基于COⅠ及RAG2基因序列的我国科鱼类分子系统研究
2017-12-18苏国茂林聪左林贞武黎幸有梁日深黄运银
苏国茂,林聪左,林贞武,黎幸有,梁日深,黄运银
( 1.仲恺农业工程学院 动物科技学院,广东 广州 510225;2.湖北生物科技职业学院,湖北 武汉 430070 )
苏国茂1,林聪左1,林贞武1,黎幸有1,梁日深1,黄运银2
( 1.仲恺农业工程学院 动物科技学院,广东 广州 510225;2.湖北生物科技职业学院,湖北 武汉 430070 )



近年来,随着分子生物技术的快速发展,应用合适的线粒体DNA及核DNA分子标记技术可在基因水平上有效解决鱼类形态分类上悬而未决的难题[9-12]。线粒体DNA中细胞色素C氧化酶亚基Ⅰ(COⅠ)基因进化适中,已被验证为鱼类物种鉴定、分子系统发育研究中可靠的遗传标记。同时,COⅠ基因5′端一段长度为648 bp的片段,能在基因水平上成功区分物种,已被广泛应用于物种的分子鉴定[13]。重组激活基因(RAGs)是脊椎动物特异性免疫反应的关键基因,它由RAG1和RAG2两个基因组成,在引起V(D)J基因重排及淋巴系细胞发育过程中起到关键性作用[14-15]。其中,RAG2是可靠的核基因遗传标记,目前已经广泛应用于硬骨鱼类系统进化分析的研究[16-18]。

1 材料与方法
1.1 材料来源

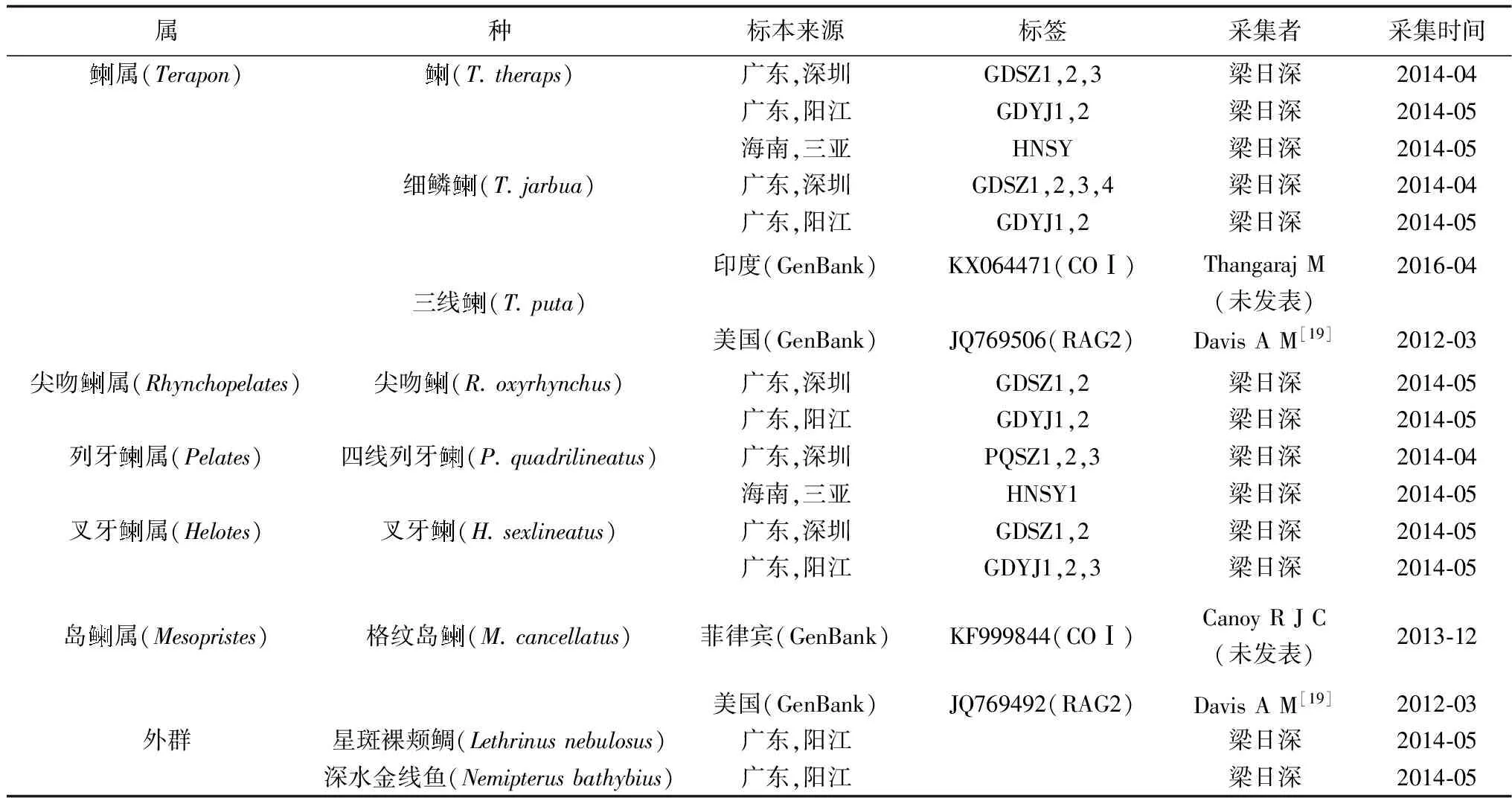
表1 试验材料的种类和采集地
1.2 总基因组DNA的提取
基因组DNA采用动物组织DNA提取试剂盒(天根生化科技有限公司)进行提取。提取的DNA溶解于100 μL灭菌水中,1%琼脂糖凝胶电泳检测提取质量,-20 ℃保存备用。
1.3 PCR扩增及序列测定
[16, 20],选择用于扩增COⅠ基因及RAG2基因片段序列的引物分别为:
COⅠ-F1: 5′-TCA ACY AAT CAY AAA GAT ATY GGC AC -3′;
COⅠ-R1: 5′-ACT TCY GGG TGR CCR AAR AAT CA -3′[20];
RAG2-F1: 5′-GAG GGC CAT CTC CTT CTC CAA-3′;
RAG2-R3: 5′-GAT GGC CTT CCC TCT GTG GGT AC-3′[16]。
PCR反应体系为50 μL,其中10×buffer 5 μL,dNTPs (各2.5 mmol/L)2 μL,上下游引物各1 μL,Ex Taq酶(1 U/μL) 2 μL。PCR反应条件为:
COⅠ基因:94 ℃ 5 min,94 ℃ 30 s,60 ℃ 60 s,72 ℃ 90 s,35个循环,72 ℃ 10 min;
RAG2基因:94℃ 5 min,94 ℃ 30 s,55 ℃ 60 s,72℃ 90 s,35个循环,72 ℃ 10 min。
PCR产物经1.5%琼脂糖凝胶电泳分离,纯化后送上海英骏生物技术有限公司进行测序。
1.4 数据分析
测序所得DNA序列利用Clustal W[21]进行比对,去除冗余序列。利用 Mega 5.0[22]计算序列的碱基组成及变异情况,基于Kimura′s 2-parameter模型计算各物种的遗传距离。利用最大似然法分别基于COⅠ、RAG2及COⅠ+RAG2整合序列,以鲈形目裸颊鲷科的星斑裸颊鲷及金线鱼科的深水金线鱼作为外类群进行分子系统进化树的构建,进化树采用重复抽样分析1000次的方法检验各分支的置信度。
2 结果与分析
2.1 序列分析

RAG2基因部分序列比对分析得到一致序列为729 bp,编码243个氨基酸。碱基A、T、G、C平均含量分别为23.9%、28.2%、21.6%和26.3%,4种碱基含量相差不大,分布平均。其中A+T的含量(52.1%)高于G+C含量(47.9%)。密码子第1、2位,4种碱基含量均相差不大,而密码子第3位,C-3含量最高,为36.1%,A-3含量最低,为16.8%,表现出反A偏倚。序列保守位点613个,约占84.4%;变异位点114个,约占15.7%;简约性信息位点85个,约11.7%。由此可知,与CO Ⅰ基因相比,RAG2基因相对保守,物种间序列差异较小。
COⅠ基因与RAG2基因序列中转换与颠换之比值分别为2.61与3.12 (Kimura 2-parameter模型),两个基因序列转换明显大于颠换,显示其位点没有突变饱和,可将两基因序列所有转换与颠换信息应用于系统发育树的构建。同时,COⅠ及RAG2两基因同质性检验分析得出P=1.0,说明两基因之间不存在异质性,可将两序列合并进行系统进化分析。
2.2 种间及种内的遗传距离

2.3 科鱼类分类及分子系统树


表2 基于 Kimura-2-Parameter模型计算7种科鱼类COⅠ基因种间遗传距离

表3 基于 Kimura-2-Parameter模型计算7种科鱼类RAG2基因种间遗传距离

图2 7种科鱼类基于COⅠ序列利用最大似然法构建的分子系统进化树

图3 7种科鱼类基于RAG2序列利用最大似然法构建的分子系统进化树
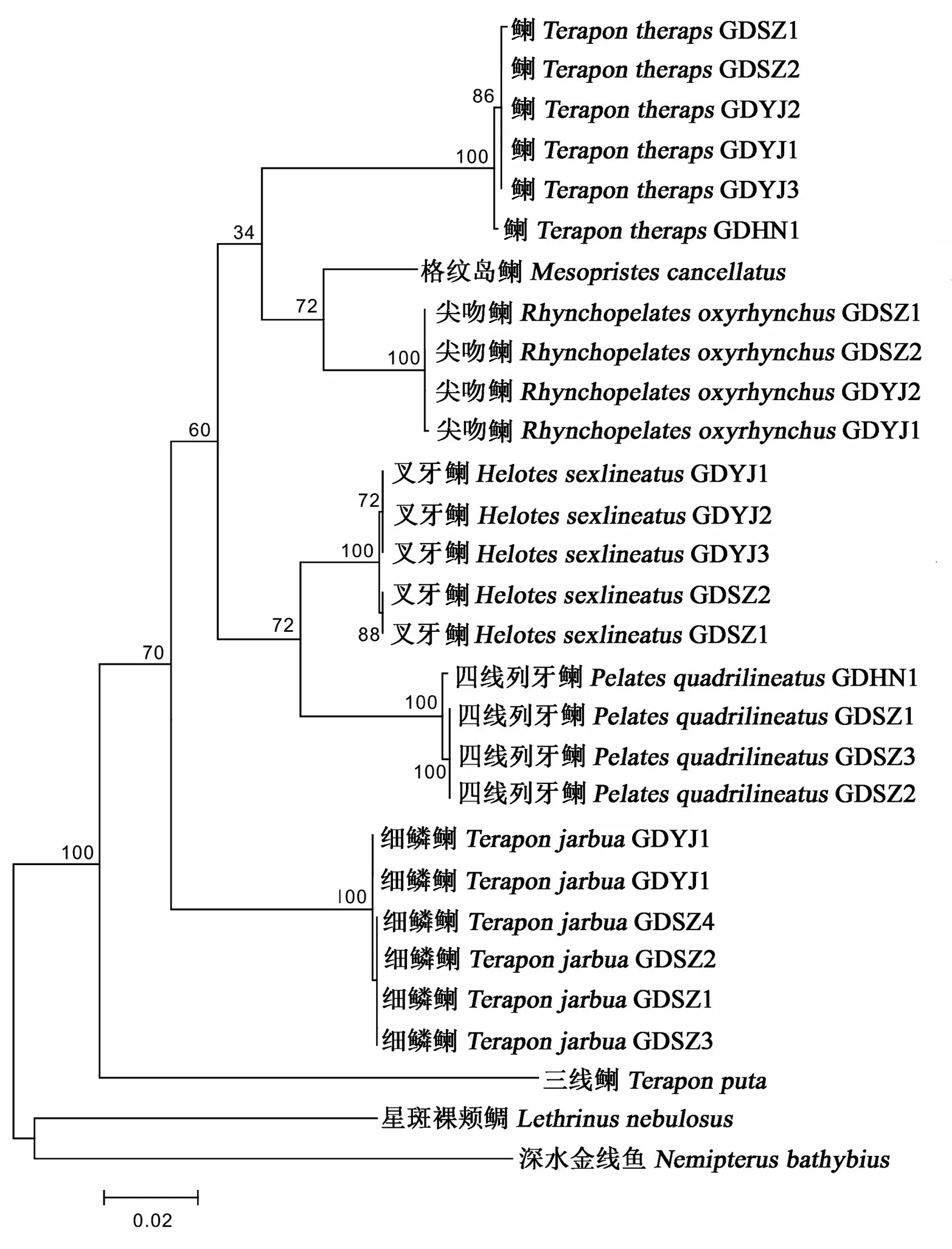
图4 7种科鱼类基于COⅠ与RAG2合并序列利用最大似然法构建的分子系统进化树
3 讨 论



参考文献:
[1] 成庆泰,郑葆珊.中国鱼类系统检索[M].北京:科学出版社,1987:339-341.
[2] 沈世杰.台湾鱼类志[M].台北:台湾大学动物学系出版,1993:360-363.
[3] 孟庆闻,苏锦祥,缪学祖.鱼类分类学[M].北京:中国农业出版社,1995, 49(2);636-640.
[4] 刘瑞玉.中国海洋生物名录[M].北京:科学出版社,2008:886-1066.
[5] Wu H L, Shao K T, Lai C F. Latin-Chinese dictionary of fishes names[M]. Taiwan:The Sueichan Press, 1999:1028.
[6] Nelson J S. Fishes of the world[M]. New York:John Wiley & Sons Inc, 2006:360-369.
[7] Fischer W, Whitehead P J P. FAO Species Identification Guide for Fishery Purposes. Eastern Indian Ocean(fishing area 57) and Western Central Pacific (fishing area 71). Volume 4[M]. Rome:Food and Agriculture Organization of the United Nations, 1974:1-12.
[8] Vari R P. The Terapon perches (Percoidei, Teraponidae), a cladistic analysis and taxonomic revision[J]. Bulletin of the American Museum of Natural History, 1978,159(5):175-340.
[9] Bohlen J,Slechtova V, Tan H H, et al. Phylogeny of the Southeast Asian freshwater fish genusPangio(Cypriniformes; Cobitidae)[J]. Molecular Phylogenetics and Evolution, 2011, 61(3):854-865.
[10] Eytan R I, Hastings P A, Holland B R. Reconciling molecules and morphology:molecular systematics and biogeography of Neotropical blennies (Acanthemblemaria)[J]. Molecular Phylogenetics and Evolution, 2012, 62(1):159-173.
[11] Bloom D D, Weir J T, Piller K R, et al. Do freshwater fishes diversify faster than marine fishes A test using state-dependent diversification analyses and molecular phylogenetics of new world silversides (atherinopsidae) [J]. Evolution, 2013, 67(7):2040-2057.
[12] Laakkonen H M, Strelkov P, Lajus D L, et al. Introgressive hybridization between the Atlantic and Pacific herrings (ClupeaharengusandC.pallasii) in the north of Europe[J]. Marine Biology, 2015, 162(1):39-54.
[13] Icolas H, Robert H, Louis B. Identification Canadian freshwater fishes through DNA barcodes[J]. PloS One, 2008, 3(6):1-8.
[14] Van G D,Mcblane J F, Ramsden D A, et al. Initiation of V( D) J recombination in a cell-free system[J]. Cell, 1995, 81(6):925-934.
[15] Market E,Papavasiliou F N. V (D) J recombination and the evolution of the adaptive immune system[J].PloS Biology, 2003,1(1):16.
[16] Rocha L A, Lindeman K C, Rocha C R, et al. Historical biogeography and speciation in the reef fish genusHaemulon(Teleostei:Haemulidae)[J]. Molecular Phylogenetics and Evolution, 2008, 48(3):918-928.
[17] Cramer C A,Bonatto S L, Reis R E. Molecular phylogeny of the Neoplecostominae and Hypoptopomatinae (Siluriformes:Loricariidae) using multiple genes[J]. Molecular Phylogenetics and Evolution. 2011, 59 (1):43-52.
[18] Martinez-Takeshita N, Purcell C M, Chabot C L, et al. A tale of three tails:cryptic speciation in a globally distributed marine fish of the genusSeriola[J]. Copeia, 2015, 103(2):357-368.
[19] Davis A M,Unmack P J, Pusey B J, et al. Marine-freshwater transitions are associated with the evolution of dietary diversification in terapontid grunters (Teleostei:Terapontidae) [J].Journal of Evolutionary Biology, 2012, 25(6):1163-1179.
[20] Ward R D, Zemlak T S, Innes B H, et al. DNA barcoding Australia′s fish species [J]. Philosophical Transactions of the Royal Society B,2005,360(1462):1847-1857.
[21] Thompson J D, Higgins D G, Gibson T J. Clustal W:improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice[J].Nucleic Acids Research,1994,22(22):4673-4680.
[22] Tamura K, Stecher G, Peterson D, et al. Mega 6:Molecular Evolutionary Genetics Analysis version 6.0[J]. Molecular Biology and Evolution, 2013, 30(12):2725-2729.
[23] Hebert P D N, Cywinska A, Ball S L, et al. Biological identifications through DNA barcodes[J]. Proceedings of the Royal Society of London, Series B, 2003, 270(1512):313-321.
[24] 彭居俐,王绪祯,王丁,等.基于线粒体COⅠ基因序列的DNA条形码在鲤科鲌属鱼类物种鉴定中的应用[J].水生生物学报,2009,33(2):271-276.
[25] 毛云涛,甘小妮,王绪祯.基于线粒体 COⅠ基因的沙鳅亚科鱼类 DNA 条形码及其分子系统发育研究[J].水生生物学报,2014,38(4):737-744.
[26] 刘璐,孙典荣,李纯厚.DNA条形码技术在鲻科鱼类鉴定中的应用[J].中国海洋大学学报:自然科学版,2016,46(11):178-186.
[27] 柳淑芳,陈亮亮,戴芳群,等.基于线粒体 CO1 基因的 DNA 条形码在石首鱼科(Sciaenidae)鱼类系统分类中的应用[J].海洋与湖沼,2016,41(2):223-232.
[28] Lee S C, Tsai M P. Molecular systematics of the thornfishes generaTeraponandPelates(Perciformes:Teraponidae) with reference to the new genusPseudoterapon[J].Zoological Studies,1999,38(3):279-286.
[30] Yearsley G K, Last P R, Hoese D F. Standard names of Australian fishes[M]. Hobart:CSIRO Marine and Atmospheric Research, 2006.
MolecularPhylogeneticAnalysisofFishesinTeraponidaefromChinaSeaBasedonMitochondrialCOⅠandNuclearRAG2GeneSequences
SU Guomao1, LIN Congzuo1, LIN Zhenwu1, LI Xingyou1, LIANG Rishen1, HUANG Yunyin2
( 1.College of Animal Technology,Zhongkai University of Agriculture and Engineering, Guangzhou 510225, China; 2.Hubei Vocational College of Bio-Technology, Wuhan 430070, China )
The 651 bp of mitochondrial DNA COⅠ sequences and 726 bp of nuclear DNA RAG2 sequences from 25 individuals of 5 Teraponidae fish collected from China Sea were determined in this study. No insertion and deletion existed in the two gene fragments. Base composition and genetic distances among and within species were calculated using Mega 5.0 and the molecular phylogenetic trees were constructed using maximum likelihood methods. The results showed that in the molecular phylogenetic tree,Teraponputawas first separated and rooted at the base of the tree, the next was speciesT.jarbua, which also formed an independent branch and located outside the other Teraponidae clusters. ThreeTeraponspecies did not formed an monophyletic group, the lastTeraponspeciesT.therapswas clustered together withRhynchopelatesoxyrhynchusandMesopristescancellatus;HelotessexlineatusandPelatesquadrilineatuswere clustered tightly as sister species, indicating that there were close phylogenetic relationships between the two species. These phylogenetic results might in accordance with the view of Lee and Tsai′s report thatT.therapsshould be removed from genusTeraponandHeloteswas combined withPelates.
Phylogeny; COⅠ gene; RAG2 gene; Teraponidae
2016-12-20;
2017-02-27.
广东省自然科学基金资助项目(2016A030310236);广东省教育厅青年创新人才类项目(2014KQNCX16);广东省大学生创新创业训练计划项目(201611347067).
苏国茂(1992-),男,硕士研究生;研究方向:水产动物种质资源.E-mail:1091109070@qq.com.通讯作者:黄运银(1972-),男,工程师,硕士;研究方向:水产养殖.E-mail:379830153@qq.com.
S917
A
1003-1111(2017)05-0652-06
