阿米巴巨大病毒及其潜在致病性研究进展
2018-02-28夏宇程钟江
夏宇程,钟江
复旦大学生命科学学院微生物学与微生物工程系,上海 200438
阿米巴是自然界中广泛分布的单细胞真核生物,通过伪足来捕捉和吞噬颗粒状物质作为食物,被称为自然界的“巨噬细胞”。许多人类病原体,如引起军团病的嗜肺军团菌,可在阿米巴细胞内繁殖。因此,阿米巴也有“人类病原体训练场”的称号[1]。
1 阿米巴巨大病毒的发现
1996年,有研究者采用与阿米巴共培养技术,从肺炎暴发地水样中分离了多种“军团菌样阿米巴内寄生菌”[2],其中一种革兰染色阳性球菌——“Bradford球菌”引起特别关注[3],因为研究者们始终无法从中扩增出16S rDNA。直到2003年,La Scola 等通过电子显微镜观察、全基因组测序及阿米巴细胞裂解周期研究发现,“Bradford球菌”并非细菌,而是一种病毒,将其命名为Mimivirus[4],意为“模拟细菌的病毒”, 中文译为拟菌病毒。这种病毒呈二十面体状,直径达400 nm,是当时报道的最大病毒,染色后可直接在光学显微镜下观察到。其基因组同样巨大,为长达1.18 Mb 的双链DNA,预测编码1 262个开放读码框。
此后,人们采用阿米巴共培养方法,从世界各地的淡水、海水、土壤、生物(昆虫、环节动物、人体等)样本中分离到马赛病毒(Marseillevirus)、Megavirus、潘多拉病毒(Pandoravirus)等多种巨大病毒[5-6]。这些病毒均具有巨大的病毒颗粒(0.3~1.5 μm)和庞大的基因组(最大的潘多拉病毒基因组达2.5 Mb,最小的马赛病毒也有0.37 Mb)。而且,一些环境微生物宏基因组研究结果证明,这些巨大病毒与其宿主阿米巴一样分布广泛[7]。
分析这些巨大病毒的基因组,发现它们均编码了其他病毒中罕见的与蛋白质合成相关的基因,包括tRNA、氨酰tRNA合成酶、转译因子等[8]。最新发现的一种巨大病毒Klosneuvirus,甚至带有全部20种氨基酸的特异性氨酰tRNA合成酶[9]。这激发了演化生物学家的广泛兴趣,也在学术界引起了一场关于病毒起源和本质的大讨论[8](表1)。
由于巨大病毒最早发现于肺炎流行地,许多研究者致力于研究此类病毒对人体的潜在感染和致病性。本文主要介绍这方面的研究进展。
表1阿米巴巨大病毒代表株
Tab.1Typicalstrainsofgiantvirusesinamoebae

YearsofsamplecollectionVirusSampletypeCharacteristicsofvirusparticleCharacteristicsofgenomeSize(Mb)CG%ORFSensitivehostsRef.2003MimivirusWaterfroma coolingtowerIcosahedral⁃like,~400nmindiameter1.1828%1262A.polyphaga[4]2009MarseillevirusWaterfrom acoolingtowerIcosahedral⁃like,~250nmindiameter0.3745%457A.polyphaga[10]2011MegavirusCoastalwaterIcosahedral⁃like,~680nmindiameter1.2625%1120A.castellanii[11]2013PandoravirusCoastalsedimentsOvoid,1μminlength,0.5μmindiameter1.91⁃2.4725%1120A.castellanii[12]2014PithovirusPermafrostsoilOvoid,1.5μminlength,0.5μmindiameter0.636%467A.castellanii[13]2015FaustovirusSewageIcosahedral⁃like,~500nmindiameter0.4636%451Vermamoeba vermiformis[14]2017KlosneuvirusSludgeIcosahedral⁃like,~300nmindiameter1.5728%355Notisolated[9]
2 人体感染阿米巴巨大病毒的血清学和分子诊断证据
表2汇总了在人体样本中用血清学和分子生物学方法进行阿米巴巨大病毒检测的结果。大多数研究在相当比例的样本中检测到了阿米巴巨大病毒特异性抗体或基因序列,但也有研究未发现该类病毒存在的证据。
2.1 拟菌病毒
La Scola等检测了376例医源性肺炎患者的血液样本,发现9.7%的样本中有抗拟菌病毒抗体,相比之下,511例健康人的血液样本中仅2.3%呈阳性[15],且抗拟菌病毒抗体水平高于抗“阿米巴相关细菌”的抗体[35];26例重症监护室(intensive care unit,ICU)源性肺炎患者血液样本中的抗体阳性率达 19.2%,从1例患者的支气管肺泡灌洗液中还检测到病毒DNA片段,而50例对照者的样本全为阴性。Berger等采用荧光定量聚合酶链反应(polymerase chain reaction,PCR)检测了157例医院/社区获得性肺炎患者的210份支气管肺泡灌洗液样品,发现19%含有与阿米巴相关的微生物,拟菌病毒阳性率为7.1%,居肺炎病因的第4位[16]。Raoult等报道了1例在法国马赛研究拟菌病毒的实验室工作人员突然患上不明原因的病毒性肺炎,其血液中抗多种拟菌病毒抗体呈阳性,且抗体水平随病情好转而降低[17]。Bousbia等对97例ICU患者的173份血液样品进行血清学研究,结果显示抗阿米巴相关微生物的IgG、IgM随ICU停留时间延长而增加,其中抗拟菌病毒抗体占10%[18];Vincent等也报道了相同现象[19]。
表2阿米巴巨大病毒的血清学及分子生物学检测结果
Tab.2SummaryoftheserologicalandmolecularbiologicaldataonMimivirusandMarseillevirus
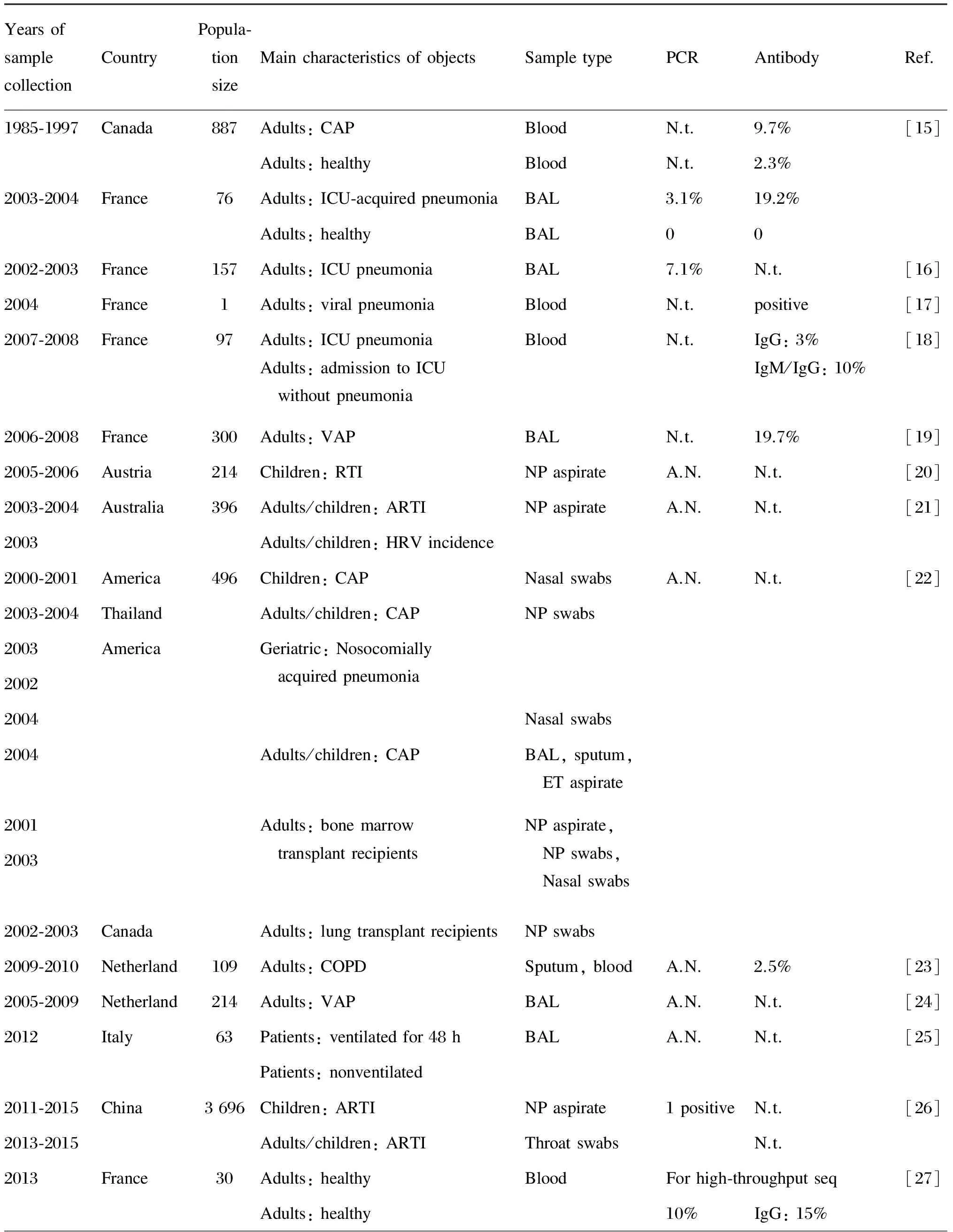
YearsofsamplecollectionCountryPopula⁃tionsizeMaincharacteristicsofobjectsSampletypePCRAntibodyRef.1985⁃19972003⁃2004CanadaFrance88776Adults:CAPAdults:healthyAdults:ICU⁃acquiredpneumoniaAdults:healthyBloodBloodBALBALN.t.N.t.3.1%09.7%2.3%19.2%0[15]2002⁃2003France157Adults:ICUpneumoniaBAL7.1%N.t.[16]2004France1Adults:viralpneumoniaBloodN.t.positive[17]2007⁃2008France97Adults:ICUpneumoniaAdults:admissiontoICUwithoutpneumoniaBloodN.t.IgG:3%IgM/IgG:10%[18]2006⁃2008France300Adults:VAPBALN.t.19.7%[19]2005⁃2006Austria214Children:RTINPaspirateA.N.N.t.[20]2003⁃20042003Australia396Adults/children:ARTIAdults/children:HRVincidenceNPaspirateA.N.N.t.[21]2000⁃20012003⁃20042003200220042004200120032002⁃2003AmericaThailandAmericaCanada496Children:CAPAdults/children:CAPGeriatric:NosocomiallyacquiredpneumoniaAdults/children:CAPAdults:bonemarrowtransplantrecipientsAdults:lungtransplantrecipientsNasalswabsNPswabsNasalswabsBAL,sputum,ETaspirateNPaspirate,NPswabs,NasalswabsNPswabsA.N.N.t.[22]2009⁃2010Netherland109Adults:COPDSputum,bloodA.N.2.5%[23]2005⁃2009Netherland214Adults:VAPBALA.N.N.t.[24]2012Italy63Patients:ventilatedfor48hPatients:nonventilatedBALA.N.N.t.[25]2011⁃20152013⁃2015China3696Children:ARTIAdults/children:ARTINPaspirateThroatswabs1positiveN.t.N.t.[26]2013France30Adults:healthyAdults:healthyBloodForhigh⁃throughputseq10%IgG:15%[27]

(续表2)
N.t.: not told in refence; A.N.: all negative; CAP: community-acquired pneumonia; ICU: intensive care unit; BAL: bronchoalveolar lavage; VAP: ventilator-associated pneumonia; RTI: respiratory tract infection; NP: Nasopharyngeal; ARTI: acute respiratory tract infection; HRV: human rhinovirus; ET: endotracheal; COPD: chronic obstructive pulmonary disease; CSF: cerebrospinal fluid.
医院环境中也检测到拟菌病毒的存在。dos Santos Silva等对巴西一家大医院中多个场所收集的242份环境擦拭子样品进行定量PCR检测[36],发现拟菌病毒阳性率为12.4%,其中取自呼吸道疾病隔离设施样品的阳性率达36.4%,且通过阿米巴培养从20%样品中分离到了病毒。以上这些研究结果提示,拟菌病毒有可能与人类肺炎等呼吸道疾病有关。
也有研究结果不支持拟菌病毒与呼吸系统疾病的关系。Larcher 等[20]、Arden等[21]及Zhang等[26]用多种病毒特异性引物对呼吸道感染患者的鼻咽吸出物/拭子进行PCR检测,检出副流感病毒、腺病毒等常见呼吸道病毒,但未检测到拟菌病毒。Dare等[22]、Vanspauwen等[23-24]及Costa等[25]对肺炎患者的支气管肺泡灌洗液进行PCR检测,也未检出拟菌病毒的核酸。
2.2 马赛病毒
马赛病毒是2009年报道的采用类似分离拟菌病毒的方式从冷却水塔内分离获得的另一种巨大病毒[11]。Popgeorgiev等对10份健康献血者的血液样品采用高通量测序进行病毒组(virome)分析,发现2.5%读出序列(reads)与马赛病毒的基因组相匹配。他们对另20份血样进行血清学和PCR检测,发现其中3份IgG阳性,2份PCR阳性,并从1份IgG和PCR均阳性的血液样本淋巴细胞中分离获得一株新病毒,命名为GBM(giant blood Marseillevirus)[27]。该作者后来进一步扩大样本容量,发现174份健康献血者样品中IgG阳性率为12.6%,PCR阳性率为4%;而22例地中海贫血患者中IgG阳性率为22.7%,PCR阳性率为9.1%。作者推测频繁输血治疗会提高感染GBM的概率[28]。他们还在1例病因不明单侧腋窝淋巴结肿大的幼儿淋巴结切片中,采用荧光原位杂交(fluorescenceinsituhybridization,FISH)和免疫组化方法检测到巨噬细胞胞质中存在马赛病毒,其血液样本中也能检测到相应抗体和病毒核酸,但13个月后检测结果转为阴性。因为该患儿曾患过卡介苗感染(BCGitis),推测该病毒对免疫紊乱者有致病性[29]。Aherfi等在1 例发热性肠炎患者血液和咽部均检测到马赛病毒DNA,患者服用环丙沙星72 h后症状消退,但4个月后出现眩晕并伴随体重减轻,血液和咽喉部仍可检测到该病毒DNA,推测该病毒的存在对神经系统有慢性影响[30]。他们还在1例霍奇金淋巴瘤患者淋巴结内检测到马赛病毒[31]。Mueller等对517例健康瑞士人的血液进行检测,发现9例为抗洛桑病毒(Lausannevirus;马赛病毒科成员)抗体阳性[32]。
同样,也有研究未在血液样本中检测到马赛病毒[33-34]。针对假阳性的质疑,Sauvage等认为部分实验在其他医院完成,不太可能存在样品污染;并指出,即使是阳性的健康人血液样品,PCR信号仍非常微弱,只有引发某些疾病(如前述儿童淋巴结病[29])时才会出现PCR强阳性,且患者病愈后检测不到病毒核酸,但抗体检测仍为阳性[33]。
3 宏基因组测序检测阿米巴巨大病毒
随着高通量测序技术的成熟和费用逐渐降低,许多研究应用这项技术来寻找巨大病毒[7]。马赛病毒病毒株GBM[27]及马赛病毒科的另一成员塞内加尔病毒(Senegalvirus)[37]就是研究者对人体血液及粪便样本进行高通量测序时发现并进一步分离获得的。Verneau等设计了MG-Digger软件,专门从宏基因组数据中搜索巨大病毒的序列信息[38],并分别从Loh等[39]报道的人血液样本数据库和Law等[40]报道的慢性肝炎患者血液样本数据库中检测到185和1 706条巨大病毒读出序列,分别占总读出序列的0.6%和0.004%。Rampelli等设计了搜索人体宏基因组内病毒信息的ViromeScan软件,分析了人体微生物组计划(Human Microbiome Project,HMP)中采自不同部位(粪便-肠道、阴道、口腔黏膜、耳后折痕区)的宏基因组库,均发现了巨大病毒的序列[41],且肠道内拟菌病毒和痘病毒科占多数,病毒成分明显不同于其他部位。其他提及巨大病毒的人体宏基因组研究如表3所示。从中可看出,巨大病毒的信息占比较低,可能是因为:病毒组研究的样品制备时往往采用0.22~0.45 μm滤膜过滤,导致巨大病毒损失[7,45];巨大病毒基因组内有很多未注释的开放读码框,加上病毒基因的多样性,妨碍了对所建库中病毒信息的注释和挖掘[7]。
4 从人体样本中分离阿米巴巨大病毒
许多研究将样本与阿米巴进行共培养,从而获得巨大病毒的分离株,详见表4。常用阿米巴为卡氏棘阿米巴(Acanthamoebacastellanii)、多噬棘阿米巴(Acanthamoebapolyphaga)和蚓状哈曼阿米巴(Vermamoebavermiformis)[52]。La Scola等从1例角膜炎患者的隐形眼镜护理液内分离出1株拟菌病毒Lentille[46-47]。Saadi等收集了196例社区获得性肺炎患者的不同呼吸道样本,从1例不明原因肺炎妇女的支气管抽吸液中分离到1株拟菌病毒LBA111[48];该作者还收集并检测了1 605份粪便样品,从其中1例不明原因肺炎患儿的粪便中分离到1株拟菌病毒Shan virus[49]。Scheid等发现,从1例角膜炎患者的隐形眼镜护理液内分离到的棘阿米巴细胞含有细菌样颗粒,会裂解棘阿米巴的滋养体[50],PCR和全基因组测序证实该胞内物是1株潘多拉病毒[51,53]。
5 阿米巴巨大病毒感染细胞及实验动物
研究表明,巨大病毒通过阿米巴细胞的胞吞作用侵入细胞[54-55]。哺乳动物中,胞吞作用一般由巨噬细胞、单核细胞、中性粒细胞等特定细胞完成[56]。Ghigo等发现,拟菌病毒可通过胞吞作用进入人髓系细胞(循环血单核细胞、单核细胞源性巨噬细胞、单核细胞系THP-1)和鼠髓系细胞(骨髓源性巨噬细胞、巨噬细胞系RAW264.7、J774A.1),但无法进入非吞噬细胞(HeLa细胞等)。拟菌病毒被巨噬细胞胞吞后,病毒DNA含量随时间延长而逐渐增加,并在30 h后引起明显的细胞病变,表明巨大病毒不仅可进入人巨噬细胞,还可在其中复制并引起细胞病变[55]。Silva等也证实,拟菌病毒能在人外周血单核细胞(peripheral blood mononuclear cell,PBMC)中复制,被感染的细胞还可产生干扰素,但细胞受干扰素激活的基因转录被病毒抑制;如果在病毒感染前预先用干扰素β1而非干扰素α2处理PBMC,拟菌病毒就无法复制。这表明干扰素系统能抑制病毒复制,而拟菌病毒编码了相应的免疫逃避分子以帮助其在宿主细胞内复制[57]。
表3宏基因组测序分析检测人体样本中阿米巴巨大病毒
Tab.3Identificationofsequencescloselyrelatedtogiantvirusesinamoebaeinhumansamplesbymetagenomicanalysis

YearsofsamplecollectionCountryPopulationsizeMaincharacteristicsofobjectsSampletypeMeanRawreadsNumberofhits(%)Ref.2013France30Adults:healthyBloodRoche45420238333(2.5)[28]2012Senegal3Adults:healthyFecesRoche45418285353(0.11)[37]2002⁃2005Middle East399Adults:AFI/ healthyBloodRoche45429117185(0.6)[38,39]2013Canada11Adults:liver disease/healthyBloodIlluminaGAII427068831706(0.004)[38,40]1978⁃2005Australia/ America12Children: diarrheaFeces“Micro⁃mass sequencing”46085(0.001)[42]2011Sweden90Adults:twins, CFSBloodRoche4543051912(0.06)[43]2004⁃2005Sweden210Adults:RTINP aspirateRoche4547037902(0.0003)[44]
AFI: acute febrile illness; CFS: chronic fatigue syndrome; RTI: respiratory tract infection; NP: nasopharyngeal.
表4人体样本中分离到的阿米巴巨大病毒株
Tab.4Typicalstrainsofgiantvirusesinamoebaeinhumansamples
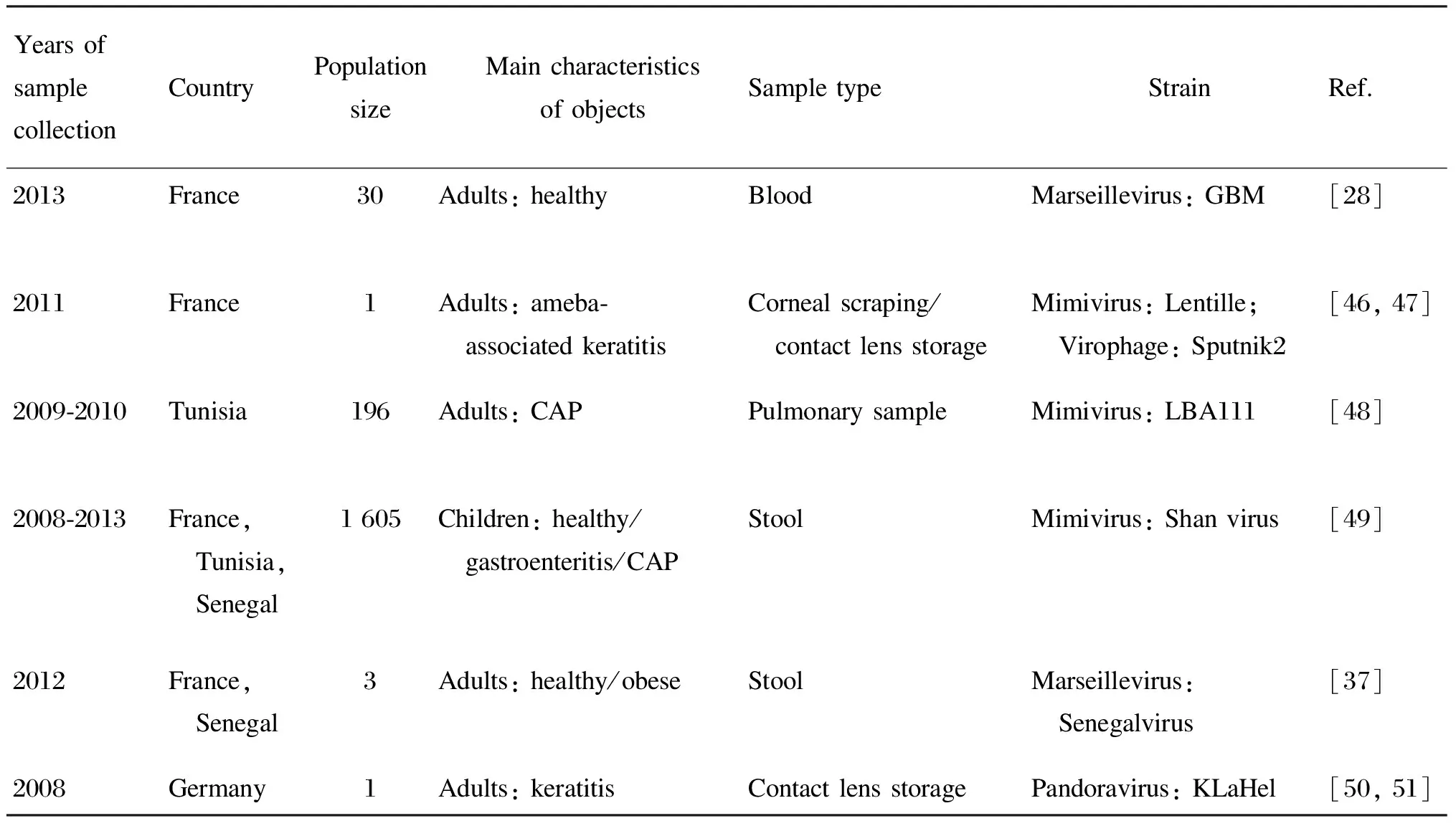
YearsofsamplecollectionCountryPopulationsizeMaincharacteristicsofobjectsSampletypeStrainRef.2013France30Adults:healthyBloodMarseillevirus:GBM[28]2011France1Adults:ameba⁃ associatedkeratitisCornealscraping/ contactlensstorageMimivirus:Lentille; Virophage:Sputnik2[46,47]2009⁃2010Tunisia196Adults:CAPPulmonarysampleMimivirus:LBA111[48]2008⁃2013France, Tunisia, Senegal1605Children:healthy/ gastroenteritis/CAPStoolMimivirus:Shanvirus[49]2012France, Senegal3Adults:healthy/obeseStoolMarseillevirus: Senegalvirus[37]2008Germany1Adults:keratitisContactlensstoragePandoravirus:KLaHel[50,51]
CAP: community-acquired pneumonia; Pulmonary sample: including bronchial aspirations,bronchoalveolar lavages,lung biopsies,pleural samples.
目前关于巨大病毒及其组分对动物的致病实验有两篇报道。Khan等通过心内注射将拟菌病毒接种至小鼠,发现75%小鼠出现急性肺炎。他们从发病小鼠的肺部和脾脏分离到拟菌病毒,表明拟菌病毒很可能是肺炎的病原体[58]。Shah等研究了拟菌病毒胶原蛋白L71(一种病毒表面蛋白)在类风湿关节炎中的潜在作用,用L71免疫胶原性关节炎模型小鼠,诱导产生的抗体可与小鼠Ⅱ型胶原蛋白反应并引起T细胞反应,导致关节炎症。他们还对100例健康人和100例类风湿关节炎患者进行酶联免疫吸附试验(enzyme-linked immunosorbent assay,ELISA)和蛋白免疫检测,发现分别有30份和36份血液样品中病毒核心蛋白L425抗体阳性,同时分别有6份和22份样品中L71抗体阳性,据此认为L71是引起类风湿关节炎的潜在致病因子[59]。
6 结语
越来越多的研究表明拟菌病毒与肺炎等炎症有关,但也有研究不支持这一观点。阴性结果的出现不能排除由技术限制所致:这些研究几乎只进行了PCR,没有开展血清学检测。目前尚缺乏合适的巨大病毒抗原,导致大规模血清学筛查难以进行。免疫沉淀与高通量测序相结合的新技术——VirScan[60]也许可丰富这方面的研究。另一方面,除拟菌病毒和马赛病毒外,其他阿米巴巨大病毒甚至非阿米巴巨大病毒也有可能对人体造成一定影响[61]。例如,用蚓状哈曼阿米巴分离获得的Faustovirus与猪瘟病毒的进化地位接近[14];有研究者在人口咽拭子中检测到一种分离自绿藻的巨大病毒Acanthocystisturfaceachlorella virus的DNA,且该病毒能在小鼠模型中引起认知功能紊乱[62]。此外,目前我国还少见此类病毒的研究。考虑到不少疾病,如20%~30%的呼吸系统疾病,还未找到致病原因[63],因此探讨巨大病毒等在常规检测中易被忽视的微生物的潜在致病性有重要现实意义。
[1] Moliner C,Fournier PE,Raoult D.Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution [J].FEMS Microbiol Rev,2010,34(3): 281-294.
[2] Birtles RJ,Rowbotham TJ,Raoult D,Harrison TG.Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison [J].Microbiology,1996,142(Pt 12): 3525-3530.
[3] La Scola B.Looking at protists as a source of pathogenic viruses [J].Microb Pathog,2014,77: 131-135.doi: 10.1016/j.micpath.2014.09.005.
[4] La Scola B,Audic S,Robert C,Jungang L,de Lamballerie X,Drancourt M,Birtles R,Claverie JM,Raoult D.A giant virus in amoebae [J].Science,2003,299(5615): 2033.
[5] Abergel C,Legendre M,Claverie JM.The rapidly expanding universe of giant viruses: Mimivirus,Pandoravirus,Pithovirus and Mollivirus [J].FEMS Microbiol Rev,2015,39(6): 779-796.
[6] Pagnier I,Reteno DG,Saadi H,Boughalmi M,Gaia M,Slimani M,Ngounga T,Bekliz M,Colson P,Raoult D,La Scola B.A decade of improvements in Mimiviridae and Marseilleviridae isolation from amoeba [J].Intervirology,2013,56(6): 354-363.
[7] Halary S,Temmam S,Raoult D,Desnues C.Viral metagenomics: are we missing the giants? [J].Curr Opin Microbiol,2016,31: 34-43.doi: 10.1016/j.mib.2016.01.005.
[8] Abrahão JS,Araújo R,Colson P,La Scola B.The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses [J].PLoS Genet,2017,13(2): e1006532.
[9] Schulz F,Yutin N,Ivanova NN,Ortega DR,Lee TK,Vierheilig J,Daims H,Horn M,Wagner M,Jensen GJ,Kyrpides NC,Koonin EV,Woyke T.Giant viruses with an expanded complement of translation system components [J].Science,2017,356(6333): 82-85.
[10] Boyer M,Yutin N,Pagnier I,Barrassi L,Fournous G,Espinosa L,Robert C,Azza S,Sun S,Rossmann MG,Suzan-Monti M,La Scola B,Koonin EV,Raoult D.Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms [J].Proc Natl Acad Sci USA,2009,106(51): 21848-21853.
[11] Arslan D,Legendre M,Seltzer V,Abergel C,Claverie JM.Distant Mimivirus relative with a larger genome highlights the fundamental features of megaviridae [J].Proc Natl Acad Sci USA,2011,108(42): 17486-17491.
[12] Philippe N,Legendre M,Doutre G,Couté Y,Poirot O,Lescot M,Arslan D,Seltzer V,Bertaux L,Bruley C,Garin J,Claverie JM,Abergel C.Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes [J].Science,2013,341(6143): 281-286.
[13] Legendre M,Bartoli J,Shmakova L,Jeudy S,Labadie K,Adrait A,Lescot M,Poirot O,Bertaux L,Bruley C,Couté Y,Rivkina E,Abergel C,Claverie JM.Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a Pandoravirus morphology [J].Proc Natl Acad Sci USA,2014,111(11): 4274-4279.
[14] Reteno DG,Benamar S,Khalil JB,Andreani J,Armstrong N,Klose T,Rossmann M,Colson P,Raoult D,La Scola B.Faustovirus,an asfarvirus-related new lineage of giant viruses infecting amoebae [J].J Virol,2015,89(13): 6585-6594.
[15] La Scola B,Marrie TJ,Auffray JP,Raoult D.Mimivirus in pneumonia patients [J].Emerg Infect Dis,2005,11(3): 449-452.
[16] Berger P,Papazian L,Drancourt M,La Scola B,Auffray JP,Raoult D.Ameba-associated microorganisms and diagnosis of nosocomial pneumonia [J].Emerg Infect Dis,2006,12(2): 248-255.
[17] Raoult D,Renesto P,Brouqui P.Laboratory infection of a technician by mimivirus [J].Ann Intern Med,2006,144(9): 702-703.
[18] Bousbia S,Papazian L,Saux P,Forel JM,Auffray JP,Martin C,Raoult D,La Scola B.Serologic prevalence of amoeba-associated microorganisms in intensive care unit pneumonia patients [J].PLoS One,2013,8(3): e58111.
[19] Vincent A,La Scola B,Forel JM,Pauly V,Raoult D,Papazian L.Clinical significance of a positive serology for mimivirus in patients presenting a suspicion of ventilator-associated pneumonia [J].Crit Care Med,2009,37(1): 111-118.
[20] Larcher C,Jeller V,Fischer H,Huemer HP.Prevalence of respiratory viruses,including newly identified viruses,in hospitalised children in Austria [J].Eur J Clin Microbiol Infect Dis,2006,25(11): 681-686.
[21] Arden KE,Mcerlean P,Nissen MD,Sloots TP,Mackay IM.Frequent detection of human rhinoviruses,paramyxoviruses,coronaviruses,and bocavirus during acute respiratory tract infections [J].J Med Virol,2006,78(9): 1232-1240.
[22] Dare RK,Chittaganpitch M,Erdman DD.Screening pneumonia patients for mimivirus [J].Emerg Infect Dis,2008,14(3): 465-467.
[23] Vanspauwen MJ,Franssen FM,Raoult D,Wouters EF,Bruggeman CA,Linssen CF.Infections with mimivirus in patients with chronic obstructive pulmonary disease [J].Respir Med,2012,106(12): 1690-1694.
[24] Vanspauwen MJ,Schnabel RM,Bruggeman CA,Drent M,van Mook WN,Bergmans DC,Linssen CF.Mimivirus is not a frequent cause of ventilator-associated pneumonia in critically ill patients [J].J Med Virol,2013,85(10): 1836-1841.
[25] Costa C,Bergallo M,Astegiano S,Terlizzi ME,Sidoti F,Solidoro P,Cavallo R.Detection of mimivirus in bronchoalveolar lavage of ventilated and nonventilated patients [J].Intervirology,2012,55(4): 303-305.
[26] Zhang XA,Zhu T,Zhang PH,Li H,Li Y,Liu EM,Liu W,Cao WC.Lack of mimivirus detection in patients with respiratory disease,China [J].Emerg Infect Dis,2016,22(11): 2011-2012.
[27] Popgeorgiev N,Boyer M,Fancello L,Monteil S,Robert C,Rivet R,Nappez C,Azza S,Chiaroni J,Raoult D,Desnues C.Marseillevirus-like virus recovered from blood donated by asymptomatic humans [J].J Infect Dis,2013,208(7): 1042-1050.
[28] Popgeorgiev N,Colson P,Thuret I,Chiarioni J,Gallian P,Raoult D,Desnues C.Marseillevirus prevalence in multitransfused patients suggests blood transmission [J].J Clin Virol,2013,58(4): 722-725.
[29] Popgeorgiev N,Michel G,Lepidi H,Raoult D,Desnues C.Marseillevirus adenitis in an 11-month-old child [J].J Clin Microbiol,2013,51(12): 4102-4105.
[30] Aherfi S,Colson P,Raoult D.Marseillevirus in the pharynx of a patient with neurologic disorders [J].Emerg Infect Dis,2016,22(11): 2008-2010.
[31] Aherfi S,Colson P,Audoly G,Nappez C,Xerri L,Valensi A,Million M,Lepidi H,Costello R,Raoult D.Marseillevirus in lymphoma: a giant in the lymph node [J].Lancet Infect Dis,2016,16(10): e225-e234.
[32] Mueller L,Baud D,Bertelli C,Greub G.Lausannevirus seroprevalence among asymptomatic young adults [J].Intervirology,2013,56(6): 430-433.
[33] Sauvage V,Livartowski A,Boizeau L,Servant-Delmas A,Lionnet F,Lefrère JJ,Laperche S.No evidence of Marseillevirus-like virus presence in blood donors and recipients of multiple blood transfusions [J].J Infect Dis,2014,210(12): 2017-2018.
[34] Phan TG,Desnues C,Switzer WM,Schneider BS,Deng XT,Delwart E.Absence of giant blood Marseille-like virus DNA detection by polymerase chain reaction in plasma from healthy US blood donors and serum from multiply transfused patients from Cameroon [J].Transfusion,2015,55(6): 1256-1262.
[35] Marrie TJ,Raoult D,La Scola B,Birtles RJ,de Carolis E; Canadian Community-Acquired Pneumonia Study Group.Legionella-like and other amoebal pathogens as agents of community-acquired pneumonia [J].Emerg Infect Dis,2001,7(6): 1026-1029.
[36] dos Santos Silva LK,Arantes TS,Andrade KR,Lima Rodrigues RA,Miranda Boratto PV,de Freitas Almeida GM,Kroon EG,La Scola B,Clemente WT,Abrahão JS.High positivity of mimivirus in inanimate surfaces of a hospital respiratory-isolation facility,Brazil [J].J Clin Virol,2015,66:62-65.doi: 10.1016/j.jcv.2015.03.008.
[37] Lagier JC,Armougom F,Million M,Hugon P,Pagnier I,Robert C,Bittar F,Fournous G,Gimenez G,Maraninchi M,Trape JF,Koonin EV,La Scola B,Raoult D.Microbial culturomics: paradigm shift in the human gut microbiome study [J].Clin Microbiol Infect,2012,18(12): 1185-1193.
[38] Verneau J,Levasseur A,Raoult D,La Scola B,Colson P.MG-Digger: an automated pipeline to search for giant virus-related sequences in metagenomes [J].Front Microbiol,2016.doi: 10.3389/fmicb.2016.00428.
[39] Loh J,Zhao G,Presti RM,Holtz LR,Finkbeiner SR,Droit L,Villasana Z,Todd C,Pipas JM,Calgua B,Girones R,Wang D,Virgin HW.Detection of novel sequences related to African Swine Fever virus in human serum and sewage [J].J Virol,2009,83(24): 13019-13025.
[40] Law J,Jovel J,Patterson J,Ford G,O’Keefe S,Wang W,Meng B,Song D,Zhang Y,Tian Z,Wasilenko ST,Rahbari M,Mitchell T,Jordan T,Carpenter E,Mason AL,Wong GK.Identification of hepatotropic viruses from plasma using deep sequencing: a next generation diagnostic tool [J].PLoS One,2013,8(4): e60595.
[41] Rampelli S,Soverini M,Turroni S,Quercia S,Biagi E,Brigidi P,Candela M.ViromeScan: a new tool for metagenomic viral community profiling [J].BMC Genomics,2016.doi: 10.1186/s12864-016-2446-3.
[42] Finkbeiner SR,Allred AF,Tarr PI,Klein EJ,Kirkwood CD,Wang D.Metagenomic analysis of human diarrhea: viral detection and discovery [J].PLoS Pathog,2008,4(2): e1000011.
[43] Sullivan PF,Allander T,Lysholm F,Goh S,Persson B,Jacks A,Evengård B,Pedersen NL,Andersson B.An unbiased metagenomic search for infectious agents using monozygotic twins discordant for chronic fatigue [J].BMC Microbiol,2011.doi: 10.1186/1471-2180-11-2.
[44] Lysholm F,Wetterbom A,Lindau C,Darban H,Bjerkner A,Fahlander K,Lindberg AM,Persson B,Allander T,Andersson B.Characterization of the viral microbiome in patients with severe lower respiratory tract infections,using metagenomic sequencing [J].PLoS One,2012,7(2): e30875.
[45] Conceição-Neto N,Zeller M,Lefrere H,de Bruyn P,Beller L,Deboutte W,Yinda CK,Lavigne R,Maes P,van Ranst M,Heylen E,Matthijnssens J.Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis [J].Sci Rep,2015.doi: 10.1038/srep16532.
[46] La Scola B,Campocasso A,N’Dong R,Fournous G,Barrassi L,Flaudrops C,Raoult D.Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry [J].Intervirology,2010,53(5): 344-353.
[47] Cohen G,Hoffart L,La Scola B,Raoult D,Drancourt M.Ameba-associated keratitis,France [J].Emerg Infect Dis,2011,17(7): 1306-1308.
[48] Saadi H,Pagnier I,Colson P,Cherif JK,Beji M,Boughalmi M,Azza S,Armstrong N,Robert C,Fournous G,La Scola B,Raoult D.First isolation of Mimivirus in a patient with pneumonia [J].Clin Infect Dis,2013,57(4): e127-e134.
[49] Saadi H,Reteno DG,Colson P,Aherfi S,Minodier P,Pagnier I,Raoult D,La Scola B.Shan virus: a new mimivirus isolated from the stool of a Tunisian patient with pneumonia [J].Intervirology,2013,56(6): 424-429.
[50] Scheid P,Zöller L,Pressmar S,Richard G,Michel R.An extraordinary endocytobiont in Acanthamoeba sp.isolated from a patient with keratitis [J].Parasitol Res,2008,102(5): 945-950.
[51] Scheid P,Balczun C,Schaub GA.Some secrets are revealed: parasitic keratitis amoebae as vectors of the scarcely described pandoraviruses to humans [J].Parasitol Res,2014,113(10): 3759-3764.
[52] Colson P,Aherfi S,La Scola B,Raoult D.The role of giant viruses of amoebas in humans [J].Curr Opin Microbiol,2016,31:199-208.doi: 10.1016/j.mib.2016.04.012.
[53] Antwerpen MH,Georgi E,Zoeller L,Woelfel R,Stoecker K,Scheid P.Whole-genome sequencing of a pandoravirus isolated from keratitis-inducing Acanthamoeba [J].Genome Announc,2015,3(2): e00115-e00136.
[54] Suzan-Monti M,La Scola B,Barrassi L,Espinosa L,Raoult D.Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga mimivirus [J].PLoS One,2007,2(3): e328.
[55] Ghigo E,Kartenbeck J,Lien P,Pelkmans L,Capo C,Mege JL,Raoult D.Ameobal pathogen mimivirus infects macrophages through phagocytosis [J].PLoS Pathog,2008,4(6): e1000087.
[56] Conner SD,Schmid SL.Regulated portals of entry into the cell [J].Nature,2003,422(6927): 37-44.
[57] Silva LC,Almeida GM,Oliveira DB,Dornas FP,Campos RK,La Scola BA,Ferreira PC,Kroon EG,Abrahão JS.A resourceful giant: APMV is able to interfere with the human type I interferon system [J].Microbes Infect,2014,16(3): 187-195.
[58] Khan M,La Scola B,Lepidi H,Raoult D.Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus [J].Microb Pathog,2007,42(2/3): 56-61.
[59] Shah N,Hülsmeier AJ,Hochhold N,Neidhart M,Gay S,Hennet T.Exposure to mimivirus collagen promotes arthritis [J].J Virol,2014,88(2): 838-845.
[60] Xu GJ,Kula T,Xu Q,Li MZ,Vernon SD,Ndung’u T,Ruxrungtham K,Sanchez J,Brander C,Chung RT,O’Connor KC,Walker B,Larman HB,Elledge SJ.Comprehensive serological profiling of human populations using a synthetic human virome [J].Science,2015,348(6239): aaa0698.
[61] Colson P,de Lamballerie X,Yutin N,Asgari S,Bigot Y,Bideshi DK,Cheng XW,Federici BA,van Etten JL,Koonin EV,La Scola B,Raoult D.“Megavirales”,a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses [J].Arch Virol,2013,158(12): 2517-2521.
[62] Yolken RH,Jones-Brando L,Dunigan DD,Kannan G,Dickerson F,Severance E,Sabunciyan S,Talbot CC,Prandovszky E,Gurnon JR,Agarkova IV,Leister F,Gressitt KL,Chen O,Deuber B,Ma F,Pletnikov MV,Van Etten JL.Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice [J].Proc Natl Acad Sci USA,2014,111(45): 16106-16111.
[63] Heikkinen T,Järvinen A.The common cold [J].Lancet,2003,361(9351): 51-59.
