植物体对硝态氮的吸收转运机制研究进展
2015-06-15张然然都韶婷
张 鹏, 张然然, 都韶婷
(浙江工商大学环境科学与工程学院,浙江杭州 310018)
植物体对硝态氮的吸收转运机制研究进展
张 鹏, 张然然, 都韶婷*
(浙江工商大学环境科学与工程学院,浙江杭州 310018)
硝态氮是高等植物重要的氮素营养,直接影响植物的生长。植物根系吸收硝态氮并向地上部转运的机制一直是研究者十分关注的问题。近几年的深入研究使得新的现象与结论被揭示,推动了我们对植物体吸收转运硝态氮生理与分子机制的认识。本文综述了近年来国内外关于植物硝态氮吸收转运的生理及分子机制的相关研究结果。通过整理归类植物硝酸盐吸收相关的生理学数据,介绍了影响植物吸收硝态氮的各种因素。基于膜转运体在植物硝态氮吸收转运过程中发挥的重要作用,本文还重点介绍参与该过程的四大基因家族的成员及功能,即硝酸盐转运体1(NRT1)、硝酸盐转运体2(NRT2)、氯离子通道(CLC)和s型阴离子通道(SLAC),以期为后续研究者提供一个较为全面的理论依据。
硝态氮; 亲和力转运系统; 吸收; 再分配; 转运蛋白
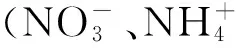
1 生理机制
1.1 硝态氮的吸收转运



图1 拟南芥中部分硝酸盐转运蛋白的功能 [3,15,48]Fig.1 Function of nitrate transporters in Arabidopsis thaliana[注(Note):HATS—高亲和吸收系 High-affinity transport systems; LATS—低亲和吸收系统 Low-affinity transport systems.]

运输到植株地上部的氮素除了参与代谢过程外,一部分又以氨基酸的形态通过韧皮部向根部转运,甚至外泌[14]。由韧皮部向下运输到根部的总氮量远远超过根系的生长需要,大部分又进入木质部同新吸收的氮素一起向上运输至地上部[15]。与此同时,硝态氮也能从老叶运输到新叶中,进行再分配[16]。再分配是所有作物提高氮素利用效率的主要因素,也是植物对硝态氮吸收转运的重要过程之一。
1.2 硝态氮吸收的影响因素
与其他矿质养分类似,植物对硝态氮的吸收也受到许多外界因素的影响。鉴于弄清植物吸收硝态氮的影响因素对于农业氮素利用有着重要意义,因而笔者从环境条件、养分元素和信号物质等对吸收过程影响较大的几个方面介绍它们对植物硝态氮吸收的影响(表1)。

表1 硝态氮吸收的影响因素

就供氮条件的变化而言,它也会极大地影响植物对硝态氮的吸收。例如,外源硝酸盐能显著促进高亲和力转运系统的转运能力。以大麦作为研究材料,在培养液中添加硝酸盐后,组成型高亲和力转运系统的活性可以增加3倍左右[23]。甚至,诱导型高亲和力转运系统具有更强的硝酸盐诱导效应[24]。介质中的铵态氮同样也会影响植物对硝态氮的吸收能力[25]。研究结果显示,铵离子浓度超过 1×10-5mol/L会抑制硝酸盐的吸收[26]。氨基酸也能直接影响植物对硝态氮的吸收,如分别给大麦和玉米幼苗供应谷氨酸和天冬氨酸等均抑制了硝酸盐的吸收[27-28]。综上所述,在实际农田使用氮肥的过程中,所选用的氮肥形态也会极大地影响植物对硝态氮的吸收效率。



2 分子机制

早在30年前,研究者就利用拟南芥这一模式植物开展了硝酸盐转运体编码基因的相关研究。目前,已经克隆了4类硝酸盐转运蛋白基因家族,主要包括NRT1、NRT2、CLC和SLAC[15]。它们均编码了受硝酸盐诱导的共运体,但差异较大,基因序列无相似性[47]。我们将上述拟南芥的硝酸盐转运体基因家族的概况作如下介绍,其命名及功能[48]见图1和表2。
2.1 NRT1转运蛋白


表2 拟南芥中硝酸盐转运蛋白的命名及功能
续表2 Table 2 continuous
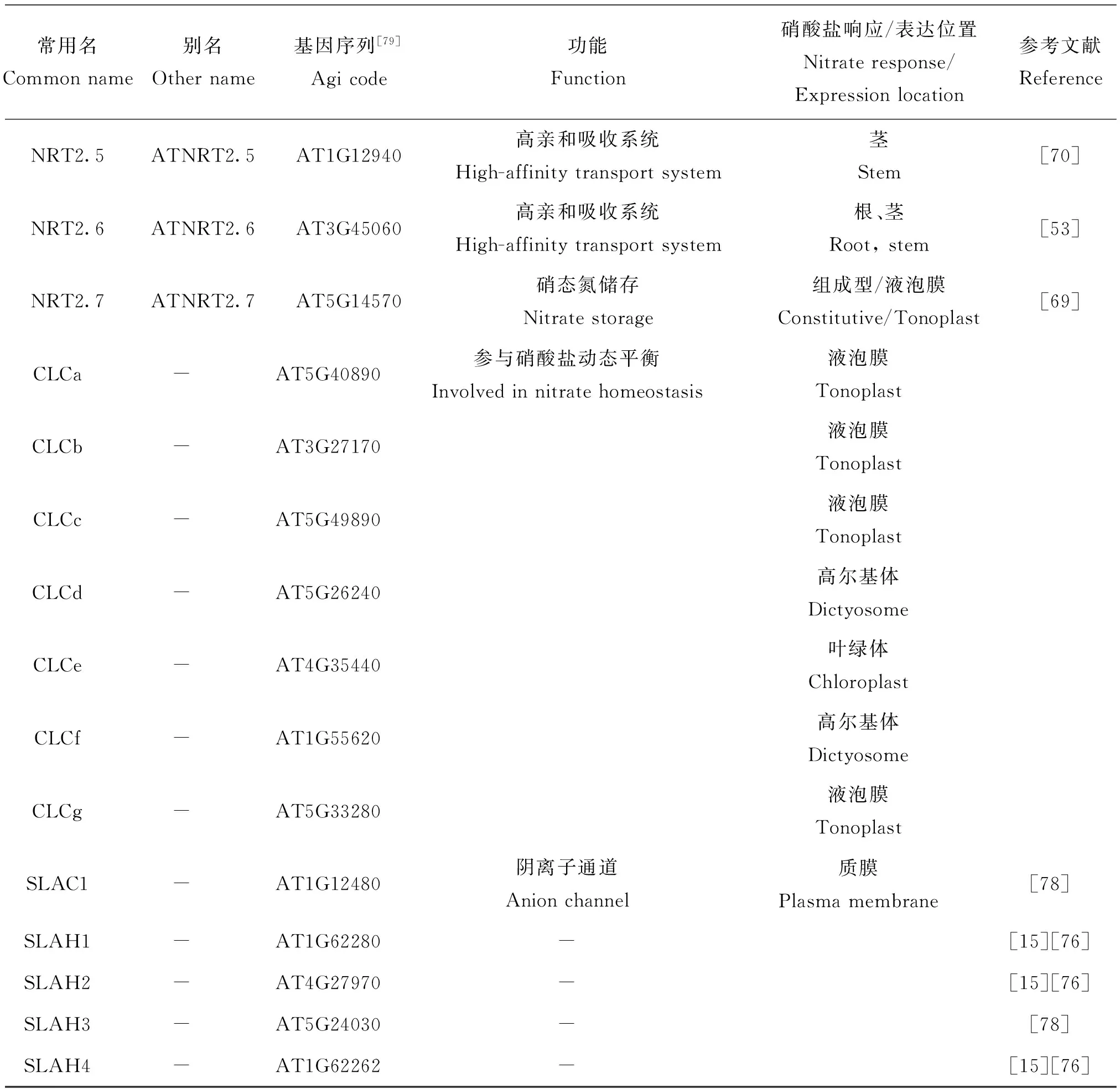
常用名Commonname别名Othername基因序列[79]Agicode功能Function硝酸盐响应/表达位置Nitrateresponse/Expressionlocation参考文献ReferenceNRT2.5ATNRT2.5AT1G12940高亲和吸收系统High-affinitytransportsystem茎Stem[70]NRT2.6ATNRT2.6AT3G45060高亲和吸收系统High-affinitytransportsystem根、茎Root,stem[53]NRT2.7ATNRT2.7AT5G14570硝态氮储存Nitratestorage组成型/液泡膜Constitutive/Tonoplast[69]CLCa-AT5G40890CLCb-AT3G27170CLCc-AT5G49890CLCd-AT5G26240CLCe-AT4G35440CLCf-AT1G55620CLCg-AT5G33280参与硝酸盐动态平衡Involvedinnitratehomeostasis液泡膜Tonoplast[15][71][74]液泡膜Tonoplast[72]液泡膜Tonoplast[75]高尔基体Dictyosome[75]叶绿体Chloroplast[75]高尔基体Dictyosome[75]液泡膜Tonoplast[75]SLAC1-AT1G12480阴离子通道AnionchannelSLAH1-AT1G62280-SLAH2-AT4G27970-SLAH3-AT5G24030-SLAH4-AT1G62262-质膜Plasmamembrane[78][15][76][15][76][78][15][76]
注(Note): “-”表示未知 Unkown.
2.2 NRT2转运蛋白
目前,在拟南芥中已报道发现7种NRT2转运蛋白,该家族中大部分成员均能在根内表达,并且都是高亲和转运蛋白。其中,NRT2.1和NRT2.2主要通过在根中表达后促使植物根部从土壤中吸收硝酸盐[64]。更有反向遗传学的研究表明,NRT2.2只有在NRT2.1缺失的情况下才显著表达[65]。因此,一般认为相较于NRT2.2,NRT2.1调控植物根系吸收硝酸盐的过程更为重要。此外,研究还发现硝酸盐浓度较低时,它们的表达量都明显减少[66]。甚至,其他氮源(铵盐和谷氨酸盐)供应不足,也会导致NRT2.1的表达量受到抑制[67]。NRT2.1的表达量还受光合作用产物的影响[66]。除了上述两个转运蛋白,NRT2.4也是备受研究者关注的对象。它是一种高亲和转运蛋白,在侧根的表皮部分和茎的韧皮部表达,在氮素(硝酸盐)缺乏的情况下,nrt2.4突变体对硝酸盐的吸收及其根韧皮部硝酸盐的外泌均减少,表明NRT2.4在低氮情况下对硝酸盐的运输起着很大的作用[68]。与NRT2.1、NRT2.2和NRT2.4不同,NRT2.7在种子细胞的液泡膜上表达,负责将硝酸盐运输到种子的液泡内储存,其表达量在种子成熟后达到一个最高峰[69]。目前为止,关于NRT2家族的其他成员如NRT2.3、NRT2.5和NRT2.6的功能研究仅有少量报道。如NRT2.5在没有外源氮素(硝酸盐和铵盐)供应的情况下,其表达量增加,当供氮恢复时表达受到抑制[53,70]。不同的是,研究发现NRT2.3和NRT2.6在根和茎中的表达,并且不受供氮浓度的影响[53]。
2.3 CLC和SLAC转运蛋白
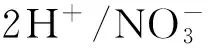
拟南芥中有5种SLAC转运蛋白:SLAC1、SLAH1、SLAH2、SLAH3和SLAH4,其中SLAH1、SLAH2、SLAH3和SLAH4都是SLAC1的同系物。SLAC1是第一个被发现的s-型阴离子通道上的转运蛋白位于质膜[76]。其后,也有学者报道了有关SLAC1的调控机制[77]。与SLAC1一样,它的同系物SLAHs也都位于质膜。研究发现当SLAC1主导气孔关闭时SLAH1和SLAH3会表现出与SLAC1相似的功能[76]。但是,SLAH2是否与SLAC1家族中其它转运蛋白功能类似尚未明确。近期一项研究通过GUS染色法在保卫细胞中仅观察到了SLAH3以及少量的SLAH2,SLAH1和SLAH4则未观察到[78]。虽然,有关SLAC转运蛋白的研究仍处起步阶段,但我们相信识别这类阴离子通道转运蛋白有助于弄清楚气孔关闭过程,并对干旱、信号传导和硝酸盐代谢有着深刻影响。
3 展望
植物对硝态氮的吸收是植物生理过程中极为关键的一个部分。近六七十年来的关于植物吸收硝态氮的生理机制很好地帮助并解决了较多硝态氮施用的农业问题。随着近一二十年的分子机制的研究,更推进了我们对植物吸收硝态氮过程的理解。尤其是负责硝酸盐吸收的转运蛋白,它们被认为在这个过程中发挥了重要的作用。蔡宜芳教授曾评价说这些转运蛋白既是硝态氮的搬运工,也是负责探测土壤硝态氮含量的守门员,它们将土壤中硝态氮含量这一信息传递到细胞的指挥中心。因此,有关硝态氮转运蛋白的研究仍应给予极大的重视。此外,除了本文介绍的拟南芥中发现的转运蛋白外,也有很多关于各种植物硝态氮转运蛋白的报道,如水稻的 OsNTR1.1、大豆的 GmNRT2、番茄的 LeNRT1、大麦的HvNRT2A、烟草的NpNRT2.1及苜蓿的MtNRT1.3[3,69,80-81]。不同物种硝态氮转运蛋白之间的差异也值得我们探究。我们相信,虽然硝态氮转运体的功能及特点尚未完全弄清,但是随着人们对植物体硝酸盐吸收途径认识的日益加深,以及分子生物学技术的发展,必将取得研究上的突破。
[1] Novoa R, Loomis R S. Nitrogen and plant production[J]. Plant and Soil, 1981, 58: 177-204.
[2] Li D D, Tian M Y, Cai Jetal. Effects of low nitrogen supply on relationships between photosynthesis and nitrogen status at different leaf position in wheat seedlings[J]. Plant Growth Regulation, 2013, 70: 257-263.
[3] Crawford N M, Glass A D M. Molecular and physiological aspects of nitrate uptake in plants[J]. Trends in Plant Science, 1998, 3(10): 389-395.
[4] Stitt M. Nitrate regulation of metabolism and growth[J]. Current Opinion in Plant Biology, 1999, 2(3): 178-186.
[5] Miller A J, Smith S J. Nitrate transport and compartmentation in cereal root cells[J]. Journal of Experimental Botany, 1996, 47: 843-854.
[6] McClure P R, Kochian L V, Spanswick R M, Shatf J E. Evidence for cotransport of nitrate and protons in maize roots. I. Effects of nitrate on the membrane potential[J]. Plant Physiology, 1990, 93: 281-289.
[7] Siebrecht S, Herdel K, Schurr U, Tischner R. Nutrient translocation in the xylem of poplar: diurnal variations and spatial distribution along the shoot axis[J]. Planta, 2003, 217: 783-793.
[8] Wegner L H, Raschke K. Ion channels in the xylem parenchyma of barley roots. A procedure to isolate protoplasts from this tissue and a patch-clamp exploration of salt passageways into xylem vessels[J]. Plant Physiology, 1994, 105: 799-813.
[9] Clarkson D T. Roots and the delivery of solutes to the xylem[J]. Philosophical Transactions of the Royal Society London B, 1993, 341: 5-17.
[10] Kohler B, Wegner L H, Osipov V, Raschke K. Loading of nitrate into the xylem: apoplastic nitrate controls the voltage dependence of X-QUAC, the main anion conductance in xylem-parenchyma cells of barley roots[J]. The Plant Journal, 2002, 30: 133-142.
[11] Gilliham M, Tester M. The regulation of anion loading to the maize root xylem[J]. Plant Physiology, 2005, 137: 819-828.
[12] Chen B M, Wang Z H, Li S Xetal. Effects of nitrate supply on plant growth, nitrate accumulation, metabolic nitrate concentration and nitrate reductase activity in three leafy vegetables[J]. Plant Science, 2004, 167(3): 635-643.
[13] Miller A J, Fan X R, Orsel Metal. Nitrate transport and signalling[J]. Journal of Experimental Botany, 2007, 58(9): 2297-2306.
[14] Pearson C J, Volk R J, Jackson W A. Daily changes in nitrate influx, efflux and metabolism in maize and pearl millet[J]. Planta, 1981, 152: 319-324.
[15] Wang Y Y, Hsu P K, Tsay Y F. Uptake, allocation and signaling of nitrate[J]. Trend in Plant Science, 2012, 17(8): 458-467.
[16] Fan S C, Lin C S, Hsu P Ketal. TheArabidopsisnitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate[J]. The Plant Cell, 2009, 21: 2750-2761.
[17] Sasakawa H, Yamamoto Y. Comparison of the uptake of nitrate and ammonium by rice seedlings[J]. Plant Physiology, 1978, 62: 665-669.

[19] Lillo C. Signalling cascades integrating light-enhanced nitrate metabolism[J]. Biochemical Journal, 2008, 415: 11-19.
[20] Jackson W A, Flesher D, Hageman R H. Nitrate uptake by dark-grown corn seedlings[J]. Plant Physiology, 1973, 51: 120-127.
[21] Matt P, Geiger M, Walch-Liu Petal. Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate[J]. Plant Cell and Environment, 2001, 24(11): 1119-1137.
[22] 都韶婷,章永松. 增施CO2降低小白菜硝酸盐积累的机理研究[J]. 植物营养与肥料学报, 2010, 16(6): 1509-1514. Du S T, Zhang Y S. Mechanisms of CO2enrichment-induced decrease of nitrate accumulation in Chinese cabbage(BrassicachinensisL.)[J]. Plant Nutrition and Fertilizer Science, 2010, 16(6): 1509-1514.
[23] Aslam M, Travis R L, Huffaker R C. Comparative kinetics and reciprocal inhibition of nitrate and nitrite uptake in roots of uninduced and induced barley seedlings[J]. Plant Physiology, 1992, 99: 1124-1133.
[24] Siddiqi M Y, Glass A D, Ruth T, Rufty T W. Studies of the uptake of nitrate in barley[J]. Plant Physiology, 1990, 93: 1426-1432.
[25] Youngdahl L J, Pacheco R, Street J J, Vlek P L G. The kinetics of ammonium and nitrate uptake by young rice plant[J]. Plant and Soil, 1982, 69: 225-232.
[26] Ohmori M, Ohmori K, Strotmann H. Inhibition of nitrate uptake by ammonia in a Blue-Green Algae,Anabaenacylindrica[J]. Archives of Microbiology, 1977, 114: 225-229.
[27] Aslam M, Travis R. Differential effect of amino acids on nitrate uptake and reduction systems in barley roots[J]. Plant Science, 2001, 160: 219-228.
[28] Padgett P E, Leonard R T. Free amino acid levels and the regulation of nitrate uptake in maize cell suspension cultures[J]. Journal of Experimental Botany, 1996, 47(7): 871-883.
[29] Ruffy T W Jr, MacKown C T, Israel D W. Phosphorus stress effects on assimilation of nitrate[J]. Plant Physiology, 1990, 94: 328-333.
[30] Neyra C A, Hageman R H. Nitrate uptake and induction of nitrate reductase in excised corn roots[J]. Plant Physiology, 1975, 56: 692-695.
[31] Blevins D G, Barnett N M, Frost W B. Role of potassium and malate in nitrate uptake and translocation by wheat seedlings[J]. Plant Physiology, 1978, 62: 784-788.
[32] Camacho-Cristóbal J J, González-Fontes A. Boron deficiency decreases plasmalemma H+-ATPase expression and nitrate uptake, and promotes ammonium assimilation into asparagine in tobacco roots[J]. Planta, 2007, 226: 443-451.
[33] Cakmak I, Marschner H. Decrease in nitrate uptake and increase in proton release in zinc deficient cotton, sunflower and buckwheat plants[J]. Plant and Soil, 1990, 129: 261-268.
[34] Heimer Y M, Wray J L, Filner P. The effect of tungstate on nitrate assimilation in higher plant tissues[J]. Plant Physiology, 1969, 44: 1197-1199.
[35] Quaggiotti S, Ruperti B, Pizzeghello Detal. Effect of low molecular size humic substances on nitrate uptake and expression of genes involved in nitrate transport in maize(ZeamaysL.)[J]. Journal of Experimental Botany, 2004, 55(398): 803-813.
[36] Piccolo A, Nardi S, Concheri G. Structural characteristics of humic substances as related to nitrate uptake and growth regulation in plant systems[J]. Soil Biology and Biochemistry, 1992, 24(4): 373-380.
[37] Albuzio A, Ferrari G, Nardi S. Effects of humic substances on nitrate uptake and assimilation in barley seedlings[J]. Canadian Journal of Soil Science, 1986, 66: 731-736.
[38] Rao K P, Rains D W. Nitrate absorption by barley[J]. Plant Physiology, 1976, 57: 55-58.

[40] Ward M R, Aslam M, Huffaker R C. Enhancement of nitrate uptake and growth of barley seedlings by calcium under saline conditions[J]. Plant Physiology, 1986, 80: 520-524.

[42] 周诗毅,高轩,何光源,Cram W J. 不同糖类对水稻硝酸盐吸收的影响[J].华中师范大学学报(自然科学版), 2009, 43(1): 124-127. Zhou S Y, Gao X, He G Y, Cram W J. The effects of different sugars on the nitrate uptake in rice[J]. Journal of Huazhong Normal University(Nat. Sci.), 2009, 43(1): 124-127.
[43] 郑冬超, 夏新莉, 尹伟伦. 生长素促进拟南芥AtNRT1.1基因表达增强硝酸盐吸收[J]. 北京林业大学学报, 2013, 35(2): 80-85. Zheng D C, Xia X L, Yin W L. Auxin promotes nitrate uptake by up-regulating AtNRT1.1 gene transcript level inArobidopsisthaliana[J]. Journal of Beijing Forestry University, 2013, 35(2): 80-85.
[44] Campbell S J. Uptake of ammonium by four species of macroalgae in Port Phillip Bay, Victoria, Australia[J]. Marine and Freshwater Research, 1999, 50(6): 515-522.
[45] Wanek W, Poertl K. Short-term15N uptake kinetics and nitrogen nutrition of bryophytes in a lowland rainforest, Costa Rica[J]. Functional Plant Biology, 2008, 35(1): 51-62.
[46] 都韶婷, 李玲玲, 章永松, 林咸永. 不同基因型小白菜硝酸盐积累差异及筛选研究[J]. 植物营养与肥料学报, 2008, 14(5): 969-975. Du S T, Li L L, Zhang Y S, Lin X Y. Nitrate accumulation discrepancies and variety selection in different Chinese cabbage (BrassicachinensisL.) genotypes[J]. Plant Nutrition and Fertilizer Science, 2008, 14(5): 969-975.
[47] Steiner H Y, Naider F, Becker J M. The PTR family: A new group of peptide transporters[J]. Molecular Microbiology, 1995, 16: 825-834.
[48] Krouk G, Crawford N M, Coruzzi G M, Tsay Y F. Nitrate signaling: adaptation to fluctuating environments[J]. Current Opinion in Plant Biology, 2010, 13: 265-272.
[49] Tsay Y F, Chiu C C, Tsai C Betal. Nitrate transporters and peptide transporters[J]. FEBS Letters, 2007, 581: 2290-2300.
[50] Huang N C, Chiang C S, Crawford N M, Tsay Y F. CHL1 encodes a component of the low-affinity nitrate uptake system inArabidopsisand shows cell type-specific expression in roots[J]. The Plant Cell, 1996, 8: 2183-2191.
[51] Tsay Y F, Schroeder J I, Feldmann K A, Crawford N M. The herbicide sensitivity gene CHL1 ofArabidopsisencodes a nitrate-inducible nitrate transporter[J]. Cell, 1993, 72: 705-713.
[52] Kanno Y, Hanada A, Chiba Yetal. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(24): 9653-9658.
[53] Okamoto M, Vidmar J J, Glass A D M. Regulation of NRT1 and NRT2 gene families ofArabidopsisthaliana: responses to nitrate provision[J]. Plant and Cell Physiology, 2003, 44(3): 304-317.
[54] Chiu C C, Lin C S, Hsia A Petal. Mutation of a nitrate transporter, AtNRT1: 4, results in a reduced petiole nitrate content and altered leaf development[J]. Plant and Cell Physiology, 2004, 45: 1139-1148.
[55] Lin S H, Kuo H F, Canivence Getal. Mutation of theArabidopsisNRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport[J]. The Plant Cell, 2008, 20: 2514-2528.
[56] Almagro A, Lin S H, Tsay Y F. Characterization of theArabidopsisnitrate transporter NRT1.6 reveals a role of nitrate in early embryo development[J]. The Plant Cell, 2008, 20: 3289-3299.
[57] Mendoza-Cózatl D G, Jobe T O, Hauser F, Schroeder J I. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic[J]. Current Opinion in Plant Biology, 2011, 14: 554-562.
[58] Li J Y, Fu Y L, Pike S Metal. TheArabidopsisnitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance[J]. The Plant Cell, 2010, 22: 1633-1646.
[59] Wang Y Y, Tsay Y F.Arabidopsisnitrate transporter NRT1.9 is important in phloem nitrate transport[J]. The Plant Cell, 2011, 23: 1945-1957.
[60] Nour-Eldin H H, Andersen T G, Burow Metal. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds[J]. Nature, 2012, 488: 531-534.
[61] Hsu P K, Tsay Y F. Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth[J]. Plant Physiology, 2013, 163: 844-856.
[62] 郑令欣. 阿拉伯芥AtNRT1.13功能分析 [D]. 台北:台湾大学博士论文,, 2010. Zheng L X. Functional study ofArabidopsisAtNRT1.13 [D]. Taibei: PhD dissertation, Taiwan University, 2010.
[63] Leran S, Varala K, Boyer J Cetal. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants[J]. Trend in Plant Science, 2014, 19(1): 5-9.
[64] Filleur S, Dorbe M F, Cerezo Metal. AnArabidopsisT-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake[J]. FEBS Letters, 2001, 489: 220-224.
[65] Li W, Wang Y, Okamoto Metal. Dissection of the AtNRT2.1: AtNRT2.2 inducible high-affinity nitrate transporter gene cluster[J]. Plant Physiology, 2007, 143: 425-433.
[66] Lejay L, Wirth J, Pervent Metal. Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis[J]. Plant Physiology, 2008, 146: 2036-2053.
[67] Nazoa P, Vidmar J J, Tranbarger T Jetal. Regulation of the nitrate transporter gene AtNRT2.1 inArabidopsisthaliana: responses to nitrate, amino acids and developmental stage[J]. Plant Molecular Biology, 2003, 52: 689-703.
[68] Kiba T, Feria-Bourrellier A B, Lafouge Fetal. TheArabidopsisnitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants[J]. The Plant Cell, 2012, 24: 245-258.
[69] Chopin F, Orsel M, Dorbe M Fetal. TheArabidopsisATNRT2.7 nitrate transporter controls nitrate content in seeds[J]. The Plant Cell, 2007, 19: 1590-1602.
[70] Dechorgnat J, Patrit O, Krapp Aetal. Characterization of the Nrt2.6 gene inArabidopsisthaliana: A link with plant response to biotic and abiotic stress[J]. Plos One, 2012, 7(8): 1-11.
[71] De Angeli A, Monachello D, Ephritkhine Getal. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles[J]. Nature, 2006, 442: 939-942.
[72] Geelen D, Lurin C, Bouchez Detal. Disruption of putative anion channel gene AtCLCa inArabidopsissuggests a role in the regulation of nitrate content[J]. The Plant Journal, 2000, 21: 259-267.


[75] Lv Q D, Tang R J, Liu Hetal. Cloning and molecular analysis of theArabidopsisthalianachloride channel gene family[J]. Plant Science, 2009, 176: 650-661.
[76] Negi J, Matsuda O, Nagasawa Tetal. CO2regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells[J]. Nature, 2008, 452: 483-486.
[77] Geiger D, Scherzer S, Mumm Petal. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106: 21425-21430.
[78] Geiger D, Maierhofer T, AL-Rasheid K A Setal. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1[J]. Science Signaling, 2011, 4(173): 1-12.
[79] http: //www.arabidopsis.org/
[80] Morère-Le Paven M C, Viau L, Hamon Aetal. Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legumeMedicagotruncatula[J]. Journal of Experimental Botany, 2011, 62(15): 5595-5605.
[81] Lin C M, Koh S, Stacey Getal. Cloning and functional characterization of a constitutively expressed nitrate transporter gene,OsNRT1, from rice[J]. Plant Physiology, 2000, 122(2): 379-388.
Research advances in nitrate uptake and transport in plants
ZHANG Peng, ZHANG Ran-ran, DU Shao-ting*
(CollegeofEnvironmentalScienceandEngineering,ZhejiangGongshangUniversity,Hangzhou310018,China)
Nitrate is one of the major nitrogen sources for higher plants, which most frequently limits the growth of plants. Recently, the mechanisms of nitrate uptake and transport to the aboveground parts in plants have
widespread attention. New phenomenons and conclusions have been revealed and thereby give us a new sight into the physiological and molecular mechanisms of nitrate uptake and transport in plants. In this review, the current worldwide research conducted on nitrate uptake and transport in plants was summarized in the physiological and molecular levels. Besides, the influence factors responsible for nitrate uptake in plants were also discussed by integrating the large amount of physiological data. Membrane bound transporters are required for nitrate uptake, allocation and storage inside the plants. Therefore, we also summarized the members and the functions of four gene families that encode the nitrate transporters, including nitrate transporter 1(NRT1), nitrate transporter 2(NRT2), chloride channel(CLC) and slow anion channel-associated(SLAC). We hope that the recent advances in molecular biology of nitrate uptake and transport can provide a more comprehensive theoretical basis for further research.
nitrate; affinity transport system; uptake; remobilize; transporter
2014-06-05 接受日期: 2014-10-13
浙江省自然科学基金项目(LY14C130001,R13C130001); 浙江省教育厅项目(Y201432146)资助。
张鹏(1989—),男,湖北黄石人,硕士研究生,主要从事污染环境的生理生态方面的研究。E-mail: zp2231989@163.com * 通信作者 Tel: 0571-28008209, E-mail: dushaoting@zjgsu.edu.cn
Q945.1
A
1008-505X(2015)03-0752-11
