Isolation and characterization of the secondary wall-related SND1 gene in hawthorn
2018-08-06CHENKeqinGUOYunnaSONGMengruDAIHongyanZHANGZhihong
CHEN Ke-qin, GUO Yun-na, SONG Meng-ru, DAI Hong-yan, ZHANG Zhi-hong
1 Group of Molecular Biology offruit Trees, College of Horticulture, Shenyang Agriculturaluniversity, Shenyang 110866, P.R.China
2 Group offruit Germplasm Evaluation & Utilization, College of Horticulture, Shenyang Agriculturaluniversity, Shenyang 110866,P.R.China
Abstract Secondary wall-associated NAC domain protein1 (SND1) is a key regulator directly regulating the expression levels of MYB46 and MYB83 in the regulation network for secondary wall synthesis, especially in plantfibres. In this study, a SND1 gene was isolated from hawthorn (Crataegus pinnatifida) and named as CpSND1 because it has a conservative N-terminal DNA-binding domain with AtSND1. Arabidopsis plants overexpressing CpSND1 had similar phenotypes as plants overexpressing AtSND1, including inhibited growth, upward-curling leaves, sepal dysplasia and sterility. In addition, overexpressing CpSND1 in Arabidopsis also induced the expression of downstream genes, including lignin, cellulose and xylan biosynthesis genes as wellas MYB genes. Our results provided functional information of CpSND1 for future genetic engineering in hawthorn.
Keywords: hawthorn, transcription factor, SND1, secondary cell wall, transcriptional regulation
1. Introduction
Secondary cell walls are largely composed of three main polymers: cellulose, hemicellulose, and lignin. These polymers are produced in sclerenchyma cells and are normally required to enable vascular plants not only to build strong xylem conduits for the transport of water and minerals but also to attain strong mechanical support for the plant body (Raven et al. 1999). The formation of the secondary cell wall is mainly regulated at the transcriptional level, and most of the regulation theory comes from the model herbaceous plant Arabidopsis thaliana. NAC (NAMATAF1/2-CUC2) and MYB act as the key master switches that control secondary cell wall deposition; these proteins also regulate a battery of downstream transcription factors(TFs) and secondary cell wall biosynthesis genes (Taylor-Teeples et al. 2015).
NAC proteins are plant-specific TFs and have been shown to function in plant development processes and abiotic and/or biotic stress responses. The NAC domain was named from the first letters of the Petunia NAM and Arabidopsis ATAF1/2 and CUC2 proteins (Aida et al. 1997).In Arabidopsis, there are two types of NAC TFs acting as on-off switches that take part in regulating secondary wall formation in vascular cells andfibre cells. the first type of NAC TFs (Vascular-related NAC-domain) VND6 and VND7 contribute to both secondary wall biosynthesis and programmed cell death of the vessels in both root and shoot tissues (Kubo et al. 2005; Yamaguchi et al. 2008).Secondary wall formation has been intensively studied during xylem cell differentiation. Seven genes from VND1 to VND7, which form a subclade, are preferentially expressed in developing vascular cells and are induced during xylem cell differentiation (Kubo et al. 2005; Yamaguchi et al.2008; Ohashi-Ito et al. 2010; Zhou et al. 2014). The expression of VND7 can restore defects in secondary cell wall formation in thefibre cells of in florescence stems of nst1/nst3 double mutants (Yamaguchi et al. 2011). The second type of NAC TFs consists of NST3 (NAC secondary wall thickening promoting factor3)/SND1 (secondary wallassociated NAC domain protein1), NST1 and NST2, which participate in thickening the secondary wall both in vascularfibre cells and secondary xylemfibre cells (Zhong and Ye 2014). Arabidopsis NST3/SND1 is specifically expressed in vascularfibres and xylemfibres, and SND1 expressing under the controlof the cauli flower mosaic virus 35S(CaMV35S) promoter can cause ectopic secondary wall deposition in non-sclerenchyma cells. Both the dominant suppression SND1 mutant and the nst1/snd1 double mutant show a significant reduction in secondary wall thickness between the vascular and xylemfibres; and in the nst1/nst2/snd1 triple mutant, the secondary cell wall infibres is completely absent in Arabidopsis (Zhong 2006; Mitsuda et al. 2007; Zhong and Ye 2015b). In addition, the process of anther dehiscence to release pollen requires a thick secondary wall in the interior cortex of the anther, and it has been found that anthers in nst1/nst2 double mutants cannot crack, while the single mutant is normal; these observations indicate that NST1 and NST2 are functionally redundant during the secondary wall formation in anthers(Mitsuda et al. 2005).
Protein binding assays have demonstrated that SND1 can bind to a DNA section with the sequence (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T), named SNBE(secondary wall NAC binding element) (Zhong et al. 2008).Among the SND1-regulated transcription factors, MYB46,MYB83, SND3, MYB103 and KNAT7 have been shown to be direct targets of NAC TFs, including SND1, NST1,NST2, VND6 and VND7 (Zhong et al. 2007; McCarthy et al.2009; Ko et al. 2014; Zhong and Ye 2014). These NAC genes were shown to be expressed infibres and vessels specifically, and dominant repression of their expression levels led to a reduction in secondary wall thickening,indicating that they play important roles involved in the regulation of secondary wall biosynthesis (Zhong and Ye 2015a). In particular, MYB46 and MYB86, functioning as another levelof molecular switches, redundantly turn on the entire secondary wall biosynthetic programme (Zhong et al. 2007; Ko et al. 2014; Zhong and Ye 2015a). In addition to targeting the downstream TFs, SND1 can also activate the synthesis enzymes of the secondary wall directly, such as CESA4, CESA7 and CESA8 (Zhong et al. 2010b).
In this study, transgenic Arabidopsis plants with overexpressed CpSND1 (a SND1 gene isolated from Crataegus pinnatifida) driven by the CaMV35S promoter presented growth inhibition, upward-curling leaves, sepal dysplasia and sterile phenotypes, which are similar to the phenotypes observed when AtSND1 is overexpressed.In addition, in transgenic Arabidopsis plants, the genes involved in lignin, cellulose and xylan biosynthesis were significantly induced.
2. Materials and methods
2.1. Plant materials and growth conditions
Trees of Crataegus pinnatifida accession H8 (hard-endocarp hawthorn) were maintained in the National Hawthorn Germplasm Repository at Shenyang, China. The cDNA of C. pinnatifida accession H8 was used as the template for CpSND1 cloning. Arabidopsis Columbia (Col-0) was used as the transgenic plants. The rice protoplast for cellular localization was obtained from the Institute of Crop Sciences,Chinese Academy of Agricultural Sciences. Arabidopsis plants were maintained in a growth chamber under short-day conditions (8 h light/16 h dark) at 23°C for 2 wk and then in long-day conditions (16 h light/8 h dark) at 23°C.
2.2. Arabidopsis thaliana transformation
To overexpress the CpSND1 gene in Arabidopsis, coding sequences of CpSND1 were inserted into the plant expression vector pRI101-AN, containing a CaMV35S promoter and a nopaline synthase (NOS) terminator.The primers used for vector construction are listed in Appendix A. The reconstructed plasmid pRI101-CpSND1 was introduced into A. thaliana (Col-0) by Agrobacterium tumefaciens (strain GV3101)-mediated transformation using the floral dip method. Arabidopsis flowers were dipped into A. tumefaciens GV3101 suspended in a 1/2 MS liquid containing 10% sucrose and 0.02% Silwet, and then the plants were covered with a black bag and incubated in a growth chamber at 23°C for 1 d andfinally allowed to grow in a growth chamber as usual. The collected seeds were screened in 1/2 MS medium supplemented with 30 mg L-1kanamycin, and transgenic seedlings were identified by PCR. Total RNA was extracted from the seedlings of the T1generation. In addition, the T1transgenic lines were used for further analyses.
2.3. Phylogenetic tree graphics and bioinformatics
The predicted amino acid sequences of NACs were aligned using ClustalX 1.83, and then a phylogenetic tree was derived from the multiple alignment using the neighbourjoining method in MEGA6. Analysis of the conservation of the NAC domain in hawthorn SND1 was determined by using the NCBI CDD tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The respective domains of NAC proteins were aligned using DNAMAN software.
2.4. Cellular localization assays
CpSND1-coding sequences were amplified from the cDNA and then inserted into the C-terminal side of green fluorescent protein (GFP) at the XbaI and XmaI sites in the vector pGPTII-GFP. The correct recombination plasmid was sequenced to ensure insert accuracy. pGPTII-GFP (positive control) and fusion constructs pGPTII-CpSND1-GFP were employed for localization studies. The constructs were transiently expressed in rice protoplasts, and then the GFP fluorescence was observed using a laser scanning confocal microscope. All the primers are listed in Appendix A. Transferred protoplasts were detected under a Nikon E600 fluorescence microscope (Nikon, Japan), and GFP fluorescence was visualized with afilter setting consisting of an excitationfilter of 450-490 nm, a dichroic mirror of 510 nm, and a barrierfilter of 520-560 nm. Images were captured with a SPOT2 Slider charge-coupled device camera and associated software (Sterling Heights, MI,USA).
2.5. Transcriptionalactivity assay in yeast
For the transactivation activity assay, the full-length CpSND1,the N-terminal NAC domain of CpSND1 (CpSND1-N) and the C-terminalof CpSND1 (CpSND1-C) were amplified and inserted into vector pGBT9 (BD) (Clontech, Palo Alto, CA,USA) at the EcoRI and SalI sites. All primers are listed in Appendix A. All the fusion constructs were introduced into the yeast strain Y2H GOLD according to manufacturer’s instructions, and the transformed yeast cells were grown on SD/-T (growth control) and SD/-T/-H/-A/+X-alpha-gal plates to check transactivation activity.
2.6. Quantitative RT-PCR
Total RNA was extracted from the Arabidopsis leaves using TRIzol Reagent and precipitating with lithium chloride and chilled absolute ethanol. First-strand cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China).The reverse transcription products of cDNA, diluted four times, were used as the template for quantitative PCR.Reactions were set up with SYBR Green Fast qPCR Mix(TaKaRa) according to the manufacturer’s instructions in a total volume of 20 μL with each primer at 0.2 μmol L-1.The amplification programme was as follows: one cycle of 30 s at 95°C followed by 40 cycles of 5 s at 95°C and 10 s at 60°C. All reactions were run in triplicate, and average values were calculated. Relative expression levels of target genes and SD values were calculated using the 2−ΔΔCTmethod (Livak and Schmittgen 2001). All the primer sequences are listed in Appendix A.
3. Results
3.1. Isolation and sequence characteristics of hawthorn SND1
From early transcriptome data of soft-endocarp and hard-endocarp hawthorns (Dai et al. 2013), we found four NAC family transcription factors that were strongly down-regulated in the fruits of soft-endocarp hawthorn compared to the fruits of hard-endocarp hawthorn. We suspected that they might participate in the biosynthesis of lignin or secondary cell walls. Therefore, according to the conservative NAC domain of these TFs, we aligned other NAC TFs in other species, which all contained a similar NAC DNA-binding domain, by the NCBI (https://www.ncbi.nlm.nih.gov/pubmed) and PLAZA (https://bioinformatics.psb.ugent.be/plaza/) web blast. Because AtSND1 was the first key switch involved in secondary wall formation,8_Unigene_BMK.37276 (log2(S7/H8)=-6.31) gene which was the most homologous to AtSND1 (Fig. 1-A), was named as CpSND1. The sequence alignment results also indicated that there were six NAC genes in Brassica rapa,two genes in Populus trichocarpa, four genes in Gossypium raimondii, one gene in Fragaria vesca, one gene in Citrus sinensis, two genes in Malus domestica and one gene in Prunus persica closer to AtNST1-AtNST3. In addition,four NAC TFs of G. raimondii, one off. vesca, one of C. sinensis, two of M. domestica, one of P. persica and two of B. rapa were closer to AtSND1. The amino acid sequence alignment showed that allof the SND1 homologous genes had a highly conserved NAC DNA-binding domain (Fig. 1-B).These bioinformatic results revealed that CpSND1 may have a similar function as AtSND1.
3.2. CpSND1 is localized in the nuclei and acts as a transcription activator in yeast cells
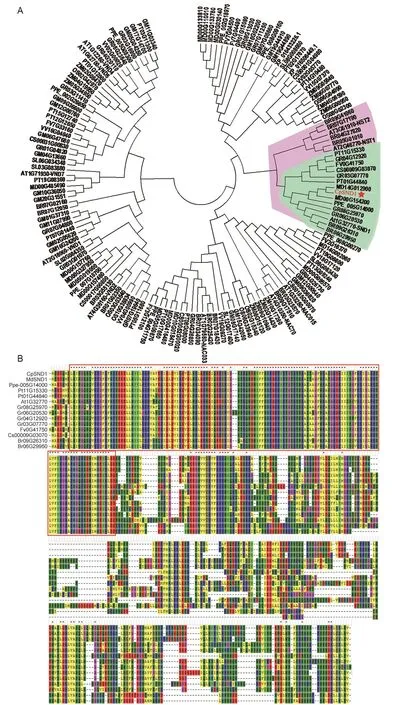
Fig. 1 Identification of CpSND1. A, alignment of the hawthorn SND1 (secondary wall-associated NAC domain protein1) transcription factor with other NAC numbers in different species. The red frame indicated the conservative N-terminalof NAC protein. B, the amino acid sequence alignment of the same branch with AtSND1. Cp, Crataegus pinnatifida; Md, Malus domestica; Ppe, Prunus persica; Pt, Populus trichocarpa; At, Arabidopsis thaliana; Gr, Gossypium raimondii; Fv, Fragaria vesca; Cs, Citrus sinensis; Br,Brassica rapa; Gm, Glycine max; Sl, Solanum lycopersicum; Zm, Zea mays; Os, Oryza sativa.
To detect whether CpSND1 is a transcription factor localized in nuclei, the coding sequence of CpSND1 was fused to GFP and expressed under the controlof the constitutive CaMV35S promoter. Merged images of GFP fluorescence showed that CpSND1-GFP localized in the nuclei in rice protoplasts, and the GFP empty vector was used as a control(Fig. 2-A). Then, we performed a yeast two-hybrid assay to detect the transcription activation of CpSND1. Constructs of the CpSND1 gene as a full-length cDNA or with N- and C-terminal deletions were tested for transcriptionalactivation activity in yeast cells (Fig. 2-B); the full-length cDNA and the C-terminalof CpSND1 exhibited transcriptionalactivity in yeast cells. These results demonstrated that the CpSND1 protein is targeted to the nucleus and is able to activate transcription in yeast cells, indicating that it is a transcription activator.
3.3. Overexpression of CpSND1 induced stunted growth in Arabidopsis
To determine the biological function of CpSND1, the recombinant plasmid pRI101-CpSND1 was introduced into A. thaliana. In transgenic Arabidopsis, the levelof CpSND1 sharply increased (Fig. 3-A). In addition, the plants that overexpressed CpSND1 appeared to have restricted growth, curled leaves, sepal dysplasia and sterility (Fig. 3-B and C). These phenotypes induced by CpSND1 were similar to those induced by AtSND1 (Zhong et al. 2006).To study whether CpSND1 could participate in secondary wall biosynthesis, RT-PCR was performed to detect the expression of the synthesis genes during secondary wall formation in transgenic Arabidopsis. In transgenic lines 1, 4 and 5, the biosynthesis genes of lignin, cellulose and xylan were significantly induced (Fig. 3-D). To determine whether CpSND1 functioned in regulating secondary wall thickening in stems, transverse sections were taken from the basal stems of wild-type (WT) and transgenic plants. Compared with WT plants, CpSND-overexpressing plants had thicker secondary cell walls in the xylem, as the red arrows indicate,and lignin accumulated in xylem cells, which were stained by sarranine (Fig. 3-E).
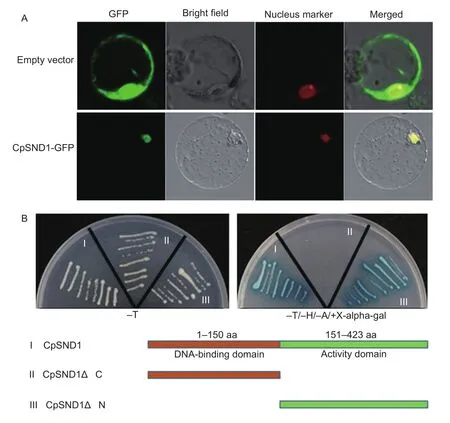
Fig. 2 Subcellular localization and transactivation assay of CpSND1. A, 35S::CpSND1-GFP fusion gene and 35S::GFP control plasmid were transformed into rice protoplasts. SND1, secondary wall-associated NAC domain protein1; GFP, green fluorescent protein. The transformed cells were imaged by confocal microscopy after transformation for 16 h. B, the full-length, N-terminaland C-terminal sequences of CDS (coding sequence) of CpSND1 were fused in-frame, separately, to the GAL4 DNA-binding domain in pGBKT7 and transformed into Y2H Gold yeast cells. The transformed cells were plated onto SD/-T (growth control)and SD/-T/-H/-A/+X-alpha-gal medium.
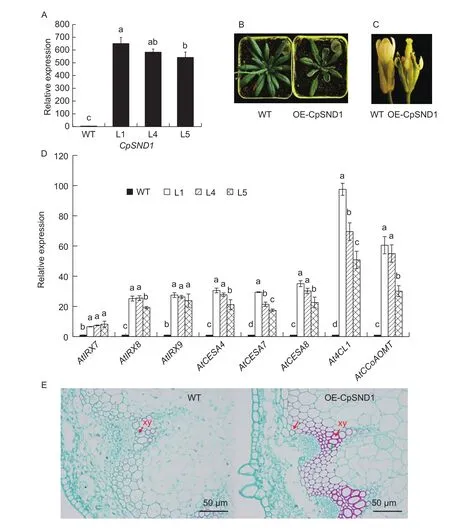
Fig. 3 Overexpression of CpSND1 in Arabidopsis. A, expression levels of CpSND1 in 3 transgenic lines, with wild type (WT) as the control. SND1, secondary wall-associated NAC domain protein1. B, the phenotype of CpSND1 overexpression lines. C, the dysplasia offlower organs in CpSND1 overexpression lines. D, the biosynthesis genes of lignin, cellulose and xylan were detected in CpSND1-transgenic Arabidopsis by RT-PCR. AtIRX7, At2G28110; AtIRX8, At5g54690; AtIRX9, At2g37090; AtCESA4, At5g44030;AtCESA7, At5g17420; AtCESA8, At4g18780; At4CL, At1g51680; AtCCoAOMT, At4g34050. Different letters mean significant differences at the 0.05 probability level. The error bars mean SD. E, the observation of transverse sections, which were taken from the basal stems of wild-type and overexpression plants by scanning electron microscopy. The red arrow indicates the thickened secondary cell wall, and the green arrow indicates the lignin deposition in the xylem (xy) stained by sarranine in transgenic plants.
3.4. The expression modelof TFs involved in secondary wall formation in CpSND1 overexpression lines
SND1, as one of the first key transcription factor switches in secondary wall formation, not only could induce the secondary wall biosynthesis genes directly but also could activate the secondary key MYB transcription factor switches. Therefore, we detected the expression of downstream MYB46, MYB83, MYB58 and MYB63 in transgenic Arabidopsis, and the results showed that the transcription levels of allof the four MYB genes greatly increased (Fig. 4-A). We also analysed the expression levels of the Arabidopsis NAC genes involved in the secondary wall formation in transgenic Arabidopsis, and the results showed that allof the five NAC transcription factors,AtSND1, AtNST1, AtNST2, AtVND6 and AtVND7, were also greatly induced (Fig. 4-B).
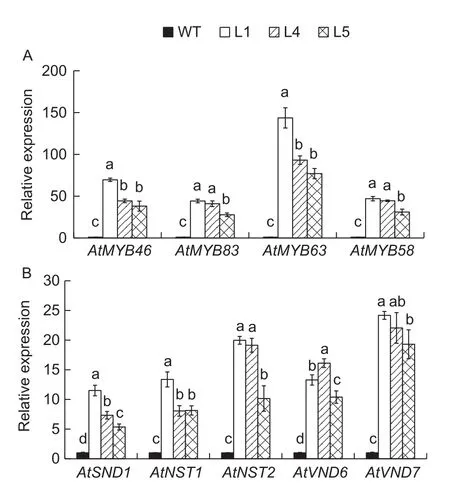
Fig. 4 The expression of MYB and NAC involved in the secondary cell wall formation in WT and CpSND1-overexpressing Arabidopsis lines. A, the expression of MYB family members. AtMYB46, AT5G12870; AtMYB83,AT3G08500; AtMYB63, AT1G79180; AtMYB58, AT1G16490.B, the expression of NAC family members. AtSND1,AT1G32770; AtNST1, AT2G46770; AtNST2, AT3G61910;AtVND6, AT5G62380; AtVND7, AT1G71930. Different letters mean significant different at the 0.05 probability level. The error bars mean SD.
4. Discussion
According to the previous reports, 28 of 57 AtSND1 transgenic Arabidopsis lines exhibited a prominent visual phenotype that showed a small rosette size and stunted growth of leaves with severely upward-curling blades; the flowers of SND1 overexpression lines were often sterile and had severely shortened sepals, petals, stamens, and carpels (Zhong et al. 2006). These phenotypes were similar to the CpSND1 overexpression lines in this study (Fig. 3-B and C). In poplar,overexpression of wood-associated NAC domain transcription factors PtrWND2B and PtrWND6B produced plants with severely curled leaves, which could also complement the secondary wall defects in the Arabidopsis snd1/nst1 double mutant. Overexpression of PtrWND2B and PtrWND6B in Arabidopsis induced the expression of secondary wall-associated transcription factors and secondary wall biosynthetic genes and caused the ectopic deposition of cellulose, xylan, and lignin. PtrWND2B and PtrWND6B were able to activate the promoter activities of a number of poplar wood-associated transcription factors and wood biosynthetic genes (Zhong et al. 2010a). The PtVNS07/PtrWND6A and PtVNS11/PtrWND1B genes contribute to the formation of xylem tissue and phloemfibres. Overexpression of the PtVNS/PtrWND genes, which are similar to AtVND7 and AtNST3/SND1, induced ectopic secondary wall thickening in Arabidopsis seedlings as wellas in poplar. Allof the PtVNS/PtrWND genes of the VND and NST groups exhibit the ability to positively regulate secondary wall thickening (Zhong et al.2010a). Wood formation in poplar is regulated by cooperative functions of the NAC domain proteins (Ohtani et al. 2011).Suppression of PtrWND1B expression led to specific inhibition offibre in secondary wall thickening in xylem and phloem tissues, and transgenic poplars were unable to grow straight.Up-regulation of PtrWND1B in poplar exhibited a phenotype of small leaves that were curled upward or downward (Zhao et al. 2014).
SND1, to gether with other secondary wall NACs,including NST1, NST2, VND6 and VND7, binds to the SNBE sites in the promoters of their direct targets, not only in downstream MYB transcription factors but also in a number of non-transcription factor genes involved in secondary wall biosynthesis that all contain multiple SNBE sites (Zhong et al. 2010b). In addition, the NAC-MYB regulation model was conserved in different species. In Arabidopsis, the SND1 protein can bind directly to its own promoter and is characterized by the critical nucleotides required for SND1 binding, which is TACXTTXXXXATGA(Zhong et al. 2010b). Promoters of AtVND6/7, AtNST1,and AtSND1 also contained the SMRE (secondary wall MYB-responsive element (ACC(A/T)A(A/C)(T/C)) binding site, which is recognized directly by MYB46 (Zhong and Ye 2014). In addition, we also found that the promoters of the AtNST1, AtNST2, AtSND1, AtVND6, AtVND7, MdSND1 and MdNST1 genes contained several MYB-binding sites,such as M46RE ((A/G)(C/T)T(A/T)GGT(A/G)) and SMRE(ACC(A/T)A(A/C)(T/C)), that are related to secondary cell wall biosynthesis (unpublished data). This may be the reason why the expression levels of AtNST1, AtNST2,AtSND1, AtVND6 and AtVND7 were increased over two-fold in the CpSND1-overexpressing Arabidopsis lines (Fig. 4-B).
5. Conclusion
CpSND1 transcription factor had similar function with AtSND1 for secondary wall synthesis in Arabidopsis. CpSND1 as the transcription activator can upregulate the expression of secondary wall biosynthesis genes and induce the formation of secondary wall in transgenic Arabidopsis. The function of SND1 is high conservative among different species.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (31170635).
Appendix associated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- ldentification and characterization of Pichia membranifaciens Hmp-1 isolated from spoilage blackberry wine
- Implications of step-chilling on meat color investigated using proteome analysis of the sarcoplasmic protein fraction of beef longissimus lumborum muscle
- Spatial-temporal evolution of vegetation evapotranspiration in Hebei Province, China
- Design of a spatial sampling scheme considering the spatialautocorrelation of crop acreage included in the sampling units
- Comparison of forage yield, silage fermentative quality, anthocyanin stability, antioxidant activity, and in vitro rumen fermentation of anthocyanin-rich purple corn (Zea mays L.) stover and sticky corn stover
- Synonymous codon usage pattern in model legume Medicago truncatula
