彭泽鲫Pcc-hira基因克隆、序列分析及时序表达
2016-02-26陈家长梁宏伟王在照
郑 尧,陈家长,梁宏伟,王在照
(1.中国水产科学研究院淡水渔业研究中心,
农业部淡水鱼类遗传育种与养殖生物学重点开放实验室,
农业部长江下游渔业资源环境科学观测实验站,
中国水产科学研究院内陆渔业生态环境与资源重点开放实验室,
南京农业大学渔业学院,江苏无锡 214081;
2.西北农林科技大学动物科技学院,陕西农业分子生物学重点实验室,陕西杨凌 712100;
3.中国水产科学研究院长江水产研究所,武汉 430223)
彭泽鲫Pcc-hira基因克隆、序列分析及时序表达
郑尧1,2,陈家长1,梁宏伟3,王在照2
(1.中国水产科学研究院淡水渔业研究中心,
农业部淡水鱼类遗传育种与养殖生物学重点开放实验室,
农业部长江下游渔业资源环境科学观测实验站,
中国水产科学研究院内陆渔业生态环境与资源重点开放实验室,
南京农业大学渔业学院,江苏无锡214081;
2.西北农林科技大学动物科技学院,陕西农业分子生物学重点实验室,陕西杨凌712100;
3.中国水产科学研究院长江水产研究所,武汉430223)
摘要:对雌核发育彭泽鲫(Carassiusauratusvar.Pengze,Pengze crucian carp,Pcc)中的Pcc-hira进行了克隆,序列分析,并对雌鱼9种组织和11个发育阶段中的表达进行了研究。结果显示,Pcc-hiraCDS全长3028 bp,编码1009个氨基酸残基,具有7个典型WD40结构域,Pcc-HIRA与鲫HIRA具有99%的同源性,跟其他鲫属鱼具有较高的亲缘关系。Pcc-hira分别在孵化后36和44 d达到最高值和最低值,且在雌鱼脑中的含量极显著高于其他8种组织。
关键词:雌核发育;HIRA;组蛋白;彭泽鲫 (Carassiusauratusvar.Pengze)
During fertilization in mammal,the sperm nucleus must decondense via the replacement of sperm-specific nuclear proteins with histones in the egg so that the paternal genome can participate in the development of the embryo.The histone H 3.3 chaperone (HIRA) is essential for chromatin assembly during male pronucleus formation[1-2],which was identified firstly inSaccharomycescerevisiaeas a negative regulator of histone gene expression[3].HIRA might be essential for vertebrate development[4],for example,gastrulation[5]and early embryo development[6-7],especially in gynogenetic and gonochoristic fish[4,8-9].
Variant histones and different histone chaperones might be differentially required for the distinct phases of differentiation pathway[10],and H3.3 specially plays important roles in the control of histone modifications at meiosis-specific gene loci and induces their transcriptional repression[11].The head of the sperm does not transform into male pronucleus in gynogenetic triploid fish,which could be obtained via meiotic[12]and mitotic gynogenesis method[13].Generally homologous sperm can undergo decondensation producing both female and male offsprings,while heterologous sperm nuclei can not decondense when incorporated in the egg.Thus the eggs produce a clonal lineage of all females by typical gynogenesis,which was supported by both gynogeneticCarassiusauratusgibelio[14]and gynogeneticC.auratusvar.Pengze[15].Recently either the defect of histone H2A variant H2af1o or acetylated histone H4 in deposition of some maternal histones for the sperm nucleus could be the reasons why heterologous sperm cannot decondense performed in theC.auratusgibelio egg[16].
Hirawas strongly expressed in mature ovaries in bothC.auratusgibelio and gonochoristic coloredC.auratus[8,16],andhiramRNA level kept stable during embryogenesis of gynogeneticC.auratusgibelio,but showed dramatic decrease shortly after fertilization till gastrulation stage in coloredC.auratus[17].The HIRA morphantC.auratusshowed delayed gastrulation,malformation of myotomes,limited yolk extension,and a twisted tail[9].Most of the mutants died during embryogenesis or shortly after hatching,suggesting that HIRA is required for early embryo development of gynogenetic fish.In the present study,the full-length cDNAs of Pcc-hirawas isolated,characterized,and Pcc-hiratranscripts for spatial and temporal distributions were analyzed.The results will provide a foundation for the further investigations on the molecular mechanism of early embryo development in Pcc.
1Materials and methods
1.1Animals
The eggs of mature gynogeneticC.auratusvar.Pengze (450 g in weight),collected originally from the Yangtze River Fisheries Research Institute of the Chinese Academy of Fishery Sciences in Wuhan,were inseminated by sperms from red common carp (Cyprinuscarpiovar.red style,600 g in weight) to activate the eggs via artificial propagation experiments.Spawning and spermiation were artificially induced by two intra-peritoneal injections with a mixture of hCG (400 IU/kg fish) and LRH-A (6 μg/kg fish) and an extraction of dry carp pituitary dissolved in the usual 0.7% NaCl solution (1 mg/kg fish,w/w).All the hormones were purchased from Shanghai Lizhu Dongfeng Biotechnology Co,LTD.,China.Fertilized eggs obtained from artificial spawning were incubated at 25±1 ℃ with a photoperiod of 14 h∶10 h (light/dark) in glass tanks (125 L) with dechlorinated tap water.Fish larvae (after 5 dph) were fedArtemianauplii at a rate of 0.1% body weight per day in our laboratory.
1.2Sampling,RNA isolation and reverse transcription (RT)
All fish were euthanized with 0.1% 2-phenoxyethanol (Sigma-Aldrich,USA) before sampling.To investigate spatial and temporal mRNA expression patterns of the target genes,samples were collected every 4 days from 20 dph until 60 dph (eleven developmental stages in total,n=9 fish/stage).Nine tissues (brain,eye,gill,hepatopancreas,intestine,kidney,muscle,ovary,spleen) of 1.5-year adult gynogenetic female Pcc (n=9,21.01±2.10 cm in length;226.23±23.29 g in weight) were dissected.The samples were frozen in liquid nitrogen and kept individually in frozen tubes at -80 ℃ until use.
Tissues of adult fish and total fish at different stages were homogenized in Trizol reagent (Invitrogen,USA) and total RNAs were extracted and treated with RNase-free DNase I (Fermentas,Canada) to remove genomic DNA contamination as previously described (Wang et al.2010).RNA quality checking and reverse transcription were done as previously described[18].
1.3cDNA cloning of Pcc-hira and sequence analysis
Primers for cDNAs cloning are shown in Table 1.PCR was performed with 2 μL cDNA template in 20 μL reaction volume containing 5 pmol of each specific primer and 1 unit of Taq DNA polymerase (TaKaRa,Japan).The PCR products were inserted into the pMD18-T vector using TA cloning kit (TaKaRa,Japan).Clones with confirmed recombinant plasmids were sequenced by Genscript Corporation (Nanjing,China) using BigDye Terminator Cycle Sequencing Kit (PE Biosystems,Foster City,CA,USA) and ABI 3730 automated DNA sequencer.The HIRAs amino acid sequences of diverse vertebrates were aligned by Clustal X sequences alignment program and subsequently a phylogenetic tree was constructed by neighbor-joining algorithms method of Mega 4.0 program (Molecular Evolutionary Genetic Analysis).
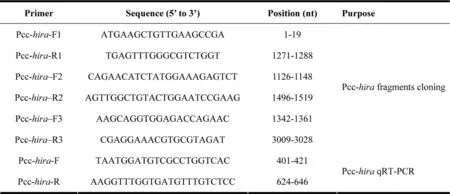
Tab.1Primers used for fragments cloning and quantitative real-time PCR
1.4qRT-PCR
The most stable reference genes (tubulinfor tissue distribution andef1afor others) for the normalization of qRT-PCR data were followed[17].The qRT-PCR was performed using CFX96 thermocycler (Bio-Rad,USA) and SYBR Premix ExTaq II kit (TaKaRa,Japan).The qRT-PCR reactions were carried out in a final volume of 25 μL,using 1×SYBRPremixExTaqTM,0.4 μM of each primer,and 2.5 μL RT reaction solution.Cycling parameters were:initial denaturation at 95 ℃ for 30 s,followed by 40 cycles of denaturation at 95 ℃ for 5 s,and annealing at 60 ℃ for 30 s.Each individual sample was run in triplicate.A melt curve analysis was performed at the end of each PCR thermal profile to verify the specificity of each amplicon.Analysis of SYBR green I density and determination of threshold cycle (Ct) values were carried out by CFX Manager software (Bio-Rad,USA).The efficiency (E) of each PCR reaction was determined on the the slope generated by 10-fold diluted cDNA series with five dilution points measured in triplicate.The equation wasE=10(-1/slope).
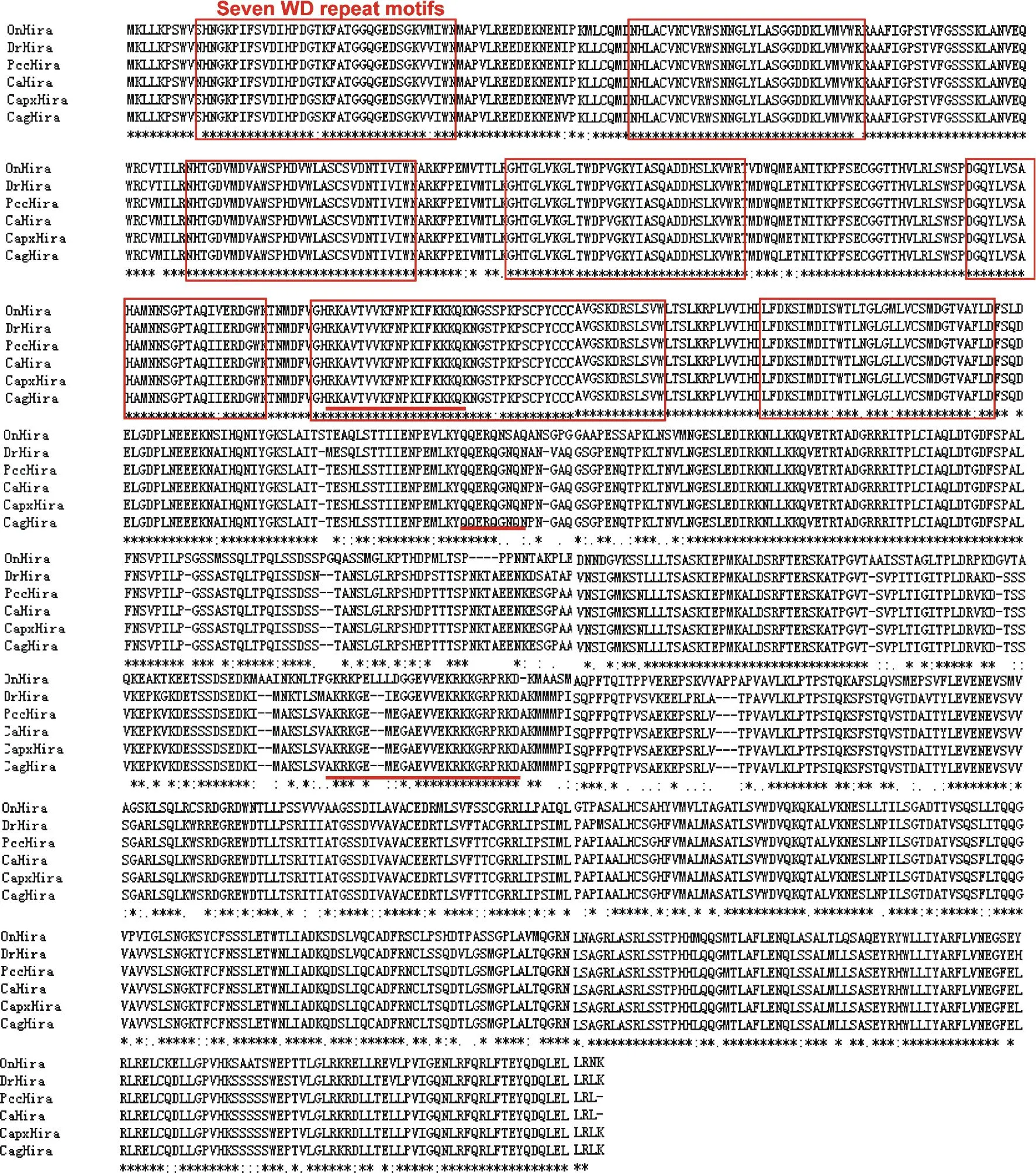
Fig.1 The match result of Pcc-HIRA compared with zebrafish and rare minnow HIRAs based on their amnio acid sequences.
GenBank IDs of the HIRAs amino acid sequences used in the alignments are XM_003444451 (Oreochromisniloticus,OnHira),HM067834 (C.auratusvar.pingxiang,CapxHira),DQ340808 (C.auratus,CaHira),DQ340807 (C.auratusgibelio,CagHira),XM_691386 (Daniorerio,DrHira).
1.5Spatial and temporal distributions of Pcc-hira
The mRNA spatial and temporal distributions of Pcc-hirawere performed by qRT-PCR.The qRT-PCR primers for Pcc-hirawere designed to span intron/exon boundaries to avoid amplification of genomic DNA.The mRNA amounts of Pcc-hirawere normalized to the most reliable reference gene.The relative mRNA levels of Pcc-hirain diverse tissues and stages were calculated using 2-△△Ctmethod[19].
1.6Statistic
All data are expressed as mean±standard deviation (SD).Data were tested for normality distribution (Kolmogorov-Smirnov test) and homogeneity of variances (Levene’s test) prior to any additional analysis.Prior to the analysis data were log transformed to meet one-way analysis of variance (ANOVA) assumptions of normality and variance homoscedasticity when necessary.In data sets where there was significant difference,Kruskal-Wallis analysis and Dunn’s post hoc test were carried out (P<0.05).
2Results
2.1Cloning and molecular charaterizations of Pcc-hira
The complete CDS of Pcc-hira(3028 bp,GenBank ID:KF373232) was obtained using RT-PCR method.Seven WD repeat conserved motifs were found in the deduced amino acid sequences of Pcc-HIRA (Fig.1).Multisequence alignment revealed that the deduced Pcc-HIRA protein shared the highest identities (99%) toC.auratusHIRA.The phylogenetic tree revealed that the Pcc-HIRA cluster together with HIRAs of crucian carps and separate from other teleosts including zebrafish and rainbow trout (Fig.2).The data suggests that Pcc-hirawas correctly isolated fromC.auratusvar.Pengze.
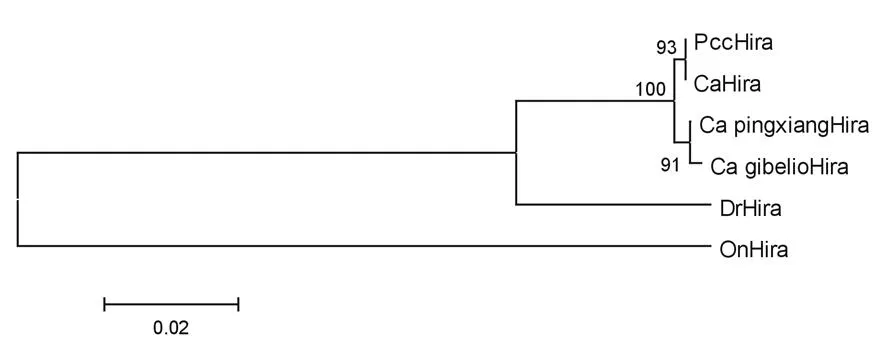
Fig.2 The phylogenetic tree based on amino acid
GenBank IDs of the HIRAs amino acid sequences used in the alignments are XM_003444451 (OnHira),HM067834 (CapxHira),DQ340808 (CaHira),DQ340807 (CagHira),XM_691386 (DrHira).
2.2Spatial gene expressions of Pcc-hira in tissues of the adult female fish
The mRNA expression patterns of Pcc-hirain nine tissues of adult female Pengze crucian carp were determined by qRT-PCR analysis.Pcc-hiramRNA was extremely significantly expressed in the brain tissue,which is the same as the studies reported in gynogeneticC.auratusgibelio (gibel carp) and coloredC.auratus(Fig.3)[16].The present study first demonstrated Pcc-hiratranscripts were detected in eye and gill tissues,and Pcc-hiratranscripts in ovary and eye were significant higher than the rest tissues (intestine,kidney,hepatopancreas,muscle and spleen).
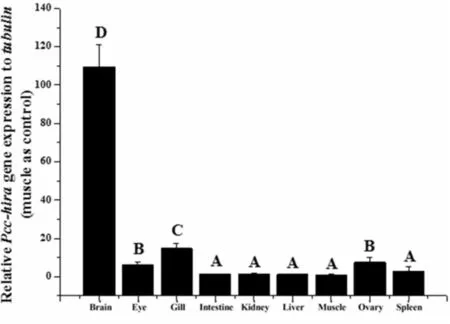
Fig.3 Tissue-specific expression profiles of Pcc-hira
The values were calibrated with the internal controltubulin.Each value is expressed as the mean±SD (n=9 for each value).The different capital letters indicate statistically significant differences (P<0.05).
2.3Temporal gene expressions of Pcc-hira during the developmental stages in larvae
The transcript level of Pcc-hirawas lowest on 44 dph (0.79±0.15) and reached peak on 36 dph (3.84±0.18,Fig.4).The mRNA level of Pcc-hirapersistently increased from 48 to 56 dph.The mRNA levels of Pcc-hiraon 36 dph and the period from 48 to 60 dph were extremely significantly higher than those on other stages (P<0.01).
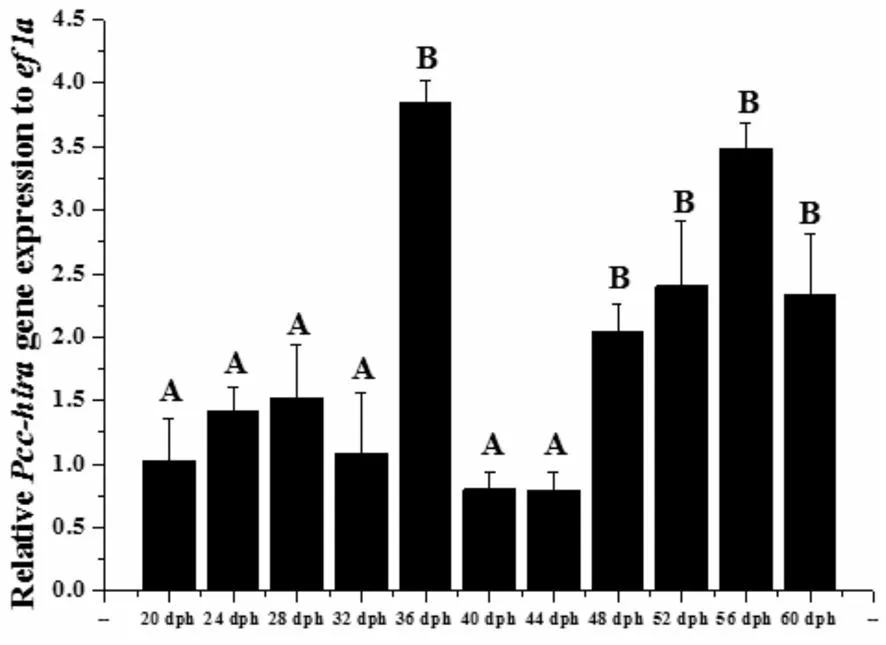
Fig.4 The mRNA expressions of Pcc-hira
The values were calibrated with the internal controlef1a.Transcript abundance is expressed relative to that of the 20 dph stage and each value is expressed as the mean±SD (n=9 for each value).
3Discussion
HIRA,belonging to the WD superfamily,contains seven WD40 domains and transcriptional regulatory elements,and WD40 could interact with β sub-unit of G protein,ubiquitin ligase E3,and transcriptional factors,TAFⅡ,etc.The functions of HIRAs were associated with endothelial gene regulation,vertebrate cell growth[20]and stress response.HIRA was identified as a histone chaperone involving in H3.3 deposition onto DNA via a DNA synthesis-independent pathway[21-22].H3.3 plays important roles in the control of histone modifications at meiosis-specific gene loci and induces their transcriptional repression[11].The seven WD40 domains were found in Pcc-HIRA,which implied Pcc-HIRA may work as other HIRAs associated with chromatin assembly through the process of histone modification.
As a chaperone to histone H3.3,HIRA has been found in various organisms,implying that it might play an important role in life.We have previously found that HIRA may play a critical role in decondensation of sperm nucleus during fertilization in Drosophila (Drosophilamelanogaster)[1-2]and fish[16].hiramRNA could be maternally deposited in the embryos,and combined with the research of high expression in mature and unfertilized eggs reported inC.auratusgibelio (gibel carp)[8].TheC.auratusgibelio (gibel carp) and pengze crucian carp belong to gynogenetic crucian carp[14].Whole-mount in situ hybridization of mouse embryos shows thatHirais ubiquitously expressed,with higher levels of transcripts appeared in the brain area.Hiraexpression was then observed mainly in head and along anterior-posterior axis inC.auratusgibelio (gibel carp) embryos,and Pcc-hiramRNA expressions were extremely significant in the brain tissues compared with other tissues,which suggesting its function in the development of body axis.In Drosophila,Northern blot analysis also showed thatHiramRNA levels are strongest in unfertilized egg and 0-3-hour-old embryos and then decrease,but can still be detected at later developmental stages[23].Pcc-hirawas continuously increased after 48 dph (Fig.3-4).The tendency of gene expression was the same as the details inC.auratusgibelio (gibel carp) and in coloredC.auratus[9],which suggesting that Pcc-hirais not only normally involved in neurogenesis,but also involved in early embryo development of pengze crucian carp.However,a reduction in Pcc-hiraexpression was found in the present study (Fig.4),and which in Wolbachia-infected male Drosophila led to detrimental effects on sperm fertility resulting in embryo lethality[23].
References:
[1]Loppin B,Bonnefoy E,Anselme C,et al.The histone H3.3chaperoneHIRAisessentialfor chromatin assembly in the male pronucleus [J].Nature,2005,437 (7063):1386-1390.
[2]Bonnefoy E,Orsi G A,Couble P,et al.The essential role of Drosophila HIRA forde novo assembly of paternal chromatin at fertilization [J].PLoS Genet,2007,3:1991-2006.
[3]Sherwood P W,Osley M A.Histone regulatory (hir) mutations suppress delta insertion alleles inSaccharomycescerevisiae[J].Genetics,1991,128 (4):729-738.
[4]Zhao Z K,Wang Y F.The function of histone chaperones during development [J].Heredity (Beijing),2010,32:41-48.
[5]Szenker E,Lacoste N,Almouzni G.A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation inXenopus[J].Cell Rep,2012,1 (6):730-740.
[6]Roberts C,Sutherland H F,Farmer H,et al.Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality [J].Mol Cell Biol,2002,22:2318-2328.
[7]Szenker E,Ray-Gallet D,Almouzni G.The double face of the histone variant H3.3 [J].Cell Res,2011,21:421-434.
[8]Du X Z,Zhou L,Zhao H B,et al.Identical sequences but different expression patterns ofHiragene in gynogenetic and gonochoristic crucian carps [J].Fish Physiol Biochem,2008,34 (2):175-184.
[9]Wang M Y,Guo Q H,Du X Z,et al.HIRAis essential for the development of gibel carp [J].Fish Physiol Biochem,2014,40 (1):235-244.
[10]Song T Y,Yang J H,Park J Y,et al.The role of histone chaperones in osteoblastic differentiation of C2C12 myoblasts [J].Biochem Biophys Res Commun,2012,423 (4):726-732.
[11]Mizuki F,Tanaka A,Hirose Y,et al.The HIRA complex subunit Hip3 plays important roles in the silencing of meiosis-specific genes inSchizosaccharomycespombe[J].PLoS One,2011,6 (4):e19442.
[12]Yamashita M,Jiang J,Onozato H,et al.A tripolar spindle formed at meiosis i assures the retention of the original ploidy in the gynogenetic triploid crucian carp,GinbunaCarassiusauratuslangsdorfii[J].Develop Growth & Differ,1993,35 (6):631-636.
[13]Kato K,Murata O,Yamamoto S,et al.Viability,growth and external morphology of meiotic- and mitotic-gynogenetic diploids in red sea bream,Pagrusmajor[J].J Appl Ichthyol,2001,17:97-103.
[14]Gui J F,Zhou L.Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploidyCarassiusauratusgibelio[J].Sci China Life Sci,2010,53:409-415.
[15]Shu H,Zhang H,Chen X.A cytological studies on gynogenesis of pengze crucian carp [J].Chinese J Zool,2000,35 (5):12-15.
[16]Zhao Z K,Li W,Wang M Y,et al.The role of HIRA and maternal histones in sperm nucleus decondensation in the gibel carp and color crucian carp [J].Mol Reprod Dev,2011,78 (2):139-147.
[17]Guo Q H,Zhao Z K,Wang Y F,et al.Expression analysis ofHiramRNA and protein during oogenesis in gynogenetic and gonochoristic crucian carps [J].Acta Hydrobiol Sin,2010,34:611-617.
[18]Zheng Y,Wang L,Li M,et al.Molecular characterization of five steroid receptors from pengze crucian carp and their expression profiles of juveniles in response to 17α-ethinylestradiol and 17α-methyltestosterone [J].Gen Comp Endocrinol,2013,191:113-122.
[19]Livak K J,Schmittgen T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ctmethod [J].Methods,2001,25:402-408.
[20]Ahmad K,Henikoff S.Histone H3 variants specify modes of chromatin assembly [J].Proc Natl Acad Sci USA,2002,99 (Suppl 4):16477-16484.
[21]Ray-Gallet D,Quivy J P,Scamps C,Martini E M,et al.Almouzni G.HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis [J].Mol Cell,2002,9:1091-1100.
[22]Tagami H,Ray-Gallet D,Almouzni G,et al.Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis [J].Cell,2004,116:51-61.
[23]Zheng Y,Ren P P,Wang J L,et al.Wolbachia-inducedcytoplasmicincompatibilityis associated with decreasedHiraexpression in male Drosophila [J].PLoS One,2011,6 (4):e19512.
(责任编辑:张潇峮)
Molecular cloning,characterization,temporal and spatial expression
patterns of Pcc-hira in Carassius auratus var.Pengze
ZHENG Yao1,2,CHEN Jia-zhang1,LIANG Hong-wei3,WANG Zai-zhao2
(1.FreshwaterFisheriesResearchCenter,
KeyOpenLaboratoryofEcologicalEnvironmentandResourcesofInlandFisheries,
ChineseAcademyofFisherySciences;KeyLaboratoryofGeneticBreedingandAquacultureBiologyofFreshwaterFishes,
ScientificObservingandExperimentalStationofFisheryResourcesandEnvironmentintheLowerReachesoftheChangjiang
River,MinistryofAgriculture;WuxiFisheriesCollege,NanjingAgriculturalUniversity,Wuxi214081,Jiangsu,China;
2.CollegeofAnimalScienceandTechnology,NorthwestA&FUniversity,
ShaanxiKeyLaboratoryofMolecularBiologyforAgriculture,Yangling712100,Shaanxi,China;
3.YangtzeRiverFisheriesResearchInstitute,ChineseAcademyofFisherySciences,Wuhan430223,China)
Abstract:In the present study,Pengze crucian carp (Pcc)-hirawas cloned and characterized in the triploid gynogenetic fish,theCarassiusauratusvar.Pengze (Pengze crucian carp).Their mRNA expression profiling was investigated in eleven juvenile developmental stages and nine tissues of the adult female fish.Results showed that the complete CDS of Pcc-hirawas 3028 bp encoding 1099 AA,containing seven classical WD40 domains.Multisequence alignment revealed that the deduced Pcc-HIRA protein shared the highest identities (99%) toCarassiusauratusHIRA.The putative Pcc-HIRA shared high identities to HIRAs of crucian carp species.Gene expression profiling in the developmental stages showed that Pcc-hirahad the highest/lowest expression at 36/44 days post hatching (dph),respectively.Pcc-hirawas predominantly expressed in brain tissue of the female fish compared with other 8 detected tissues.
Key words:gynogenetic;HIRA;histone;Carassiusauratusvar.Pengze
中图分类号:S917.4
文献标识码:A
文章编号:1000-6907-(2016)01-0018-06
Fund:National Natural Science Foundation of China (No.31270547)
First author:Zheng Yao (1986-),male,phD,Research Assistant,major in aquatic toxicology,zhengy@ffrc.cn
Corresponding author:Wang Zai-zhao,zzwang@nwsuaf.edu.cn;Chen Jia-zhang,chenjz@ffrc.cn.
