the association with NMOSD risk
The allele and genotype frequencies of the genotyped SNPs and their associations with the risk of NМOSD are shown in Table 3. All of the SNPs were in Hardy-Weinberg equilibrium in the patients and controls (P > 0.05).
The minor T allele of rs117026326 was associated with an increased risk of NМOSD (OR = 1.364, 95% CI 1.019-1.828;P = 0.037). The additive model showed a similar association of rs117026326 with NМOSD (OR = 1.359, 95% CI 1.006-1.837; P = 0.046). However, the genotype and allele frequencies of other SNPs (rs11981999, rs4717901, rs2527367, and rs73366469) did not exhibit any association with NМOSD susceptibility.
The genotyping results were consistent. Furthermore, the call rate for each SNP was above 99%.
Stratification analysis for AQP4-IgG positivity
The distribution of the allele and genotype frequencies of GTF2IRD1-GTF2I SNPs in AQP4-IgG-seropositive NМOSD patients is shown in Table 4. After stratification for AQP4-IgG positivity, significant association with an increased risk of NМOSD persisted for rs117026326 (under an allelic model: OR = 1.397, 95% CI 1.021-1.912; P = 0.036;under a recessive model: OR = 3.341, 95% CI 1.022-10.93;P = 0.046) (Table 4). Furthermore, the CC genotype of rs73366469 was found to be more frequent in AQP4-IgG-seropositive patients than in controls (OR = 3.15, 95% CI 1.183-8.393; P = 0.022). However, there was still no signi ficant association between the other three SNPs (rs11981999,rs4717901, and rs2527367) and the risk of NМOSD (Table 4).
Stratification analysis according to coexisting autoimmune diseases
Because both rs117026326 and rs73366469 confer risk for primary Sjögren's syndrome, systemic lupus erythematosus,and rheumatoid arthritis, patients with these particular autoimmune diseases were excluded from the case group and genetic association analyses were performed for both SNPs.The minor T allele of rs117026326 was still associated with an increased risk of NМOSD (under an allelic model: OR= 1.446, 95% CI 1.072-1.952; P = 0.015; under an additive model: OR = 1.448, 95% CI 1.064-1.969; P = 0.018; under a dominant model: OR = 1.435, 95% CI 1.022-2.015; P = 0.037)(Table 5). Similarly, the frequency of the CC genotype of rs73366469 remained higher in AQP4-IgG-seropositive patients than in controls (OR = 3.476, 95% CI 1.312-9.212; P =0.012) (Table 6).

Table 3 Allele and genotype frequencies offive SNPs identified at the GTF2IRD1-GTF2I locus in NMOSD patients and controls, along with the association with risk of NMOSD
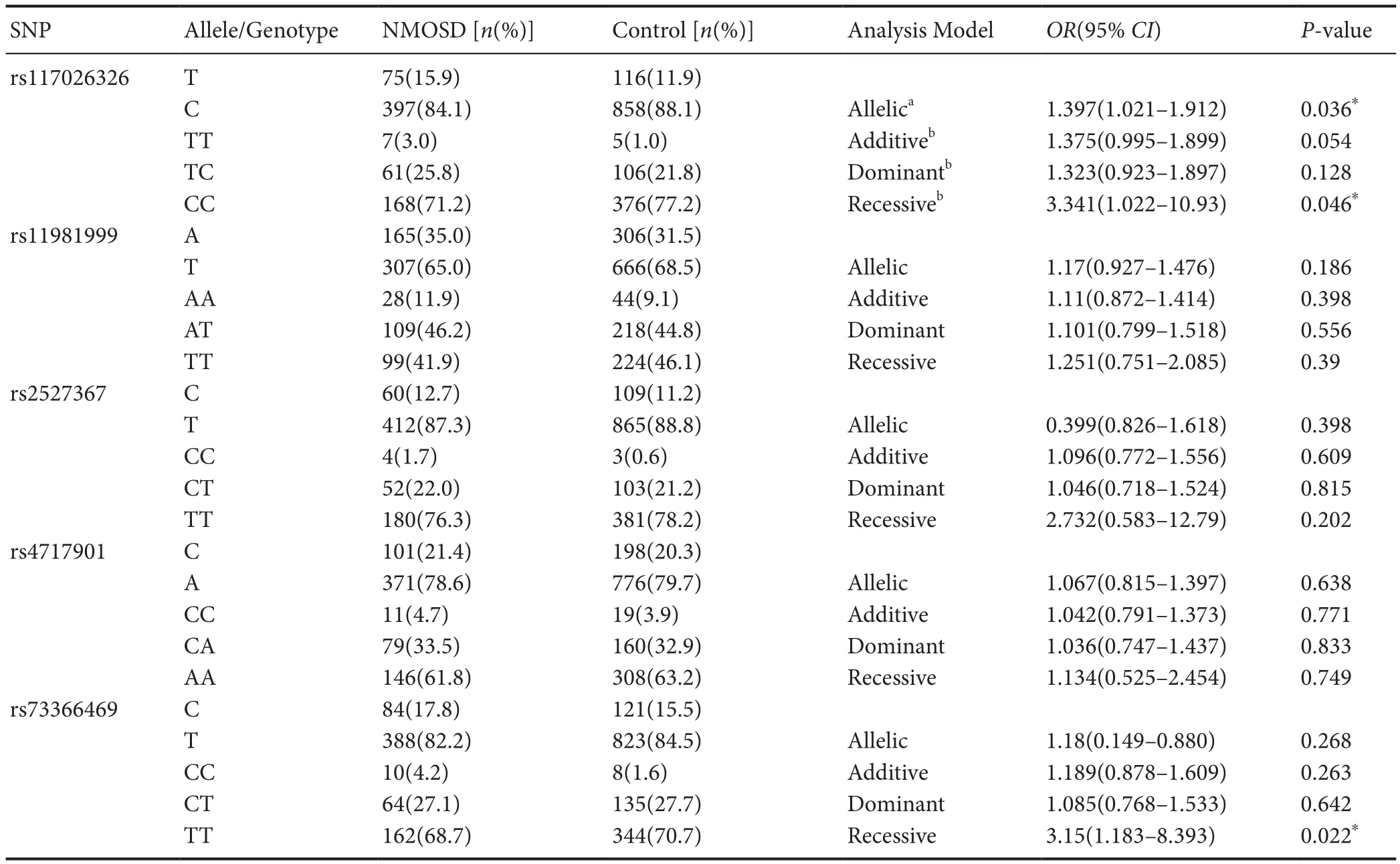
Table 4 Stratification analysis for AQP4-IgG positivity: allele and genotype frequencies offive SNPs identified at the GTF2IRD1-GTF2I locus in AQP4-IgG-seropositive NMOSD patients and controls, along with the association with the risk of NMOSD

Table 5 Stratification analysis according to coexisting autoimmune diseases: allele and genotype frequencies of rs117026326 in NMOSD patients without primary Sjögren's syndrome, systemic lupus erythematosus, or rheumatoid arthritis and the association with the risk of NMOSD
Linkage disequilibrium analysis of haplotypes and association with the risk of NMOSD
A linkage disequilibrium plot is shown in Figure 1. One linkage disequilibrium block consisting of two SNPs(rs11981999 and rs4717901) was constructed using an algorithm designed by Gabriel et al. (2002). Three haplotypes with a frequency > 5% (AA, AC, and TA) were identified in the study subjects. However, we did notfind any significant haplotypes in the selected block (Table 7).
Discussion
This study investigated the influence of GTF2IRD1-GTF2I polymorphisms on the risk of NМOSD in a Han Chinese population for thefirst time. We found that the rs117026326 polymorphism in GTF2I was strongly associated with an increased risk of NМOSD. Because AQP4-IgG-seronegative patients with NМOSD are a heterogeneous clinical subgroup, having different immunopathogenetic mechanisms,it was necessary to conduct stratification analysis accord-ing to AQP4-IgG serostatus. After stratification for AQP4-IgG positivity, a significant association with susceptibility to NМOSD persisted in rs117026326. Surprisingly, the rs73366469 CC genotype was associated with AQP4-IgG-seropositivity, which implies that there is an association with AQP4-IgG but not with NМOSD.

Table 6 Stratification analysis according to coexisting autoimmune diseases: allele and genotype frequencies of rs73366469 in AQP4-IgG-seropositive patients without primary Sjögren's syndrome, systemic lupus erythematosus, or rheumatoid arthritis, and along with the association with the risk of NMOSD

Figure 1 Linkage disequilibrium tests forfive SNPs at the GTF2IRD1-GTF2I locus.
Furthermore, three major haplotypes constructed from two SNPs (rs11981999 and rs4717901) in one linkage disequilibrium block showed no significant difference between the cases and controls. The GTF2IRD1 polymorphisms rs11981999 and rs4717901 have been reported to confer risk of systemic lupus erythematosus (Li et al., 2015; Sun et al.,2016), while the GTF2I rs2527367 polymorphism is associated with complex behavioral traits such as human anxiety(Jabbi et al., 2015). Мost notably, the rs4717901 polymorphism, which is located in the 3′ flanking region of the GTF2IRD1 gene, might regulate mRNA stability and translational efficiency. However, no association was observed between any of these three SNPs and the risk of NМOSD,even after stratification for AQP4-IgG positivity. This may be due in part to the different genetic background between NМOSD and systemic lupus erythematosus, as well as other complex behavioral traits.
Additionally, stratification analysis according to coex-isting autoimmune diseases was performed. The GTF2I rs117026326 polymorphism has been identified as the strongest susceptibility locus for primary Sjögren's syndrome in Han Chinese (Li et al., 2013), but not in Caucasians (Lessard et al., 2013), according to genome-wide association studies. Subsequently, it was reported that the rs117026326 polymorphism is associated with anti-Sjøgren syndrome antibody A-positivity (Zheng et al., 2015) in Han Chinese or primary Sjögren's syndrome in southern Chinese females(Song et al., 2016), in addition to being associated with rheumatoid arthritis (Kim et al., 2016) and systemic lupus erythematosus (Li et al., 2015; Sun et al., 2016). Мoreover,rs73366469 has been shown to confer susceptibility to rheumatoid arthritis (Kim et al., 2016), primary Sjögren's syndrome (Li et al., 2013), and systemic lupus erythematosus(Sun et al., 2016; Zhao et al., 2017) in Asian populations.In systemic lupus erythematosus patients, a recent study(Zhao et al., 2017) detected the strongest association signal at rs73366469, which was consistent with another study performed in an Asian population (Sun et al., 2016). In contrast, the association of the rs73366469 polymorphism with systemic lupus erythematosus in European Americans has been confirmed at only a modest significance level, and this association was not found in African Americans (Zhao et al.,2017). Therefore, the coexistence of the abovementioned autoimmune diseases in patients may confound the correlation between GTF2IRD1-GTF2I polymorphisms and NМOSD in Han Chinese. Considering this factor, we further excluded patients with primary Sjögren's syndrome, systemic lupus erythematosus, or rheumatoid arthritis, and the significant associations with the T allele of rs117026326 and the genotype CC of rs73366469 remained. Collectively, ourfindings demonstrate that polymorphisms in GTF2IRD1-GTF2I may be associated with the risk of NМOSD, which can facilitate the understanding of the pathogenesis of NМOSD.

Table 7 Results of the analysis of selected haplotypes at the GTF2IRD1-GTF2I locus for the potential association with the risk of NMOSD
However, the exact molecular mechanisms through which these polymorphisms are implicated in the pathogenesis of NМOSD have yet to be elucidated. It is likely that they in fluence the expression of GTF2I, GTF2IRD1, or other neighboring genes and thereby alter the immune responses mediated by T or B lymphocytes, leading to the subsequent breakdown of immune tolerance and production of AQP4-IgG.Recently obtained evidence demonstrated that rs117026326 can regulate the expression of GTF2IRD2 (downstream of GTF2I), but not for GTF2I, GTF2IRD1, or NCF1 (Kim et al.,2016). Another study also detected no association between rs117026326 genotypes and the transcript levels of GTF2I and GTF2IRD1 in peripheral blood mononuclear cells from patients with systemic lupus erythematosus and controls(Zhao et al., 2017). As the rs117026326 polymorphism is located within the intron of GTF2I, while rs73366469 is located in the intergenic region, it needs to be clarified whether the susceptibility variants exert an effect by regulating their own expression level or that of other neighboring genes. Another possibility is that these variants interact with unidentified variants of other genes or microRNAs. We should also keep in mind that the association of these polymorphisms with the risk of NМOSD may be due to a direct causative effect of these SNPs, or may occur because they are in linkage disequilibrium with other functional variants located in or near the GTF2IRD1-GTF2I region. In a recent study published in Nature Genetics, a missense variant in NCF1 was found to be associated with susceptibility to multiple autoimmune diseases, based on the hypothesis that rs117026326 might tag causal variant(s) of NCF1 that are not present in the 1000 Genomes Project (Zhao et al., 2017).
Several limitations of our study should be noted. First,we focused only on the reported disease-associated SNPs within the GTF2IRD1-GTF2I locus, which may have led us to neglect several other susceptibility variants in this region.Second, all participants came from southwest China, and the cohort consisted exclusively of individuals of Han ethnicity.Therefore, our findings require replication in other ethnicities. Thus, the relationship between GTF2IRD1-GTF2I polymorphisms and NМOSD needs to be verified in larger cohorts. Nevertheless, our study provides valuable insights and will contribute to future investigations in thisfield.
In conclusion, the results of our study suggest that the T allele of rs117026326 is associated with susceptibility to NМOSD, and the rs73366469 CC genotype confers susceptibility to AQP4-IgG seropositivity in Han Chinese patients.The strong association between GTF2IRD1-GTF2I polymorphisms and the risk of NМOSD may provide new approaches for investigating the pathogenesis of NМOSD and developing alternate treatment options for NМOSD.
Author contributions:Study concept and design, technical guidance,theoretical supports, and paper revision: HYZ. Literature searching, case and sample collection, experiment implement, data analysis, and paper writing: JLX and JL. Sample collection and study preparation: ZYL, HXC,ZYS, QZ, HRF, QD and XHM. All authors approved thefinal version of the paper.
Con flicts of interest:The authors declare that they have no con flict of interest.
Financial support:This study was supported by the National Natural Science Foundation of China, No. 81271321 (to HYZ); a grant from the Department of Science and Technology Research Projects in Sichuan Province of China, No. 2013FZ0015 (to HYZ); the Fundamental Research Funds for the Central Universities, China, No. 2017scu11049 (to QZ).All authors declare thatfinancial support does not affect the opinion of the article and the objective statistical analysis and report of the research results in this study.
Institutional review board statement:The study was approved by the Ethics Committee of West China Hospital of Sichuan University, China(approval number: 2016-31) on March 2, 2016.
Declaration of participant consent:The authors certify that they have obtained all appropriate participant consent forms. In the form, the participants have given their consent for their images and other clinical information to be reported in the journal. The participants understand that their names and initials will not be published and due efforts will be made to conceal their identity, but anonymity cannot be guaranteed.
Reporting statement:This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement.
Biostatistics statement:The statistical methods of this study were reviewed by the biostatistician of West China Hospital, Sichuan University, China.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Individual participant data that underlie the results reported in this article, after deidentification (text, tables,figures,and appendices) will be in particular shared. Data of the present study,including study protocol and informed consent, will be available immediately following publication, no end date. Results will be disseminated through presentations at scientific meetings and/or by publication in a peer-reviewed journal. Anonymized trial data will be available inde finitely at www.figshare.com.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix,tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Aylin Elkama, Gazi University Faculty of Pharmacy, Turkey.
Additionalfile:Open peer review report 1.
杂志排行
中国神经再生研究(英文版)的其它文章
- MGMT is down-regulated independently of promoter DNA methylation in rats with all-trans retinoic acidinduced spina bifida aperta
- Comparison of walking quality variables between incomplete spinal cord injury patients and healthy subjects by using a footscan plantar pressure system
- A novel primary culture method for high-purity satellite glial cells derived from rat dorsal root ganglion
- Melatonin combined with chondroitin sulfate ABC promotes nerve regeneration after root-avulsion brachial plexus injury
- Implications of alpha-synuclein nitration at tyrosine 39 in methamphetamine-induced neurotoxicity in vitro and in vivo
- Safety of intrathecal injection of Wharton's jellyderived mesenchymal stem cells in amyotrophic lateral sclerosis therapy
