Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots
2018-08-06WANGLingshuangCHENQingshanXINDaweiQIZhaomingZHANGChaoLISinanJINYangmeiLIMoMEIHongyaoSUAnyuWUXiaoxia
WANG Ling-shuang, CHEN Qing-shan, XIN Da-wei, QI Zhao-ming, ZHANG Chao, LI Si-nan, JIN Yang-mei, LI Mo, MEI Hong-yao, SU An-yu, WU Xiao-xia
1 College of Agriculture, Northeast Agriculturaluniversity/Key Laboratory of Soybean Biology, Ministry of Education, Harbin 150030, P.R.China
2 College of Resources and Environment, Northeast Agriculturaluniversity, Harbin 150030, P.R.China
Abstract Glycogen synthase kinase 3 (GSK3) is a kind of serine/threonine kinase widely found in eukaryotes. Many plant GSK3 kinases play important roles in regulating stress responses. This study investigated BRASSINOSTEROID-INSENSITIVE 2(GmBIN2) gene, a member of the GSK3 protein kinase family in soybean and an orthologue of Arabidopsis BIN2/AtSK21.GmBIN2 expression was increased by salt and drought stresses, but was not significantly affected by the ABA treatment. To examine the function of GmBIN2, transgenic Arabidopsis and transgenic soybean hairy roots were generated. Overexpression of GmBIN2 in Arabidopsis resulted in increased germination rate and root length compared with wild-type plants under salt and mannitol treatments. Overexpression of GmBIN2 increased cellular Ca2+ content and reduced Na+ content, enhancing salt tolerance in transgenic Arabidopsis plants. In the soybean hairy root assay, overexpression of GmBIN2 in transgenic roots also showed significantly higher relative root growth rate than the control when subjected to salt and mannitol treatments. Measurement of physiological indicators, including proline content, superoxide dismutase (SOD) activity, and relative electrical conductivity, supported this conclusion. Furthermore, we also found that GmBIN2 could up-regulate the expression of some stress-related genes in transgenic Arabidopsis and soybean hairy roots. Overall, these results indicated that GmBIN2 improved tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots.
Keywords: GmBIN2 gene, abiotic stress, transgenic Arabidopsis, soybean hairy roots
1. Introduction
In the natural environment, plants are often exposed to drought, cold damage, high soil salinity, or other abiotic stresses (Wang et al. 2003) that can greatly restrict plant growth and development and cause yield reduction. During the evolution of plant life, a complex regulatory system has formed to combat the impact of adverse environments(Vij and Tyagi 2007). More specifically, plants will trigger stress-responsive mechanisms to regulate associated genes at the molecular and cellular levels to withstand these environmental stresses (Ingram and Bartels 1996;Thomashow 1998).
To date, many plant protein kinases associated with abiotic stresses have been studied. For instance, the Ca2+-dependent protein kinases (CDPKland CDPKla) have been found to regulate a promoter region that was induced by drought and salt stresses in maize protoplasts (Sheen et al. 1996). Plant mitogen-activated protein kinase (MAPK)has been shown to participate in mediating various stress responses such as freezing, high salinity, and drought(Jonak et al. 1996; Kiegerl et al. 2000). Another example is SRK2C, a member of the SNF1-related protein kinase 2 family, that has been shown to enhance drought tolerance and regulate many stress-related genes in Arabidopsis(Umezawa et al. 2004).
Glycogen synthase kinase 3 (GSK3) is a kind of serine/threonine kinase widely found in eukaryotes. The mammalian GSK3s, GSK3α, and GSK3β, have high amino acid sequence homology to each other and exhibit redundancy and specificity in function (Doble et al. 2007;Lee et al. 2007). Conversely, plants possess more divergent GSK3 isoforms, with 10 GSK3 kinases identified in Arabidopsis and nine in rice that were further parsed into four subgroups (Jonak and Hirt 2002). Recent studies have indicated that plant GSK3 proteins are involved in diverse biochemical processes, including cell differentiation(Tavares et al. 2002), apoptosis (Charrier et al. 2002),flower development (Dornelas et al. 2000), stomatal cell development (Kim et al. 2012), and brassinosteroid signal transduction (He et al. 2002; Li and Nam 2002; Perez-Perez et al. 2002). Furthermore, plant GSK3 has been shown to be highly involved in stress response. One study found that,the majority of Arabidopsis GSK3-encoding genes could be induced by stress signals such as NaCland polyethylene glycol (PEG) at the transcriptional level (Charrier et al. 2002).Additionally, AtGSK1 has been shown to be involved in NaCland abscisic acid (ABA) stress responses and transgenic Arabidopsis overexpressing AtGSK1, showing a stronger salt tolerance than the controls (Piao et al. 1999; Piao et al.2001). The AtSK11 gene product has been shown to be post-translationally activated by the NaCl treatment, followed by an increase in phosphorylation of the substrate (Dal Santo et al. 2012). In rice, knockout mutations of OsGSK1 have been demonstrated to enhance tolerance to abiotic stresses(Koh et al. 2007). Overexpression of MsK4, a Medicago sativa GSK3 gene, has shown improved tolerance to salt stress by participating in carbohydrate metabolism (Kempa et al. 2007). Similarly, overexpression of the wheat GSK3 gene, TaGSK1, enhanced salt tolerance in Arabidopsis (He et al. 2012). Despite the extensive research on plant-derived GSK3s, there is little research on the characterization and function of GSK3 genes in soybean.
Here we report GmBIN2, screened under different concentrations of 2,4-dichlorophenoxyacetic acid (2,4-D)treatment in Dongnong 50 to determine the differentially expressed genes in the previous work of our laboratory.The 2,4-D is an auxin analogue for induction of callus formation. Currently, it is reported that 2,4-D is also involved in inducing stress-associated genes. For example, in Valencia sweet orange, six stress-associated proteins were induced by 2,4-D treatment in embryogenic callus, of which three were related to osmotic stress, one related to oxidative stress, and one protective protein that was resistant to UV stress (Pan et al. 2010). In addition,the herbicide 2,4-D induced associated oxidative stress responses in yeast (Teixeira et al. 2004). Combined with the correlation of GSK3 kinase with stress responses,this study mainly focuses on the regulation of GmBIN2 in response to abiotic stresses.
GmBIN2 is an orthologue of Arabidopsis BIN2/AtSK21,derived from the Glycine max cultivar Dongnong 50 and the objectives in this study were to transfer GmBIN2 into Arabidopsis and soybean hairy roots to evaluate the role of GmBIN2 in response to salt and drought stresses.
2. Materials and methods
2.1. Plant growth and stress treatments
Seeds of the soybean cultivar of Dongnong 50 were used for expression pattern analysis and the soybean hairy root assay and were obtained from the Key Laboratory of Soybean Biology of Ministry of Education of China, Harbin.Plants were cultivated under a cycle of 16 h light (26°C)/8 h dark (18°C) with a relative humidity of (70±10)%. Prior to stress treatments, the roots of soybean seedlings in the first trifoliate leaf stage were immersed into 1/4 Murashige and Skoog (MS) liquid medium for 24 h. For GmBIN2 expression patterns analysis under abiotic stress, 1/4 MS liquid medium was added to 150 mmol L-1NaCl, 5% (w/v)PEG 6000 or 100 μmol L-1ABA, respectively. Plants treated with only 1/4 MS liquid medium were used as the controls.Leaves were harvested after treatments of 0, 2, 4, 8, 12, 24,or 48 h abiotic stress exposure. For the tissue expression pattern analysis, different soybean seedling organs were harvested including roots, stems, flowers, leaves, and seeds and stored in -80°C.
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used for transformation. The homozygous T3generation were used for all experiments.
2.2. Isolation of the GmBIN2 gene and sequence analysis
A PCR was conducted to amplify the GmBIN2-coding sequence using specific primers GmBIN2-F/R (Appendix A). The PCR reaction conditions were as follows: 94°C for 4 min, followed by 30 cycles at 94°C for 30 s, 60°C for 30 s,72°C for 75 s, then 72°C for 10 min. The PCR products were cloned into the EZ-TTMvector for sequencing. Sequence alignment was carried out with DNAMAN software (http://www.lynnon.com/). For predicting the molecular weight and isoelectric point of the GmBIN2 protein, Swiss Institute of Bioinformatics Compute pI/Mw Tool (http://web.expasy.org/compute_pi/) was used. Conservative domain analysis was performed on the GmBIN2 protein using the BLAST Program(http://www.ncbi.nlm.nih.gov/blast). For constructing the phylogenetic tree for GmBIN2 and its homologues, MEGA 6 Software was used (http://www.megasoftware.net). The three-dimensional structure of GmBIN2 was analyzed based on the Phyre 2 Program (http://www.sbg.bio.ic.ac.uk/phyre2) and the protein structure was generated using the RasMol Software 2.7.4.2 (http://www.OpenRasMol.org/Copyright.html).
2.3. RNA extraction and GmBIN2 gene expression analysis
Total RNA was extracted from Arabidopsis plants or soybean organs, and were converted into cDNA using a First Strand cDNA Synthesis Kit (TaKaRa, Japan). A qRT-PCR was performed to analyze the expression levelof GmBIN2 and stress-responsive genes using a realtime PCR kit (TaKaRa, Japan) on a Roche Light Cycler 480 Instrument (Roche Molecular Biochemicals, USA)according to the manufacturers’ instruction. Gene-specific primers GmBIN2-qF/R were used to perform qRT-PCR reactions. The soybean internal gene Actin4 (GenBank accession no. AF049106) and Arabidopsis internal gene actin8 (GenBank accession no. X16077) served as the qRT-PCR controls. The 2-ΔΔCtmethod was used to analyze the relative expression levels of GmBIN2 in the various qRTPCR assays (Livak and Schmittgen 2001). Each experiment was performed using three biological replicates from RNA samples extracted from three independent plants having their respective three technical replicates. The primers for qRT-PCR of GmBIN2-qF/R, internal genes, and stressresponsive genes are listed in Appendix A.
2.4. Vector construction and generating transgenic Arabidopsis
The GmBIN2-coding sequence was digested with BglII and BstEII restriction enzymes from the recombinant vector of EZ-TTM, and inserted into the plant expression vector pCAMBIA3301 to generate a 35S::GmBIN2 recombinant plasmid. The resulting pCAMBIA3301-GmBIN2 construct was transferred into Agrobacterium tumefaciens strain LBA4404 for Arabidopsis genetic transformation. An Agrobacterium-mediated floral-dip method was used for A. thaliana transformation as previously described (Clough and Bent 1998). The transgenic lines were selected with phosphinothricin and confirmed by PCR amplification with the primer pairs Bar-F/R (Appendix A). Resistant plants were further selected until the T3generation was reached to obtain homozygous lines.
2.5. Evaluation of transgenic Arabidopsis stress tolerance
Seeds of wild-type and transgenic lines (50 each) were treated with various concentrations of NaCl (0, 75, 100,and 150 mmol L-1) and mannitol (0, 100, 150, and 200 mmol L-1) on MS agar medium and vernalized at 4°C for 72 h before being cultured at 22°C. The germination rates were recorded daily and typical lines were selected to show the germination phenotype. The 3-d-old seedlings of wild-type and transgenic lines (50 each) were transferred to mediums with NaCl (0, 75, 100, and 150 mmol L-1) and mannitol(0, 100, 150, and 200 mmol L-1), and the root lengths were measured 7 d after transplanting. Typical lines were selected to show the root growth phenotype.
Seeds of wild-type and transgenic lines were grown on MS plates for 10 d and subsequently transplanted to soil in a greenhouse at 22°C for 2 wk before being treated with 100 mmol L-1NaCl for ion determination. After 10 d, the Na+, K+,and Ca2+contents were determined by an atomic absorption spectrometer as previously described (Zhu et al. 1998).
2.6. Transformation of soybean hairy roots and stress tolerance analysis
The pCAMBIA3301-GmBIN2 recombinant construct was transferred into the Agrobacterium rhizogenes K599 for soybean hairy roots transformation as previously described(Qi et al. 2014). Briefly, A. rhizogenes K599 containing the recombinant plasmid or empty vector was infected into soybean cotyledons with a 0.3-cm depth hole made by blade. After 2 wk, cotyledons with roots emerged from the cut hole were cultured with 150 mmol L-1NaClor 200 mmol L-1mannitol, and root-inducing mediums without NaClor mannitol served as an untreated control. The fresh weight of the hairy root mass was weighed before treatments and after treatments that lasted for 1 wk and the relative growth rate was calculated as previously described (Hao et al.2011). Meanwhile, RNA was isolated from the hairy roots and a qRT-PCR was performed to analyze the GmBIN2 expression level.
The Evil One had then lost all power to take him, but so long as he had the contract he could compel him to meet him in the forest each day at a certain time, where the evil spirits then scourged32 him till he bled
2.7. Determination of stress-related physiological indicators in soybean hairy roots
Relative electrical conductivity of soybean hairy roots under each treatments was determined using a digital conductivity meter as previously described (Model DDS-11A, Shanghai Scientific Instruments, China) (Feng et al.2005). The proline contents of the controland transgenic hairy roots were determined as previously described (Bates et al. 1973). The proline content was expressed as μg g-1.Superoxide dismutase (SOD) activity was determined by the hydroxylamine method with a SOD Assay Kit A001-2(Nanjing Jiancheng Bioengineering Institute, China).
2.8. Statisticalanalysis
All experiments were repeated at least three times and the graphic data were shown as mean±SD for each treatment.The data were subjected to a Student’s t-test analysis using SPSS software (ver. 20.0).
3. Results
3.1. GmBIN2 characterization
GmBIN2 (GenBank accession no. MF457588), located on chromosome 13, was found to be composed of 11 exons.The full-length-coding sequence of the GmBIN2 gene was 1 125 bp, encoding a 374-amino acid (aa) polypeptide with a 42.4-kDa molecular weight and 8.84 isoelectric point. The phylogenetic tree of 22 soybean GSK3 kinase homologues, including GmBIN2 with other plant GSK3 kinase was developed with the MEGA 6 Program. It has been shown that the plant GSK3 family can be divided into four subgroups (Charrier et al. 2002). Our results showed thatfive GSK3 members had a close relationship with GmBIN2 and allof these belonged to Subgroup II (Fig. 1-A).
A homology analysis of GmBIN2 and several Subgroup II GSK3 kinase sequences indicated that AtBIN2 had the highest homology with GmBIN2 at 88% identity (Fig. 1-B).The predicted three-dimensional structure indicated that the GmBIN2 protein contained 18 α-helices that were mostly located at the C-terminaland 16 β-sheets mostly located at the N-terminal (Fig. 1-C). The GmBIN2 structure contained a conserved domain of 285 amino acids at residues 34-318 aa and represented a Serine/Threonine protein kinase catalytic domain (Fig. 1-D).
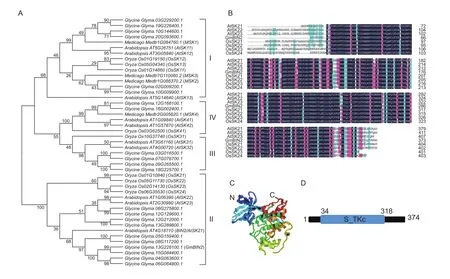
Fig. 1 Characterization of GmBIN2. A, phylogenetic tree of soybean glycogen synthase kinase 3 (GSK3) members with other plant GSK3 kinases. Evolutionary analysis was conducted in MEGA6 with neighbor-joining method. B, alignment of GmBIN2 amino acid sequences with severalother plant GSK3 homologues of Subgroup II. C, the predicted three-dimensional structure of GmBIN2. D, the conserved domain of GmBIN2 protein.
3.2. GmBIN2 expression analysis
Tissue-specific expression of GmBIN2 in Dongnong 50 soybean was performed by qRT-PCR. Total RNA from different organs was isolated including the roots, stems,flowers, leaves, and seeds. The results showed that GmBIN2 showed the highest expression in seed tissues,followed by stem and root tissues (Fig. 2-A).
The relative GmBIN2 mRNA transcript levels under different abiotic stresses were determined by qRT-PCR.Under the NaCl treatment, the GmBIN2 gene transcripts gradually increased and reached a maximum at 12 h, then declined (Fig. 2-B). The PEG treatment caused a significant change in GmBIN2 expression at 2-48 h, with the maximum transcript level reached at 8 h (Fig. 2-C). Under the ABA treatment, GmBIN2 expression levels did not change significantly compared to the control (Fig. 2-D). The above results showed that GmBIN2 transcript levels were regulated differently under different environmental stresses, however,we also showed that GmBIN2 does have a participatory role in soybean abiotic stress response.
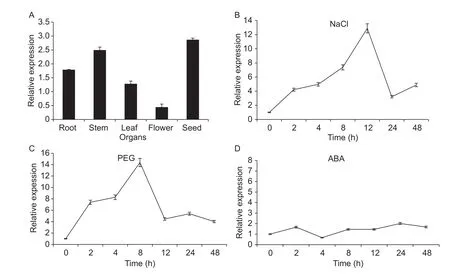
Fig. 2 Expression patterns analysis of GmBIN2 by quantitative real-time PCR. A, transcript abundances of GmBIN2 gene in different organs of Dongnong 50 soybean. B-D, the transcript pattern of GmBIN2 in response to various abiotic stresses. Exogenous chemicals were NaCl (150 mmol L-1) (B), polyethylene glycol (PEG, 5%) (C), and abscisic acid (ABA, 100 μmol L-1) (D). Leaves were harvested after treatments of 0, 2, 4, 8, 12, 24, or 48 h abiotic stress exposure. In this experiment, transcript abundances were normalized against the soybean reference gene Actin4. The data were the mean±SD of three replicates.
3.3. Stress tolerance of transgenic GmBIN2-expressing Arabidopsis
Our expression analysis results indicated that GmBIN2 transcription responds abiotic stress exposure, so in order to further investigate the function of GmBIN2, we generated transgenic Arabidopsis expressing this gene. The Bar gene used for selection was amplified in transgenic Arabidopsis plants but not in wild type (Fig. 3-A). From 14 progenies of the T3generation, those designated L2, L3, L4, L5,L6, L8, L9, L10, L13, and L14 were determined to be homozygous transgenic Arabidopsis lines. In lines L2, L4,and L5, expression levels of GmBIN2 gene were detected via RT-PCR and qRT-PCR. The expression levels of the three independent lines (L2, L4, and L5) were much higher than those of wild-type plants (Fig. 3-B). Seed germination assays showed that both wild type and transgenic seeds were able to germinate successfully in control MS medium.When salt was added, the germination was inhibited significantly (Fig. 3-C), but the inhibitory effect in wildtype was much greater than that of the transgenic lines with increasing salt concentrations after treatment for 4 d(Fig. 3-D). Furthermore, the root growth differed between wild-type and transgenic seeds under the salt treatment after 7 d (Fig. 3-E) and the transgenic plants showed longer root lengths than wild type under different concentrations of NaCl treatment (Fig. 3-F). When exposed to different concentrations of mannitol, transgenic seeds showed higher germination rates and longer root lengths than wild type (Fig. 3-G-J). These results indicated that transgenic Arabidopsis plants overexpressing the GmBIN2 gene could increase the resistance to salt and drought stresses.
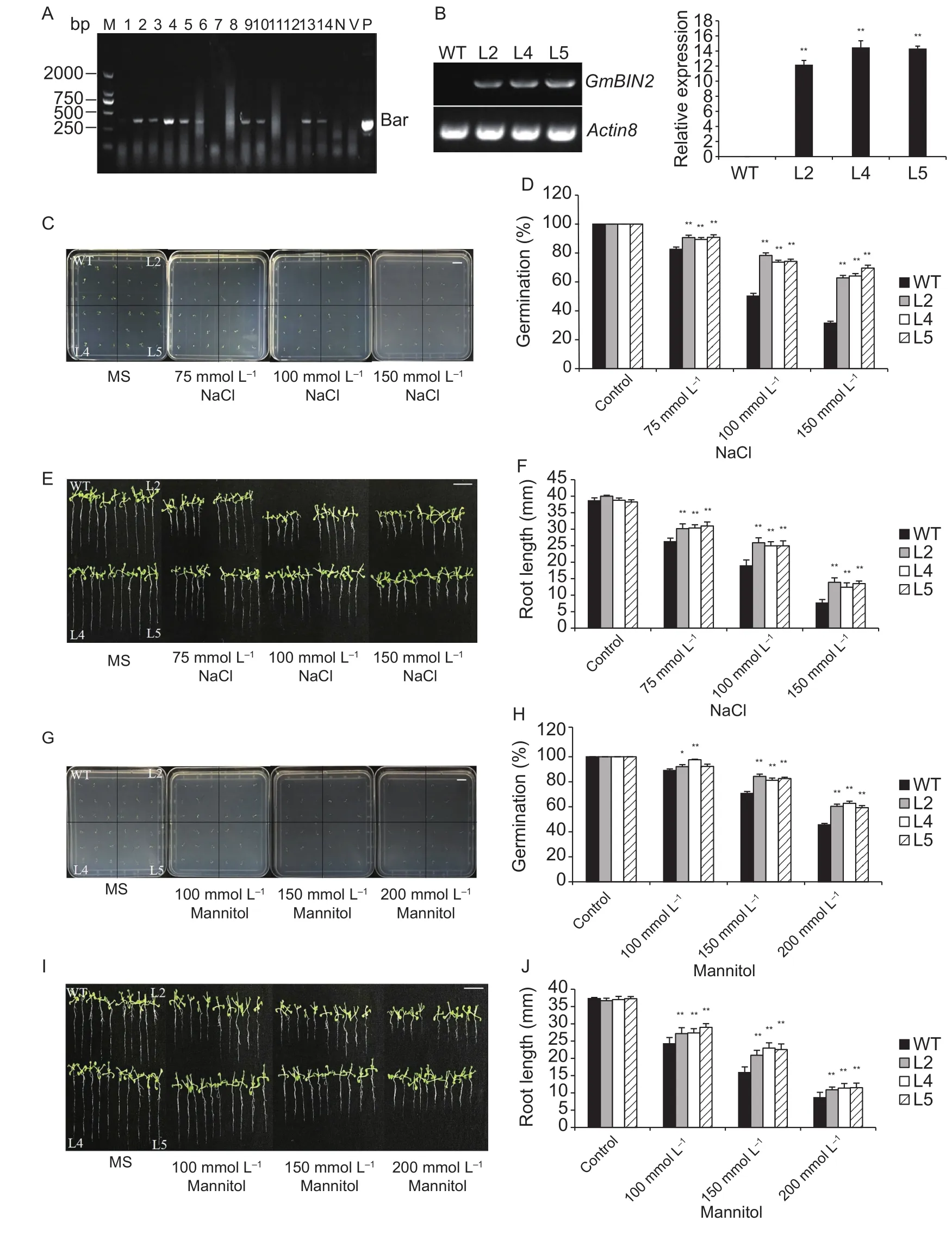
Fig. 3 Germination and root growth assay of transgenic Arabidopsis plants to salt and drought stresses. A, identification of T3 transgenic Arabidopsis by PCR amplification. M, DNA marker; lanes 1-14, transgenic plants; lane 15, negative control (N); lane 16, empty vector (V); lane 17, positive control plasmid (P). The PCR target fragment was 366 bp. B, identification of transgenic Arabidopsis lines using RT-PCR (left) and qRT-PCR (right). Transcript levels were normalized against the Arabidopsis reference gene Actin8. Wild type, WT. C and D, germination and germination rates of WT and Arabidopsis transgenic lines under 0, 75,100, and 150 mmol L-1 NaCl treatment for 4 d. Scale bar=1.0 cm. E and F, root growth and root lengths of WT and Arabidopsistransgenic lines measured 7 d after the transfer of 3-d-old seedlings from Murashige and Skoog (MS) plates to new mediums supplemented with 0, 75, 100, and 150 mmol L-1 NaCl. G and H, germination and germination rates of WT and Arabidopsistransgenic lines under 0, 100, 150, and 200 mmol L-1 mannitol treatment for 4 d. Scale bar=1.0 cm. I and J, root growth and root lengths of WT and Arabidopsis-transgenic lines measured 7 d after the transfer of 3-d-old seedlings from MS plates to new medium supplemented with 0, 100, 150, and 200 mmol L-1 mannitol. Values are mean±SD (n=50). Each experiment was performed three times with similar results. Asterisks indicate significant differences (*, P<0.05; **, P<0.01).
To clarify the physiological changes of transgenic GmBIN2-expressing Arabidopsis under salt stress, cation contents were determined. The Na+content was similar in both transgenic lines and wild-type Arabidopsis without a NaCl treatment. After treatment with 100 mmol L-1NaCl,the Na+content increased significantly, but the transgenic seedlings accumulated much lower Na+than wild type(Fig. 4-A). In the absence of NaCl, the K+content of the transgenic lines was similar to wild type. After the NaCl treatment, the K+concentrations in wild-type and transgenic lines all decreased, but transgenic seedlings contained lower K+content compared to wild type (Fig. 4-B). Transgenic lines accumulated 15-34% higher Ca2+than wild-type plants without NaCl treatment. After the induction of salt stress, Ca2+content decreased in all plants, but transgenic plants still contained greater Ca2+levels than wild-type plants (Fig. 4-C). The above results demonstrated that GmBIN2 may activate a Ca2+-dependent signaling pathway to enhance the ability of transgenic Arabidopsis plants to resist salt stress.
3.4. Expression analysis of stress-responsive genes in transgenic Arabidopsis
Our results showed that transgenic GmBIN2-expressing Arabidopsis exhibited a high resistance to salt and drought stresses. In order to clarify the mechanism of the increased stress tolerance, the expression levels of stress-responsive genes RD29A, AtCBL1, and KIN1 were determined. The transcript abundance of RD29A in transgenic plants was higher than that in wild-type plants under both stressed and non-stressed conditions (Fig. 5-A). Expression of AtCBL1 in transgenic Arabidopsis was up-regulated under NaCl stress compared to wild type (Fig. 5-B). There was no significant change in KIN1 transcripts between transgenic plants and wild-type plants in the presence or absence of NaCl stress(Fig. 5-C).
3.5. Stress tolerance of transgenic GmBIN2-expressing soybean hairy roots
To further investigate the role of the soybean GmBIN2 gene in response to environmental stresses, the soybean hairy root assay was conducted. Expression levels of GmBIN2 gene in transgenic soybean hairy roots were confirmed by qRT-PCR. Compared with the control roots infected with wild-type A. rhizogenes K599, transgenic roots overexpressing GmBIN2 (GmBIN2-oe) showed an 8- to 11-fold increase in GmBIN2 expression (Fig. 6-A). In the control mediums without treatment, all roots grew well(Fig. 6-B). However, when subjected to NaCland mannitol treatments, the growth of the control (K599) and transgenic roots were significantly inhibited, but roots transformed with GmBIN2 showed higher relative root growth rate than the control roots (Fig. 6-C). The results indicated that transgenic hairy roots overexpressing the GmBIN2 gene conferred high resistance to salt and drought stresses.
3.6. Changes in stress-related physiological indicators and gene expression in transgenic hairy roots
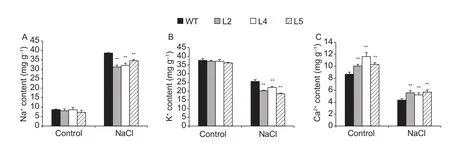
Fig. 4 Cellular ion content in wild type (WT) and transgenic Arabidopsis under salt stress. The WT and transgenic Arabidopsis plants were under normalor salt stress conditions (subjected to 100 mmol L-1 NaCl treatment for 10 d). Values are mean±SD(n=5). Each experiment was performed three times with similar results. Asterisks indicate significant differences (**, P<0.01).
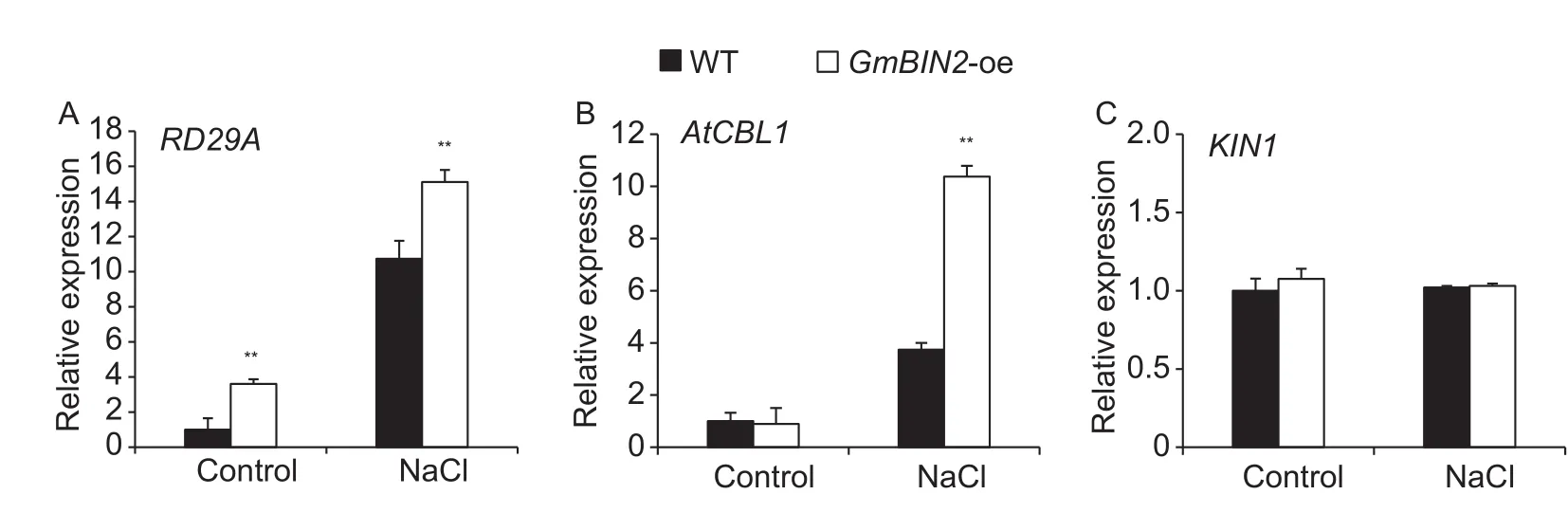
Fig. 5 Relative transcript levels of NaCl stress-responsive genes in transgenic and wild-type (WT) plants. Relative expression of RD29A, AtCBL1, and KIN1 was examined by qRT-PCR analysis in transgenic and WT plants under normaland salt stress conditions (subjected to 100 mmol L-1 NaCl treatment for 6 h) in 14-d-old seedlings. Transcript levels were normalized against the Arabidopsis reference gene Actin8. The data are the mean±SD of three replicates (n=5). Asterisks indicate significant differences(*, P<0.05; **, P<0.01).
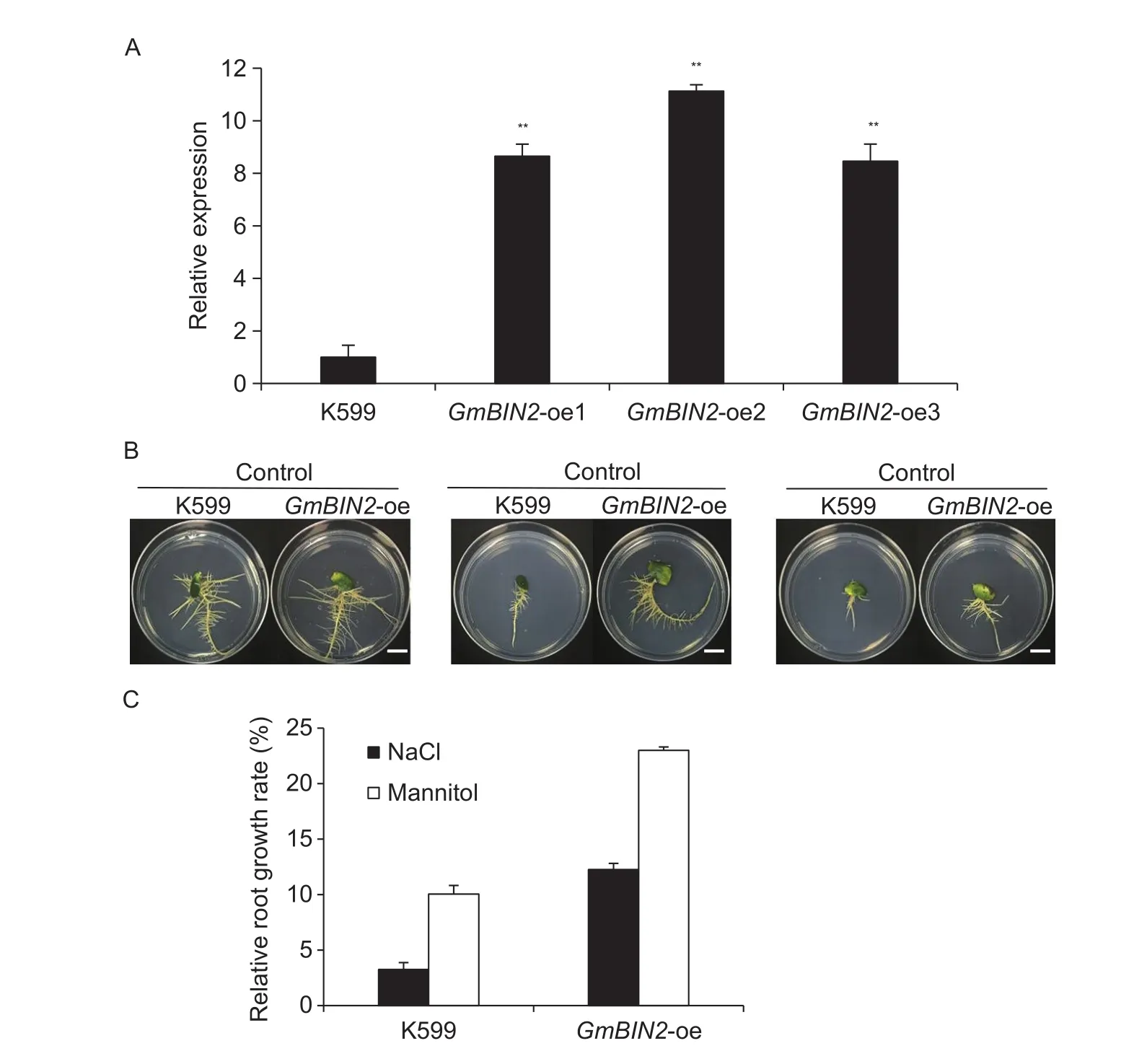
Fig. 6 Analysis the role of GmBIN2 under salt and drought stresses using the hairy roots system. A, relative GmBIN2 expression in overexpressed GmBIN2 (GmBIN2-oe) roots and control roots (K599) as revealed by qRT-PCR analysis. Transcript abundances were normalized against the soybean reference gene Actin4. B, growth of controland transgenic hairy roots in root-inducing mediums with 150 mmol L-1 NaClor 200 mmol L-1 mannitol for 1 wk. A totalof 30 samples were analyzed, and the typical lines were selected to show the growth of roots. Scale bar=1 cm. C, relative growth rate of transgenic hairy roots under 150 mmol L-1 NaClor 200 mmol L-1 mannitol for 1 wk. Values are mean±SD (n=30). Asterisks indicate significant differences (**, P<0.01).
Under salt and drought treatments, the relative electrical conductivity in the controland transgenic roots both increased gradually, but the increase trend of the control roots was higher than that of the transgenic roots (Fig. 7-A).Without any treatments, the proline contents in both control(K599) and transgenic hairy roots were nearly the same(Fig. 7-B). Under NaCland mannitol conditions for 7 d,proline content accumulated sharply in transgenic roots and was about one- to two-fold higher than that in control roots. Consistent with thefinding of higher proline contents in transgenic hairy roots, the SOD activity in transgenic roots was dramatically higher than that of in the control roots under drought and salt stress conditions (Fig. 7-C).In addition, we investigated GmCBL1 gene expression,a homolog of AtCBL1 in soybean hairy roots. As a result,transcript abundance of GmCBL1 in transgenic roots was higher than that in wild-type roots under salt and drought stresses (Fig. 7-D). The analysis above implied that transgenic roots overexpressing GmBIN2 suffered less damage to cell membranes and accumulated more proline and SOD to resist the damage caused by stresses. It was also demonstrated that the overexpression of GmBIN2 in transgenic soybean hairy roots may activate a stresssignaling pathway through the induction of the GmCBL1 gene.
4. Discussion
In the present study, we found that the GmBIN2 gene,a member of GSK3 protein kinase family in soybean,enhanced the resistance to salt and drought stresses in Arabidopsis and soybean hairy roots when overexpressed.It has been shown that plants have more divergent GSK3 isoforms compared with mammalian GSK3s, with 10 identified in Arabidopsis and nine identified in rice (Yoo et al.2006). Youn and Kim (2015) investigated the phylogenetic relationships, gene structure, and the diverse cellular signal transduction pathways of the GSK3 kinase in Arabidopsis and rice to understand the versatile functions of plant GSK3 kinases. It has been demonstrated that plant GSK3 kinases play a prominent part in multiple signaling pathways in response to diverse environmental stimuli, however, the specific functions of GSK3 kinase in soybean remain largely unknown.
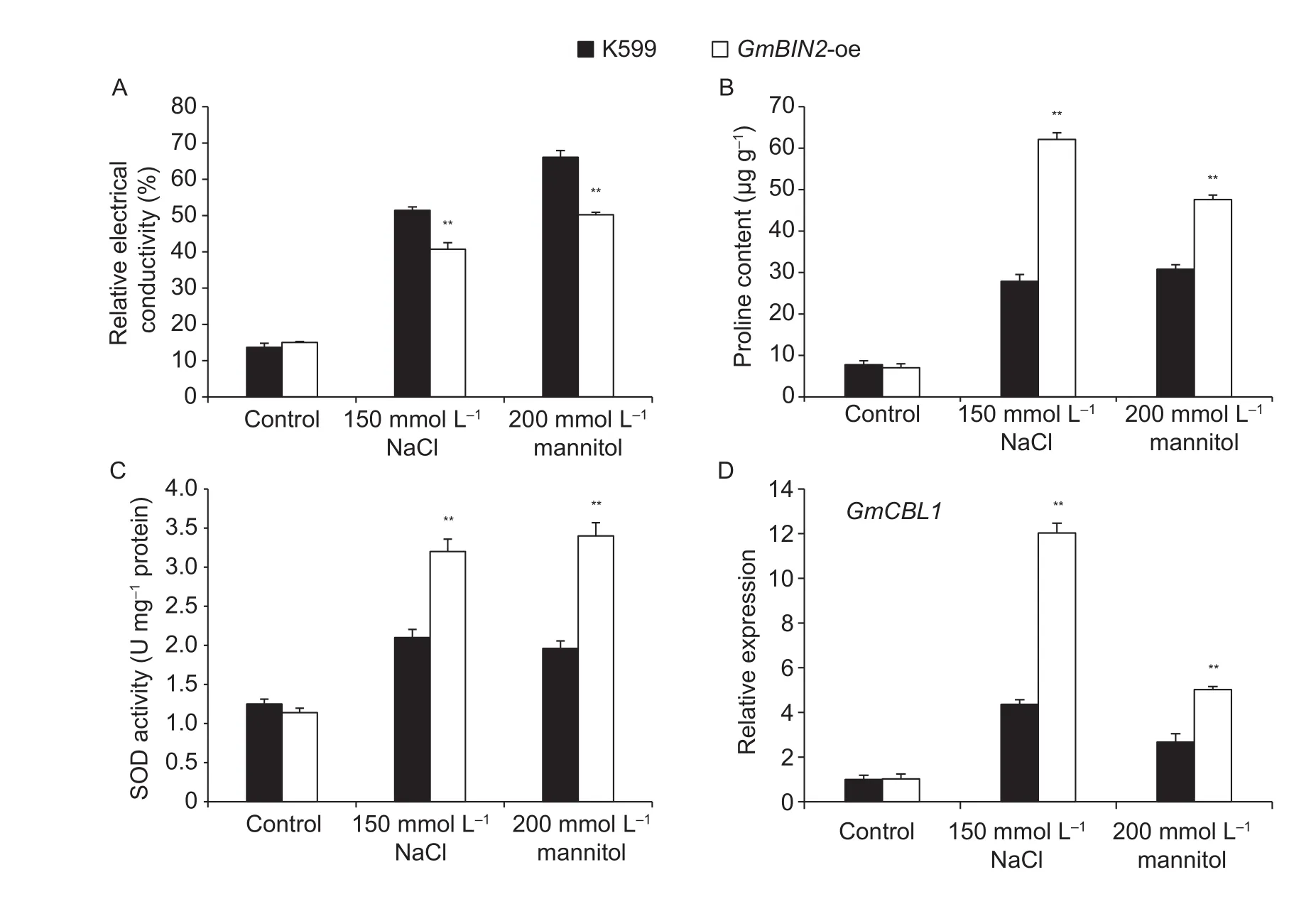
Fig. 7 Altered physiological indicators and gene expression in soybean hairy roots. A, relative electrical conductivity between K599 and GmBIN2-oe (transgenic roots) with and without 150 mmol L-1 NaClor 200 mmol L-1 mannitol treatments for 7 d. B,proline contents between K599 and GmBIN2-oe with and without 150 mmol L-1 NaClor 200 mmol L-1 mannitol treatments for 7 d.C, superoxide dismutase (SOD) activities between K599 and GmBIN2-oe with and without 150 mmol L-1 NaClor 200 mmol L-1 mannitol treatments for 7 d. D, expression of GmCBL1 in K599 and GmBIN2-oe with and without 150 mmol L-1 NaClor 200 mmol L-1 mannitol treatments for 2 d. Transcript abundances were normalized against the soybean reference gene Actin4. Values are mean±SD (n=5). Each experiment was performed three times with similar results. Asterisks indicate significant differences (**,P<0.01).
According to the data analyzed in the Phytozome database, 22 soybean GSK3 kinases existed in the soybean genome andfive of these GSK3 members exhibited a close relationship with GmBIN2 protein. In the previous work of our laboratory, GmBIN2 was screened under different 2,4-D treatment concentrations in the soybean Dongnong 50 cultivar to determine the differentially expressed genes.A close association has been proposed between 2,4-D and stress response (Cho et al. 2000; Teixeira et al. 2006;Pan et al. 2010). Therefore, this study mainly focused on the regulation of GmBIN2 in response to stress tolerance.GmBIN2 was shown to contain a serine/threonine protein kinase catalytic domain (Fig. 1). The protein structure,amino acid sequence and evolutionary relationship of the GmBIN2 protein indicated that GmBIN2 belonged to GSK3 kinase family. GmBIN2 expression was increased by salt and drought stresses (Fig. 2). When subjected to different NaCland mannitol concentrations, transgenic Arabidopsis seeds observably increased in germination rate and root length compared to wild-type seeds (Fig. 3). Our results indicated that transgenic Arabidopsis plants showed a high resistance to salt and drought stresses. Further examination of Na+, K+, and Ca2+contents revealed that GmBIN2 could reduce cellular Na+content and increase cellular Ca2+content in transgenic plants under NaCl treatment (Fig. 4).We know Ca2+is an important messenger in plants, and plays an important role in regulating plant resistance to salt stress. It has been shown that increased Ca2+content may restrict the Na+in flux and K+efflux to maintain cellular K+/Na+homeostasis (Amtmann and Sanders 1999; White and Davenport 2002; Shabala et al. 2006). Additionally,GmBIN2 may trigger a Ca2+-dependent pathway to control the ion content to enhance salt tolerance in transgenic Arabidopsis (Liu and Zhu 1998; Pardo et al. 1998).
In addition, two NaCl stress-responsive genes, RD29A and AtCBL1, showed altered expression in transgenic Arabidopsis plants (Fig. 5). RD29A is involved in response to salt, drought, and cold stresses (Yamaguchi-Shinozaki and Shinozaki 1993), whereas AtCBL1 is reported to function as Ca2+sensor in osmotic stress signaling and could also be induced by salt and drought stresses (Kudla et al. 1999;Cheong et al. 2003). In the present study, expression levels of RD29A and AtCBL1 genes were higher in transgenic Arabidopsis than in wild-type plants under NaCl stress and overexpression of GmBIN2 up-regulated RD29A expression even in the absence of NaCl stress, suggesting that GmBIN2 may play different roles in inducing these stressresponsive genes under different conditions and is involved in a complicated regulatory stress response network. To date, there are 10 AtSK genes in the GSK3/SHAGGY-like protein kinases family (Dornelas et al. 1998). We know that AtSK22 and AtSK23 are the two closest homologues of AtBIN2 (AtSK21). Like in mammals, Arabidopsis GSK3 functions both redundantly and specifically to regulate plant development. The specificity of GSK3 has been exhibited in several studies. AtBIN2 has been shown to have a primary role in the BR signaling pathway and is not responsive to abiotic stress (Charrier et al. 2002; Yan et al.2009). In addition, overexpression of GmBIN2 in transgenic Arabidopsis did not cause a differential expression of AtBIN2 compared to wild type (Appendix B). However, AtGSK1(AtSK22) has been shown to be involved in the NaClstressed signal pathway, and overexpression of AtGSK1 enhanced NaCl tolerance in Arabidopsis. When transgenic plants were under NaCl stress, AtGSK1 was seen to activate a Ca2+-dependent signaling pathway and several small Ca2+-binding proteins, including AtCBL1, that may activate downstream components of NaCl stress signaling and enhance NaCl tolerance (Piao et al. 2001). In our study,we posit that the GmBIN2 gene may encode protein that is functionally homologous to AtGSK1 and may act through similar pathways to regulate the stress signaling.
Furthermore, overexpression of GmBIN2 in soybean hairy roots also showed significantly higher relative root growth rate than control roots under stress treatments(Fig. 6). Under salt and drought conditions, transgenic hairy roots showed a lower relative electrical conductivity compared to control roots, meanwhile, transgenic roots accumulated more proline and SOD (Fig. 7). When plant tissues are under stress conditions, the structure of the cell membrane is destroyed, and the membrane permeability is greatly increased, resulting in an increase of the relative electrical conductivity of the tissue exudate(Feng et al. 2005). The lower relative electrical conductivity of transgenic hairy roots meant a lower degree of damage to the cell membrane. SOD is an antioxidant enzyme used to eliminate oxygen free radicals and counteract the toxic effects on cells. For example, overexpression of tomato Cu/Zn superoxide dismutase could enhance oxidative stress defense in transgenic potato (Perl et al. 1993). Ourfindings showed that SOD activities in GmBIN2-overexpression hairy roots were higher than in control roots after treatment with salt and drought (Fig. 7-B). This might be the result of reducing the toxic effects of superoxide radicals and enhancing stress tolerance. Plants accumulate a large amount of proline in response to environmental stresses(Saradhi and Mohanty 1997) and high proline content can result from injury to the plants (Liu and Zhu 1997; Wang et al.2003). Therefore, proline accumulation may be a signalof stress tolerance in plants. The present results inferred that the high proline contents of transgenic soybean hairy roots may be an indicator for salt and drought stresses,and high proline accumulations in soybean hairy roots could protect transgenic lines against injury from salt and drought stresses.
Moreover, we investigated the expression of the GmCBL1 gene, a homolog of AtCBL1 in soybean hairy roots. CBL1 is a calcineurin B-like calcium sensor and plays a critical role in plant responses to abiotic stress signaling pathways.Importantly, CBL1 functions as a positive regulator of salt and drought responses (Cheong et al. 2003). It has been reported that overexpression of GmCBL1 in Arabidopsis enhances tolerances to both salt and drought stresses in transgenic plants. Thus, GmCBL1, like AtCBL1, may function as a positive regulator in salt and drought stress signaling pathways (Li et al. 2012). As shown in Fig. 7-D,transgenic lines showed high GmCBL1 mRNA levels under salt and drought stresses, indicating that GmBIN2 may be involved in the induction of the GmCBL1 gene in the stresssignaling pathways in response to salt and drought stresses.
5. Conclusion
Here, we characterized the soybean GmBIN2 gene, a member of the GSK3/SHAGGY-like kinase family. The GmBIN2 transcripts were able to be induced by salt and drought stresses. Our data clearly demonstrated that GmBIN2 could enhance salt and drought tolerance in Arabidopsis and soybean hairy roots. At present, research regarding GmBIN2 is at an elementary stage and additional investigations are needed to elucidate its exact mechanism during stress response. Further study may focus on the molecular level including probable cross-talk among various signaling pathways to clarify its involvement in environmentalabiotic stress response.
Acknowledgements
This research was supported by the funding from the Creative Research Groups of Heilongjiang Province of China (JC2016004), the National Key R&D Program of China (2016YFD0100201-21), the Project of Outstanding Academic Leaders in Harbin, China (2015RQXXJ018),and the China Agriculture Collaborative Creation Research System of Miscellaneous Grain Crops.
Appendices associated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- ldentification and characterization of Pichia membranifaciens Hmp-1 isolated from spoilage blackberry wine
- Implications of step-chilling on meat color investigated using proteome analysis of the sarcoplasmic protein fraction of beef longissimus lumborum muscle
- Spatial-temporal evolution of vegetation evapotranspiration in Hebei Province, China
- Design of a spatial sampling scheme considering the spatialautocorrelation of crop acreage included in the sampling units
- Comparison of forage yield, silage fermentative quality, anthocyanin stability, antioxidant activity, and in vitro rumen fermentation of anthocyanin-rich purple corn (Zea mays L.) stover and sticky corn stover
- Synonymous codon usage pattern in model legume Medicago truncatula
