新型抗菌肽
——表面活性素、伊枯草菌素和丰原素
2018-02-28金清肖明
金清,肖明
上海师范大学生命与环境科学学院,上海 200234
抗菌肽(antimicrobial peptide)具有抗菌谱广、作用迅速强大、不易产生耐药性等优点,在医药、化妆品、食品工业中具有广阔的应用前景[1-3]。表面活性素(surfactin)、伊枯草菌素(iturin)和丰原素(fengycin)是芽胞杆菌产生的主要活性物质,可抑制农作物病害[4-6]。近年来发现这3类物质在医药领域有重要应用前景,具有抗病毒[5,7]、抗肿瘤[8-12]、抗细菌[13-15]、抗真菌[16-18]、抗支原体[19]、抗炎[20]作用,展示出极大的临床应用潜力[21],是一种新型抗菌肽[22-26],但人们对它们在医药领域中的研究进展所知甚少。因此,本文就表面活性素、伊枯草菌素和丰原素的发现历史、结构特点、作用机制、生物合成及应用价值进行综述。
1 发现历史
表面活性素于1968年首次在枯草芽胞杆菌(Bacillussubtilis)的培养液中发现,由4种异构体(枯草菌素A~D)组成,表现出各种生理活性,包括作为纤维蛋白凝固抑制剂和细胞裂解物[27]。伊枯草菌素于1957年首次在从土壤中分离到的枯草芽胞杆菌中发现[28],丰原素则首次在枯草芽胞杆菌F29-3中发现[29]。
表面活性素、伊枯草菌素和丰原素具有广泛的工业应用价值,如制造洗涤剂[30-31]、抑制植物病害[32-34]、提高油采收率[31]等。近年来,这3种新型抗菌肽的医疗应用研究也取得重大突破,在医药领域中扮演着越来越重要的角色。
与其他菌肽的生产类似,这3种抗菌肽也是通过发酵生产。目前研究者从各方面努力,包括优良菌种选育[5]、发酵过程优化[35-36]、高效分离纯化方式的探索等来提高产率[37-38],取得了一系列成果。
2 结构特点及作用机制
表面活性素、伊枯草菌素和丰原素是一类主要由革兰阳性芽胞杆菌产生的抗菌肽,一般由1个疏水的脂肪烃链以羧基、羰基或氨基与亲水的由7~10个氨基酸构成的肽链以酰胺键或内酯的形式连接构成环肽,其结构上的差异主要在于脂肪链中碳原子的个数、氨基酸的种类及脂肪酸链与肽链连接键的不同(图1)。
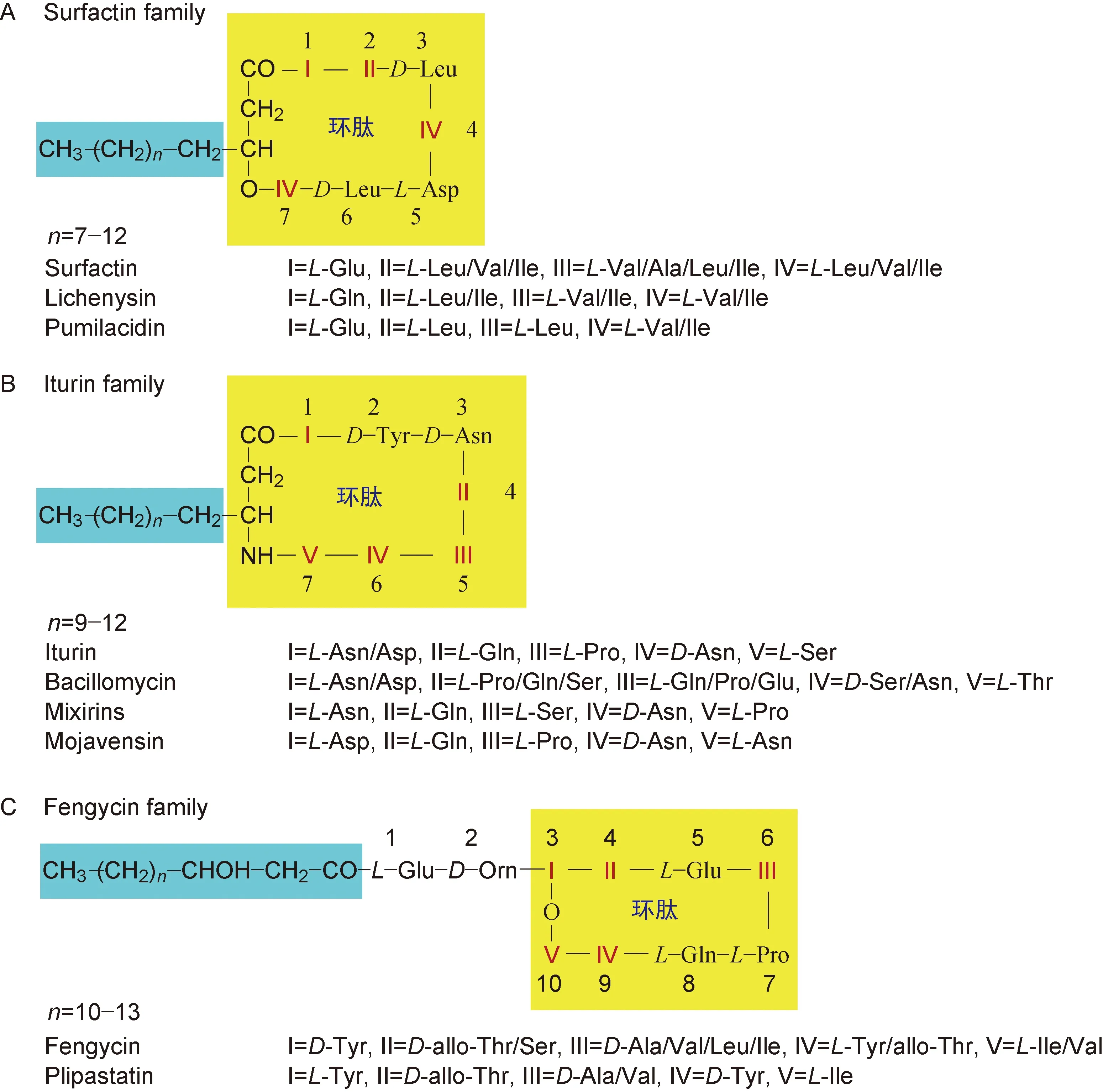
A: Surfactin is formed by β-hydroxy fatty acids (12-17 carbon atoms) with peptide chain through lactone bonds.The molecular peptide chain is composed of seven α-amino acids.B: Iturin is a ring formed by β-amino fatty acids (14-17 carbon atoms) with peptide chain through amide bond.The molecular peptide chain is composed of seven α-amino acids.C: Fengycin is composed of β-hydroxy fatty acids (14-17 carbon atoms) and peptide chain through lactone bond.Fengycin is different from surfactin and iturin.The macrocyclic ring is composed of peptide chain of ten α-amino acids.The Arabic numerals represent the position of the amino acid.
图1表面活性素、伊枯草菌素和丰原素的分子结构
Fig.1Molecularstructuresofsurfactin,iturinandfengycin
表面活性素由β-羟基脂肪酸与肽链以内酯键结合而成[39-41]。多数细菌代谢产生的表面活性素的肽链为七元肽,即分子中的肽链由7个α-氨基酸组成,再与带有12~17个碳原子的β-羟基脂肪酸构成一个大内酯环,相对分子质量(Mr)为 1 000 左右。多肽中典型的氨基酸顺序为[27,42-43]:L-Glu-L-Leu-D-Leu-L-Val-L-Asp-D-Leu-L-Leu(图1A)。表面活性素家族常见的有surfactin A[44]、surfactin C[44]、lichenysin[44-46]、pumilacidin等[46]。
伊枯草菌素由β-氨基脂肪酸与肽链以酰胺键成环[47]。多数细菌代谢产生的伊枯草菌素的肽链也为七元肽,与带有14~17个碳原子的β-氨基脂肪酸构成一个大环,Mr 为 1 000 左右。多肽中典型的氨基酸顺序为[47-48]:L-Asn-D-Tyr-D-Asn-L-Gln-L-Pro-D-Asn-L-Ser(图1B)。伊枯草菌素家族种类较多,如iturin A[49-54]、iturin C[49,51]、iturin D[51]、iturin E[51-53]、iturin F[53]、杆菌霉素(bacillomycin)D[49-52]、bacillomycin F[49-51]、bacillomycin L[49-51]、mixirins[9,51-52]、mojavensin[52]、抗霉枯草菌素(mycosubtilin)[49-51,53-54]和subtulene[51]等。
丰原素由β-羟基脂肪酸与肽链以内酯键结合而成。与表面活性素不同的是,丰原素的大环由肽链自行构成,β-羟基脂肪酸并不参与[54-55]。多数细菌代谢产生的丰原素的肽环为十元肽,第三位的D-Tyr和最后一位的L-Ile以内酯键结合成环,再与带有14~17个碳原子的β-羟基脂肪酸以酯键结合,Mr 为 1 500 左右[54-56]。多肽中典型的氨基酸顺序为[54-58]:L-Glu-D-Orn-D-Tyr-D-allo-Thr-L-Glu-D-Ala-L-Pro-L-Gln-L-Tyr-L-Ile(图1C)。丰原素家族常见的有fengycin A[54-58]、fengycin B[54-55,57-58]、fengycin C[54-55,58]、fengycin S[55]和制磷脂菌素(plipastatin)[54-55,57-59]。
表面活性素可通过溶解和破坏细胞膜发挥抗菌作用,其与膜中的极性头部和疏水酰基链均可相互作用,高浓度时引起磷脂双分子层高度不稳定,中等浓度时在细胞膜中形成离子传导孔与Ca2+结合,有助于膜渗透;其还可通过有机屏障驱动其他单价和二价阳离子,导致cAMP磷酸二酯酶活性被抑制[60-62]。伊枯草菌素能迅速引起细胞膜损伤,细胞通透性改变,细胞内物质外泄,从而达到抑制真菌孢子萌发、菌丝体生长的效果;其还可与细胞内靶点(如细胞DNA)相互作用,破坏细胞内钙稳态,导致细胞死亡[63-65]。丰原素对细胞膜有显著扰乱作用,通过基于丰原素浓度的两态跃迁过程使磷脂双分子层损伤,从而导致细胞死亡。高浓度时,丰原素作为洗涤剂促进细胞膜溶解,该过程主要由其对脂质双分子层的吸引力等物理化学性质所驱动;低浓度时,丰原素聚合形成孔洞,导致膜的渗透率发生变化[61,66-68]。
3 生物合成
芽胞杆菌能分泌多种肽类及由肽类衍生的抗菌活性物质,按合成途径分为核糖体肽和非核糖体肽[4]。非核糖体肽Mr较小,一般为 3 000 以下,通过非核糖体肽链合成酶(non-ribosomal peptide synthetase,NRPS)来合成,多发生于菌体生长停止之后;而核糖体肽Mr较大,大多于菌体快速生长时期合成[4,69]。
非核糖体途径合成的脂肽类抗菌活性物质合成于菌体生长停止之后,属于微生物次级代谢产物,能绕开核糖体,不以mRNA 为模板,也不需tRNA 作为运载工具,而是通过NRPS识别特定的氨基酸并连接成多肽链[4,70]。表面活性素、伊枯草菌素和丰原素就是NRPS合成的次级代谢产物,由胞内游离氨基酸经活化后结合到合成酶系特定的结构域,从而实现肽链的延长和环化[71-72](图2)。
NRPS是由多个功能模块组成的复合酶体系,按功能分为必不可少模块和可供选择模块,各模块负责活化不同的氨基酸使肽链延长。必不可少模块有氨基酸激活结构域(amino acid activating domain)、氨酰载体结构域(acyl carrier)、缩合结构域(condensation domain)和硫酯酶结构域(thioesterase domain)。氨基酸激活结构域由550个氨基酸残基构成,负责识别和腺苷酰化特定的氨基酸,又称为腺苷酰化结构域(adenylation domain);氨酰载体结构域负责运载氨基酸,又称为巯基化结构域T或肽酰载体蛋白( peptidyl carrier protein,PCP);缩合结构域负责肽键形成;硫酯酶结构域负责释放多肽和肽的环化。可供选择模块包括环化结构域(cyclization domain)、甲基转移酶结构域(methyltransferase domain)、差向异构酶结构域(epimerization domain)等。差向异构酶结构域负责将被激活的L-氨基酸转化为D-氨基酸。全酶由多个模块按特定的空间顺序排列而成,模块的数量、种类及排列次序决定了氨基酸种类、顺序和最终产物肽链的长短[73-74](图2)。

A: amino acid activating domain;PCP: peptidyl carrier protein;C: condensation domain;E: epimerization domain;TE: thioesterase domain;MCT: monocarboxylate transporter.
图2表面活性素、伊枯草菌素和丰原素的代谢通路
Fig.2Metabolicpathwaysofsurfactin,iturinandfengycin
NRPS合成多肽的一般过程为:首先,氨基酸激活结构域选择并结合特定的氨基酸,在ATP 作用下激活氨基酸(腺苷化),形成氨酰腺苷酸。氨酰腺苷酸与氨酰载体结构域上的4-磷酸泛酰巯基辅基以共价键形式结合,形成氨酰载体复合体。然后,携带有活化氨基酸的氨酰载体与缩合结构域特定部位结合,在其合成酶的作用下,按相邻合成酶各组成模块的顺序依次向前形成肽键。肽键形成后,进入下一循环,即肽链延伸过程。最后,硫酯酶结构域终止肽链合成,将肽链从磷酸泛酰巯基辅基释放下来,并进行环化[75-77]。
3.1 表面活性素
表面活性素合成酶包括3 个亚单位:SrfA、ComA(SrfB)和SrfC[4,26,78-86]。编码表面活性素合成酶的基因srfA-A、srfA-B、srfA-C共同构成srfA操纵子(长约25 kb),分别负责编码Mr为 401 000、402 000 和 144 000 的3个合成单体酶E1A、E1B和E2。srfA-C编码的第一个硫酯酶结构域负责终止肽链延伸并释放多肽产物(图2A)。
sfp基因(约4.5 kb)是参与表面活性素合成的第二调控元件,位于srfA操纵子下游30.5 kb处,与srfA-D末端相距约4 kb。sfp基因编码的SFP 酶具有编码磷酸泛酰巯基乙胺基转移酶(phosphopantetheinyl transferase,PPTase)的功能,可催化非核糖体肽和载铁蛋白前体的形成,并借此将表面活性素合成酶激活,属PPTase超家族。有研究认为sfp和srfA-C-TE基因共同发挥主导作用,还有研究认为sfp基因在表面活性素合成中有更直接的调节作用[85]。在基因图谱中,srfA与sfp相邻,而与srfB相隔,srfB功能等同于comA基因,即激活srfA启动子的转录。
SrfA-A负责组装前3位氨基酸;SrfA-B负责组装第4~6位氨基酸;SrfA-C负责组装第7位氨基酸,并将羟基脂肪酸转移到蛋白SrfA-A上。
3.2 伊枯草菌素
伊枯草菌素通过聚酮合酶(polyketide synthase,PKS)-NRPS杂合体系合成[78,87-88]。
编码伊枯草菌素合成酶的基因ituD、ituA、ituB和ituC共同构成itu操纵子(长约38 kb)。ituD负责编码丙二酰辅酶A转酰酶,对伊枯草菌素的形成起重要作用,被破坏可导致iturin A产生特异性缺陷;其还可调控伊枯草菌素产量,可能与脂肪酸合成有关。ituA编码Mr为 449 000 的蛋白,与脂肪酸合成酶、氨基酸转移酶和肽合成酶具有同源性,可能与β-氨基脂肪酸形成有关;部分基因与聚酮合酶相关,其编码的聚酮合酶参与脂肽类分子碳链合成的最终步骤,并为肽段部分氨基酸分子的组装做好准备。ituB编码Mr为 609 000 的具有4个氨基酸腺苷酸化结构域的肽合成酶。ituC编码Mr为 297 000 的另一种肽合成酶,具有2个腺苷酸化结构域、1个差向异构酶结构域和硫酯酶结构域,这可能有助于肽环化。ItuA负责组装第1位氨基酸,ItuB负责组装第2~5位氨基酸,ItuC负责组装第6和7位氨基酸(图2B)。
3.3 丰原素
编码丰原素合成酶的基因fenC、fenD、fenE、fenA、fenB共同构成fen操纵子(长约37 kb),分别编码5个亚基,即5个单体酶——FenC、FenD、FenE、FenA和FenB。每个单体酶一般含1~3个氨基酸激活模块,且每个模块具有接受特定氨基酸及形成相应肽键的功能。丰原素合成从肽单体酶FenC开始,途经FenD、FenE和FenA,终止于肽单体酶FenB[60,89-91]。
FenC的Mr为 287 000,负责活化并组装第1和2位氨基酸;FenD的Mr为 290 000,负责活化第3和4位氨基酸;FenE的Mr为 286 000,活化第5和6位氨基酸;FenA的Mr为 406 000,负责活化第7~9位氨基酸;FenB的Mr为 146 000,负责组装最后一位氨基酸。FenB含有能中断肽链合成的硫酯酶结构域,具有释放肽链的功能,在其下游也发现了与脂肪酸代谢有关的基因(图2C)。
4 应用价值
研究表明,表面活性素对新城疫病毒(Newcastle disease virus,NDV)不仅具有直接灭活作用,还可阻断其对细胞的吸附;随着浓度升高还可抑制NDV的生物合成,具有明显的量-效关系。表面活性素的治疗指数为 12.16,高于病毒唑的 9.70,有望开发成为一种有效的抗病毒药物,这对养殖业生产及防治NDV感染所致人类疾病具有重要意义[7]。另有研究表明,表面活性素对人乳腺癌细胞Michigan Cancer Foundation-7(MCF-7)表现出较强的抑制作用。噻唑蓝(methylthiazolyldiphenyl-tetrazolium bromide,MTT)法显示,表面活性素能抑制MCF-7增殖,呈现浓度与时间依赖关系,细胞处理24 h后的半抑制浓度(half maximal inhibitory concentration,IC50)为10 μg/mL。随着发酵期间表面活性素含量升高(0.3~48.2 mg/kg),SEC(surfactin extractions of cheonggukjang)(100 μg/mL)的抗癌活性逐渐升高(20.3%~54.7%)[8]。表面活性素除具有抗病毒、抗肿瘤作用,还可抗细菌[13-15]、抗真菌[16-17]、抗支原体[19]、抗炎[20],抗菌谱较广,同时具有不易产生耐药性、可被动物消化酶降解、无残留等优点,均显示其应用于医药业的潜力。
伊枯草菌素具有强烈的抗真菌、抗肿瘤作用,还具有低毒、低残留、低过敏性和抗菌谱广的特点,是一种潜在的具有极大开发应用价值的医药产品。研究表明,伊枯草菌素对红色毛癣菌具有较强抑制作用[18]。Mixirins A、B和C具有抗肿瘤作用,可抑制人结肠癌细胞HCT-116的生长,其IC50分别为 0.68、1.6 和 1.3 μg/mL[9]。
丰原素具有显著的抗肿瘤效果,对肿瘤细胞及肿瘤组织具有较好的选择性,对肿瘤凋亡相关蛋白有明显影响,而对正常造血系统和白细胞无影响,为寻找新型抗肿瘤药物提供了方向。研究表明,与对照组相比,丰原素浓度达20 μg/mL时即可抑制人结肠癌细胞HT-29的生长,并呈浓度与时间依赖关系。蛋白免疫印迹分析发现,加入丰原素后,HT-29细胞中的Bax、Caspase-3和Caspase-6表达明显增加,而Bcl-2和CDK4/cyclin D1表达降低,表明丰原素可通过影响人HT-29细胞周期的G1期停滞和诱导细胞凋亡而对其产生抑制作用[10]。此外,大量研究也表明丰原素可抑制人结肠癌HCT-15细胞增殖[11],调节人肺癌95D细胞G0/G1期引起细胞周期停滞和促进细胞凋亡来抑制癌细胞生长[12],显示其具有极大的抗肿瘤潜力。
综上所述,抗菌肽是极具价值的新一代抗菌药物,能作为抗病原体药物,并可能发展成为抗肿瘤药物,在免疫调节、促进伤口愈合等方面有应用价值。多数文献表明,表面活性素、伊枯草菌素和丰原素在医疗领域有着巨大价值与广阔前景,具备低毒、抗菌谱广、不易产生耐药性等优势及工业化生产潜力,但临床应用还不广泛。随着对抗菌肽研究的不断深入及技术的不断改进,如何将其应用于临床、应用于人类医疗事业将成为研究的主要方向。
[1] Zhang LJ,Gallo RL.Antimicrobial peptides [J].Curr Biol,2016,26(1): R14-R19.
[2] Sadredinamin M,Mehrnejad F,Hosseini P,Doustdar F.Antimicrobial peptides (AMPs) [J/OL].Novelty Biomed,2016,4(2): 70-76.http://journals.sbmu.ac.ir/nbm/article/view/9158.
[3] Ramesh S,Govender T,Kruger HG,de la Torre BG,Albericio F.Short antimicrobial peptides (SAMPs) as a class of extraordinary promising therapeutic agents [J].J Pept Sci,2016,22(7): 438-451.
[4] Leclère V,Béchet M,Adam A,Guez JS,Wathelet B,Ongena M,Thonart P,Gancel F,Chollet-Imbert M,Jacques P.Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism’s antagonistic and biocontrol activities [J].Appl Environ Microbiol,2005,71(8): 4577-4584.
[5] Wei YH,Wang LC,Chen WC,Chen SY.Production and characterization of fengycin by indigenous Bacillus subtilis F29-3 originating from a potato farm [J].Int J Mol Sci,2010,11(11): 4526-4538.
[6] Furuya S,Mochizuki M,Aoki Y,Kobayashi H,Takayanagi T,Shimizu M,Suzuki S.Isolation and characterization of Bacillus subtilis KS1 for the biocontrol of grapevine fungal diseases [J].Biocontrol Sci Technol,2011,21(6): 705-720.
[7] 黄现青,魏战勇,高晓平,韩庆功,崔艳红.表面活性素体外抗新城疫病毒活性研究 [J].浙江农业科学,2008,1(5): 630-632.
[8] Lee JH,Nam SH,Seo WT,Yun HD,Hong SY,Kim MK,Cho KM.The production of surfactin during the fermentation of cheonggukjang by potential probiotic Bacillus subtilis CSY191 and the resultant growth suppression of MCF-7 human breast cancer cells [J].Food Chem,2012,131(4): 1347-1354.
[9] Zhang HL,Hua HM,Pei YH,Yao XS.Three new cytotoxic cyclic acylpeptides from marine Bacillus sp [J].Chem Pharm Bull (Tokyo),2004,52(8): 1029-1030.
[10] Cheng W,Feng YQ,Ren J,Jing D,Wang C.Anti-tumor role of Bacillus subtilis fmbJ-derived fengycin on human colon cancer HT29 cell line [J].Neoplasma,2016,63(2): 215-222.
[11] Sivapathasekaran C,Das P,Mukherjee S,Saravanakumar J,Mandal M,Sen R.Marine bacterium derived lipopeptides: characterization and cytotoxic activity against cancer cell lines [J].Int J Pept Res Ther,2010,16(4): 215-222.
[12] Yin H,Guo C,Wang Y,Liu D,Lv Y,Lv F,Lu Z.Fengycin inhibits the growth of the human lung cancer cell line 95D through reactive oxygen species production and mitochondria-dependent apoptosis [J].Anticancer Drugs,2013,24(6): 587-598.
[13] Korenblum E,de Araujo LV,Guimarães CR,de Souza LM,Sassaki G,Abreu F,Nitschke M,Lins U,Freire DM,Barreto-Bergter E,Seldin L.Purification and characterization of a surfactin-like molecule produced by Bacillus sp.H2O-1 and its antagonistic effect against sulfate reducing bacteria [J].BMC Microbiol,2012,12: 252.doi: 10.1186/1471-2180-12-252.
[14] Das P,Mukherjee S,Sen R.Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans [J].J Appl Microbiol,2008,104(6): 1675-1684.
[15] Loiseau C,Schlusselhuber M,Bigot R,Bertaux J,Berjeaud JM,Verdon J.Surfactin from Bacillus subtilis displays an unexpected anti-legionella activity [J].Appl Microbiol Biotechnol,2015,99(12): 5083-5093.
[16] Liu X,Ren B,Gao H,Liu M,Dai H,Song F,Yu Z,Wang S,Hu J,Kokare CR,Zhang L.Optimization for the production of surfactin with a new synergistic antifungal activity [J].PLoS One,2012,7(5): e34430.
[17] Qi G,Zhu F,Du P,Yang X,Qiu D,Yu Z,Chen J,Zhao X.Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway [J].Peptides,2010,31(11): 1978-1986.
[18] Cotta SR,da Mota FF,Tupinambá G,Ishida K,Rozental S,E Silva DO,da Silva AJ,Bizzo HR,Alviano DS,Alviano CS,Seldin L.Antimicrobial activity of Paenibacillus kribbensis POC 115 against the dermatophyte Trichophyton rubrum [J].World J Microbiol Biotechnol,2012,28(3): 953-962.
[19] Vollenbroich D,Pauli G,Ozel M,Vater J.Antimycoplasma properties and application in cell culture of surfactin,a lipopeptide antibiotic from Bacillus subtilis [J].Appl Environ Microbiol,1997,63(1): 44-49.
[20] Hwang MH,Lim JH,Yun HI,Rhee MH,Cho JY,Hsu WH,Park SC.Surfactin C inhibits the lipopolysaccharide-induced transcription of interleukin-1beta and inducible nitric oxide synthase and nitric oxide production in murine RAW 264.7 cells [J].Biotechnol Lett,2005,27(20): 1605-1608.
[21] Selvam R,Maheswari P,Kavitha P,Ravichandran M,Sas B,Ramchand CN.Effect of Bacillus subtilis PB6,a natural probiotic on colon mucosal inflammation and plasma cytokines levels in inflammatory bowel disease [J].Indian J Biochem Biophys,2009,46(1): 79-85.
[22] Shaligram NS,Singhal RS.Surfactin—a review on biosynthesis,fermentation,purification and applications [J].Food Technol Biotechnol,2010,48(2): 119-134.
[23] Patel S,Ahmed S,Eswari JS.Therapeutic cyclic lipopeptides mining from microbes: latest strides and hurdles [J].World J Microbiol Biotechnol,2015,31(8): 1177-1193.
[24] Inès M,Dhouha G.Lipopeptide surfactants: Production,recovery and pore forming capacity [J].Peptides,2015,71: 100-112.
[25] Mnif I,Ghribi D.Review lipopeptides biosurfactants: Mean classes and new insights for industrial,biomedical,and environmental applications [J].Biopolymers,2015,104(3): 129-147.
[26] Pratap SS,Seema G,Neetu P,Nimisha S.Surfactin:a review on novel microbial surfactant [J/OL].Int J Bioassays,2013,2(5): 740-745.http://ijbio.com/index.php/ijb/article/view/390.
[27] Chen WC,Juang RS,Wei YH.Applications of a lipopeptide biosurfactant,surfactin,produced by microorganisms [J].Biochem Eng J,2015,103: 158-169.
[28] Goodman SA,Orr C,Warner PJ.Development of yeast inhibitory compounds for incorporation into silage inoculants [J].Cell Biol,1992,65: 479-485.
[29] Wei YH.Enhanced fengycin production with an indigenous isolate Bacillus subtilis F29-3 [J].J Biotechnol,2010,150: S345.doi:10.1016/j.jbiotec.2010.09.376.
[30] Taira T,Yanagisawa S,Nagano T,Tsuji T,Endo A,Imura T.pH-induced conformational change of natural cyclic lipopeptide surfactin and the effect on protease activity [J].Colloids Surf B Biointerfaces,2017,156: 382-387.
[31] Onaizi SA,Nasser MS,Al-Lagtah NM.Self-assembly of a surfactin nanolayer at solid-liquid and air-liquid interfaces [J].Eur Biophys J,2016,45(4): 331-339.
[32] Alvarez F,Castro M,Príncipe A,Borioli G,Fischer S,Mori G,Jofré E.The plant-associated Bacillus amyloliquefaciens strains MEP2 18 and ARP2 3 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease [J].J Appl Microbiol,2012,112(1): 159-174.
[33] Arrebola E,Jacobs R,Korsten L.Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens [J].J Appl Microbiol,2010,108(2): 386-395.
[34] Fan H,Ru J,Zhang Y,Wang Q,Li Y.Fengycin produced by Bacillus subtilis 9407 plays a major role in the biocontrol of apple ring rot disease [J].Microbiol Res,2017,199: 89-97.
[35] Mizumoto S,Shoda M.Medium optimization of antifungal lipopeptide,iturin A,production by Bacillus subtilis in solid-state fermentation by response surface methodology [J].Appl Microbiol Biotechnol,2007,76(1): 101-108.
[36] Rangarajan V,Dhanarajan G,Sen R.Bioprocess design for selective enhancement of fengycin production by a marine isolate Bacillus megaterium [J].Biochem Eng J,2015,99: 147-155.
[37] Yi G,Liu Q,Lin J,Wang W,Huang H,Li S.Repeated batch fermentation for surfactin production with immobilized Bacillus subtilis BS-37:two-stage pH control and foam fractionation [J].J Chem Technol Biotechnol,2017,92(3): 530-535.doi: 10.1002/jctb.5028.
[38] Willenbacher J,Zwick M,Mohr T,Schmid F,Syldatk C,Hausmann R.Evaluation of different Bacillus strains in respect of their ability to produce surfactin in a model fermentation process with integrated foam fractionation [J].Appl Microbiol Biotechnol,2014,98(23): 9623-9632.
[39] Romano A,Vitullo D,Di Pietro A,Lima G,Lanzotti V.Antifungal lipopeptides from Bacillus amyloliquefaciens strain BO7 [J].J Nat Prod,2011,74(2): 145-151.
[40] Huszcza E,Burczyk B.Surfactin isoforms from Bacillus coagulans [J].Z Naturforsch C,2006,61(9-10): 727-733.
[41] Kwon JW,Kim SD.Characterization of an antibiotic produced by Bacillus subtilis JW-1 that suppresses Ralstonia solanacearum [J].J Microbiol Biotechnol,2014,24(1): 13-18.
[42] Liu JF,Mbadinga SM,Yang SZ,Gu JD,Mu BZ.Chemical structure,property and potential applications of biosurfactants produced by Bacillus subtilis in petroleum recovery and spill mitigation [J].Int J Mol Sci,2015,16(3): 4814-4837.
[43] Huang X,Lu Z,Bie X,Lü F,Zhao H,Yang S.Optimization of inactivation of endospores of Bacillus cereus by antimicrobial lipopeptides from Bacillus subtilis fmbj strains using a response surface method [J].Appl Microbiol Biotechnol,2007,74(2): 454-461.
[44] Aleti G,Lehner S,Bacher M,Compant S,Nikolic B,Plesko M,Schuhmacher R,Sessitsch A,Brader G.Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus [J].Environ Microbiol,2016,18(8): 2634-2645.
[45] Shaligram S,Kumbhare SV,Dhotre DP,Muddeshwar MG,Kapley A,Joseph N,Purohit HP,Shouche YS,Pawar SP.Genomic and functional features of the biosurfactant producing Bacillus sp.AM13 [J].Funct Integr Genomics,2016,16(5): 557-566.
[46] Naruse N,Tenmyo O,Kobaru S,Kamei H,Miyaki T,Konishi M,Oki T.Pumilacidin,a complex of new antiviral antibiotics.Production,isolation,chemical properties,structure and biological activity [J].J Antibiot (Tokyo),1990,43(3): 267-280.
[47] Yang H,Li X,Li X,Yu H,Shen Z.Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin,fengycin,and surfactin by RP-HPLC [J].Anal Bioanal Chem,2015,407(9): 2529-2542.
[48] Kawagoe Y,Shiraishi S,Kondo H,Yamamoto S,Aoki Y,Suzuki S.Cyclic lipopeptide iturin A structure-dependently induces defense response in Arabidopsis plants by activating SA and JA signaling pathways [J].Biochem Biophys Res Commun,2015,460(4): 1015-1020.
[49] Maget-Dana R,Peypoux F.Iturins,a special class of pore-forming lipopeptides: biological and physicochemical properties [J].Toxicology,1994,87(1-3): 151-174.
[50] Nasir MN,Besson F.Conformational analyses of bacillomycin D,a natural antimicrobial lipopeptide,alone or in interaction with lipid monolayers at the air-water interface [J].J Colloid Interface Sci,2012,387(1): 187-193.
[51] Thasana N,Prapagdee B,Rangkadilok N,Sallabhan R,Aye SL,Ruchirawat S,Loprasert S.Bacillus subtilis SSE4 produces subtulene A,a new lipopeptide antibiotic possessing an unusual C15 unsaturated beta-amino acid [J].FEBS Lett,2010,584(14): 3209-3214.
[52] Ma Z,Wang N,Hu J,Wang S.Isolation and characterization of a new iturinic lipopeptide,mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A [J].J Antibiot (Tokyo),2012,65(6): 317-322.
[53] Son S,Ko SK,Jang M,Kim JW,Kim GS,Lee JK,Jeon ES,Futamura Y,Ryoo IJ,Lee JS,Oh H,Hong YS,Kim BY,Takahashi S,Osada H,Jang JH,Ahn JS.New cyclic lipopeptides of the iturin class produced by saltern-derived Bacillus sp.KCB14S006 [J].Mar Drugs,2016,14(4).doi: 10.3390/md14040072.
[54] Pathak KV,Keharia H,Gupta K,Thakur SS,Balaram P.Lipopeptides from the banyan endophyte,Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins [J].J Am Soc Mass Spectrom,2012,23(10): 1716-1728.
[55] Sang-Cheol L,Kim SH,Park IH,Chung SY,Chandra MS,Yong-Lark C.Isolation,purification,and characterization of novel fengycin S from Bacillus amyloliquefaciens LSC04 degrading-crude oil [J/OL].Biotechnol Bioproc Eng,2010,15(2): 246-253.https://link.springer.com/article/10.1007/s12257-009-0037-8.
[56] Benitez LB,Velho RV,Lisboa MP,Medina LF,Brandelli A.Isolation and characterization of antifungal peptides produced by Bacillus amyloliquefaciens LBM5006 [J].J Microbiol,2010,48(6): 791-797.
[57] Bie X,Lu Z,Lu F.Identification of fengycin homologues from Bacillus subtilis with ESI-MS/Cid [J].J Microbiol Methods,2009,79(3): 272-278.
[58] Villegas-Escobar V,Ceballos I,Mira JJ,Argel LE,Orduz Peralta S,Romero-Tabarez M.Fengycin C produced by Bacillus subtilis EA-CB0015 [J].J Nat Prod,2013,76(4): 503-509.
[59] Honma M,Tanaka K,Konno K,Tsuge K,Okuno T,Hashimoto M.Termination of the structural confusion between plipastatin A1 and fengycin IX [J].Bioorg Med Chem,2012,20(12): 3793-3798.
[60] Deleu M,Lorent J,Lins L,Brasseur R,Braun N,El Kirat K,Nylander T,Dufrêne YF,Mingeot-Leclercq MP.Effects of surfactin on membrane models displaying lipid phase separation [J].Biochim Biophys Acta,2013,1828(2): 801-815.
[61] Patel H,Huynh Q,Bärlehner D,Heerklotz H.Additive and synergistic membrane permeabilization by antimicrobial (lipo)peptides and detergents [J].Biophys J,2014,106(10): 2115-2125.
[62] Heerklotz H,Seelig J.Leakage and lysis of lipid membranes induced by the lipopeptide surfactin [J].Eur Biophys J,2007,36(4/5): 305-314.
[63] Zhang B,Dong C,Shang Q,Han Y,Li P.New insights into membrane-active action in plasma membrane of fungal hyphae by the lipopeptide antibiotic bacillomycin L [J].Biochim Biophys Acta,2013,1828(9): 2230-2237.
[64] Zhang B,Qin Y,Han Y,Dong C,Li P,Shang Q.Comparative proteomic analysis reveals intracellular targets for bacillomycin L to induce Rhizoctonia solani Kühn hyphal cell death [J].Biochim Biophys Acta,2016,1864(9): 1152-1159.
[65] Rajendran L,Ramjegathesh R,Shanthiyaa V,Raguchander T,Samiyappan GK.Biocontrol potential and mode of action of the strains EPC 5 and EPC 8 of endophytic bacterium,Bacillus subtilis [J/OL].Indian Phytopathol,2012,65(2): 122-127.http://epubs.icar.org.in/ejournal/index.php/IPPJ/article/view/18572/9363.
[66] Deleu M,Paquot M,Nylander T.Effect of fengycin,a lipopeptide produced by Bacillus subtilis,on model biomembranes [J].Biophys J,2008,94(7): 2667-2679.
[67] Deleu M,Paquot M,Nylander T.Fengycin interaction with lipid monolayers at the air-aqueous interface—implications for the effect of fengycin on biological membranes [J].J Colloid Interface Sci,2005,283(2): 358-365.
[68] Patel H,Tscheka C,Edwards K,Karlsson G,Heerklotz H.All-or-none membrane permeabilization by fengycin-type lipopeptides from Bacillus subtilis QST713 [J].Biochim Biophys Acta,2011,1808(8): 2000-2008.
[70] Iannazzo L,Laisné G,Fonvielle M,Braud E,Herbeuval JP,Arthur M,Etheve-Quelquejeu M.Synthesis of 3′-fluoro-tRNA analogues for exploring non-ribosomal peptide synthesis in bacteria [J].ChemBioChem,2015,16(3): 477-486.
[71] Tapi A,Chollet-Imbert M,Scherens B,Jacques P.New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction [J].Appl Microbiol Biotechnol,2010,85(5): 1521-1531.
[72] Dunlap CA,Bowman MJ,Schisler DA.Genomic analysis and secondary metabolite production in Bacillus amyloliquefaciens AS 43.3: A biocontrol antagonist of Fusarium head blight [J].Biol Control,2013,64(2): 166-175.doi: 10.1016/j.biocontrol.2012.11.002.
[73] Stevens BW,Lilien RH,Georgiev I,Donald BR,Anderson AC.Redesigning the PheA domain of gramicidin synthetase leads to a new understanding of the enzyme’s mechanism and selectivity [J].Biochemistry,2006,45(51): 15495-15504.
[74] Hur GH,Vickery CR,Burkart MD.Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology [J].Nat Prod Rep,2012,29(10): 1074-1098.
[75] Singh M,Chaudhary S,Sareen D.Non-ribosomal peptide synthetases: Identifying the cryptic gene clusters and decoding the natural product [J].J Biosci,2017,42(1): 175-187.
[76] Wei M,Wang S,Shang G.Biosynthetic pathways and engineering for bioactive natural products [J].Curr Org Chem,2010,14(14): 1433-1446.doi: 10.2174/ 138527210791616759.
[77] Schaffer ML,Otten LG.Substrate flexibility of the adenylation reaction in the Tyrocidine non-ribosomal peptide synthetase [J].J Mol Catal B Enzym,2009,59(1/3): 140-144.
[78] Das P,Mukherjee S,Sen R.Genetic regulations of the biosynthesis of microbial surfactants: an overview [J].Biotechnol Genet Eng Rev,2008,25: 165-185.
[79] Hwang YH,Kim MS,Song IB,Park BK,Lim JH,Park SC,Yun HI.Subacute (28 day) toxicity of surfactin C,a lipopeptide produced by Bacillus subtilis,in rats [J].J Health Sci,2009,55(3): 351-355.
[80] Weber T,Marahiel MA.Exploring the domain structure of modular nonribosomal peptide synthetases [J].Structure,2001,9(1): R3-R9.
[81] Savadogo A,Tapi A,Chollet M,Wathelet B,Traoré AS,Jacques P.Identification of surfactin producing strains in Soumbala and Bikalga fermented condiments using polymerase chain reaction and matrix assisted laser desorption/ionization-mass spectrometry methods [J].Int J Food Microbiol,2011,151(3): 299-306.
[82] Koglin A,Löhr F,Bernhard F,Rogov VV,Frueh DP,Strieter ER,Mofid MR,Güntert P,Wagner G,Walsh CT,Marahiel MA,Dötsch V.Structural basis for the selectivity of the external thioesterase of the surfactin synthetase [J].Nature,2008,454(7206): 907-911.
[83] Apao MMN,Teves FG,Madamba MRSB.Sequence analysis of putative swrW gene required for surfactant serrawettin W1 production from Serratia marcescens [J/OL].Afr J Biotechnol,2012,11(57): 12040-12044.http://www.academicjournals.org/journal/AJB/article-full-text-pdf/0854F0E34791.
[84] Satpute SK,Bhuyan SS,Pardesi KR,Mujumdar SS,Dhakephalkar PK,Shete AM,Chopade BA.Molecular genetics of biosurfactant synthesis in microorganisms [J].Adv Exp Med Biol,2010,672: 14-41.
[85] Hsieh FC,Li MC,Lin TC,Kao SS.Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR [J].Curr Microbiol,2004,49(3): 186-191.
[86] Wang D,Liu Y,Lin Z,Yang Z,Hao C.Isolation and identification of surfactin producing Bacillus subtilis strain and its effect of surfactin on crude oil [J].Acta Microbiol Sin,2008,48(3): 304-311.
[87] Zhao P,Quan C,Jin L,Wang L,Guo X,Fan S.Sequence characterization and computational analysis of the non-ribosomal peptide synthetases controlling biosynthesis of lipopeptides,fengycins and bacillomycin D,from Bacillus amyloliquefaciens Q-426 [J].Biotechnol Lett,2013,35(12): 2155-2163.
[88] Moyne AL,Cleveland TE,Tuzun S.Molecular characterization and analysis of the operon encoding the antifungal lipopeptide bacillomycin D [J].FEMS Microbiol Lett,2004,234(1): 43-49.
[89] Wu CY,Chen CL,Lee YH,Cheng YC,Wu YC,Shu HY,Götz F,Liu ST.Nonribosomal synthesis of fengycin on an enzyme complex formed by fengycin synthetases [J].J Biol Chem,2007,282(8): 5608-5616.
[90] Yaseen Y,Gancel F,Drider D,Béchet M,Jacques P.Influence of promoters on the production of fengycin in Bacillus spp [J].Res Microbiol,2016,167(4): 272-281.
[91] Samel SA,Wagner B,Marahiel MA,Essen LO.The thioesterase domain of the fengycin biosynthesis cluster: a structural base for the macrocyclization of a non-ribosomal lipopeptide [J].J Mol Biol,2006,359(4): 876-889.
