多位点序列分型(MLST)在艾伯特埃希菌鉴定中的应用
2015-02-14许彦梅邓建平孙松松熊衍文
刘 祥,许彦梅,王 斌,邓建平,肖 波,孙松松,周 阳,熊衍文,王 红
多位点序列分型(MLST)在艾伯特埃希菌鉴定中的应用
刘 祥1,许彦梅2,王 斌1,邓建平1,肖 波1,孙松松1,周 阳1,熊衍文2,王 红1
目的 探讨多位点序列分型技术在新病原艾伯特埃希菌发现与鉴定中的应用。方法 采用MLST技术对生鲜肉中分离的30株疑似艾伯特埃希菌的7个管家基因进行PCR扩增、测序、DNAstar软件分析,核酸序列上传至大肠杆菌MLST数据库进行比对,确定其等位基因编号及基因序列型(ST型),并利用MEGA6.0软件Neighbor-joining法构建遗传进化树,与大肠埃希菌、志贺氏菌、艾伯特埃希菌和伤寒沙门氏菌参考菌株进行亲缘性分析。结果 30株菌可分为7种新序列型(16株)和4种已知序列型(14株),N-J分析显示与艾伯特埃希菌参考菌株具有高度亲缘性,而与大肠埃希菌、志贺氏菌、弗格森埃希菌和鼠伤寒沙门氏菌存在较大差异。结论 MLST在管家基因水平上准确地确定艾伯特埃希菌的种属关系,首次在国内发现艾伯特埃希菌的存在。
多位点序列分型技术(MLST);艾伯特埃希菌;新病原
艾伯特埃希菌是一种新发现的致泻性病原菌,它可引起人类的散发感染和食物中毒[1-7]。该菌最初从孟加拉腹泻儿童的粪便中分离,被鉴定为蜂房哈夫尼菌[1],通过生化表型特征、16S rDNA序列和DNA杂交分析,Huys等将此类蜂房哈夫尼菌命名为埃希氏菌属种的一个新种——艾伯特埃希菌(Escherichiaalbertii)[8]。由于缺乏艾伯特埃希菌的生化特性,迄今仍无该菌种的商品化鉴定系统。常规的细菌鉴定系统经常将艾伯特埃希菌鉴定为哈夫尼菌、沙门氏菌、鲁氏耶尔森菌,最常见的确定为大肠埃希菌。缺乏动力、不发酵木糖、不发酵乳糖和产生β-D葡萄糖醛酸酶被认为是艾伯特埃希菌常见生化特性,并且携带有编码紧密粘附素(Intimin)基因eae,此外菌株还通常携带编码细胞肿胀坏死毒素(cytolethal distending toxin,CDT)基因cdtABC[3,13]。
多位点序列分型(MLST)通过对7个管家基因核酸序列的分析,将每个基因的序列与MLST数据库比对,得到相应的等位基因编号,按照指定排列顺序形成ST型。ST型可用于国际实验室间比对、溯源以及分子流行病学、遗传进化等研究;同时根据等位基因图谱使用配对差异矩阵方法构建系统进化树,与参考菌株的相应序列进行亲缘性分析,从而准确地研究菌株遗传进化关系,以确定种属[9-11]。通过MLST分析,艾伯特埃希菌明显不同于肠杆菌科及埃希菌属中的其它种[3-8]。
本研究对四川省自贡市农贸市场生鲜肉食品中分离出的30株疑似艾伯特埃希菌进行多位点序列分型,通过与NCBI数据库收录的大肠埃希菌、志贺氏菌、艾伯特埃希菌和伤寒沙门氏菌的相应序列进行亲缘性分析,为确定艾伯特埃希菌提供依据。
1 材料与方法
1.1 菌株来源与鉴定 从2013年3月-2014年7月,在中国四川省自贡市的超市和农贸市场共收集446份生肉,包含92份牛肉、41份猪肉、22份羊肉、53份鸡肉、189份鸡肠、21份鸭肉、28份鸭肠。根据艾伯特埃希菌相关研究资料,取生肉样品25 g,接种225 mL EC,20 ℃增菌24~36 h,取增菌液进行eae基因PCR检测[3]。eae阳性增菌液接种麦康凯琼脂平板,36 ℃培养过夜,取不发酵乳糖的白色菌落进行eae基因检测。最后分离eae阳性且不发酵乳糖、不发酵木糖、动力阴性、cdtB基因阳性的疑似艾伯特埃希菌30株。
1.2 仪器与试剂
(1)主要仪器:PCR仪(SensoQuest Labcycler)、Bio-Rad凝胶成像系统(GEL DocXR+)。
(2)主要试剂:2×Taq MasterMix(含染料)、DNA Marker(DM1000)、去离子水:购自康为世纪公司;核酸染料(GeneGreen):购自天根生化科技(北京)有限公司;PCR引物合成及测序:由生工生物工程(上海)有限公司完成。
1.3 DNA模板制备及反应体系 挑取少许疑似艾伯特埃希菌纯培养物接种至EC营养肉汤,36 ℃培养18~24 h。吸取1 mL菌悬液于1.5 mL无菌离心管,8 000 r/min,离心10 min,用500 μL无菌去离子水重悬并煮沸10 min,将重悬液10 000 r/min离心5 min,取上清液作为PCR模板。PCR体积为50 μL,2×Taq MasterMix 25 μL,ddH2O 20μL,上、下游引物(10 μmol/L)各2 μL,模板DNA1 μL。
1.4 MLST引物及反应程序 管家基因位点选择参照大肠杆菌MLST数据库(http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/documents/primersColi_html)提供的分型方案,PCR引物序列、退火温度及产物大小详见参考文献[12]。反应程序为94℃ 5min预变性,按照94 ℃ 30 s、退火30 s、72 ℃ 45 s进行30个循环,最后一个循环结束后72 ℃延伸7 min。每次扩增均以无DNA模板的体系作为空白对照。
1.5 结果观察 取PCR产物5 μL以1.5%琼脂糖凝胶(含1/10 000核酸染料)进行电泳,通过凝胶成像系统观察是否扩增出目的大小片段,将PCR产物送至上海生工生物工程公司双向测序。
1.6 数据分析 利用SeqMan软件对PCR产物序列与数据库中的管家基因标准序列进行拼接和校正,校正后的7个管家基因序列同时上传至E.coliMLST数据库,确定菌株的等位基因型和序列类型。并根据Ooka等人[3]描述,基于7个管家基因的串联序列,利用MEGA6.0软件Neighbor-joining法构建系统进化树,与国际数据库中已收录的大肠杆菌、志贺氏菌、艾伯特埃希菌和伤寒沙门氏菌进行亲缘性比对,探究该30株菌的种属关系。
2 结 果
2.1 MLST分型结果 通过不断优化扩增条件、选取引物,所有菌株的7个管家基因序列均得到双向峰图完好的序列,与MLST数据库中的等位基因剪切比对,结果显示14株菌分别为4种已知ST型,而其余16株菌的各等位基因双向序列通过互联网上传至数据库,被接受并确认为7种新ST型,见表1。
2.2 MLST聚类图谱分析 根据7个管家基因首尾串联整合成的3 423 bp核苷酸序列图谱分析,表明研究中的30株菌与大肠埃希菌、志贺氏菌、弗格森大肠埃希菌、鼠伤寒沙门氏菌存在较大差异。

表1 30株菌MLST型别Tab.1 MLST STs of 30 strains
30株艾伯特埃希菌中鉴别出11种序列型,均与艾伯特埃希菌LMG20976、KF1具有高度亲缘关系,其中SP140602和SP140701两株菌的序列型与艾伯特埃希菌KF1一致,见图1。结果表明30株菌全部鉴定为艾伯特埃希菌。
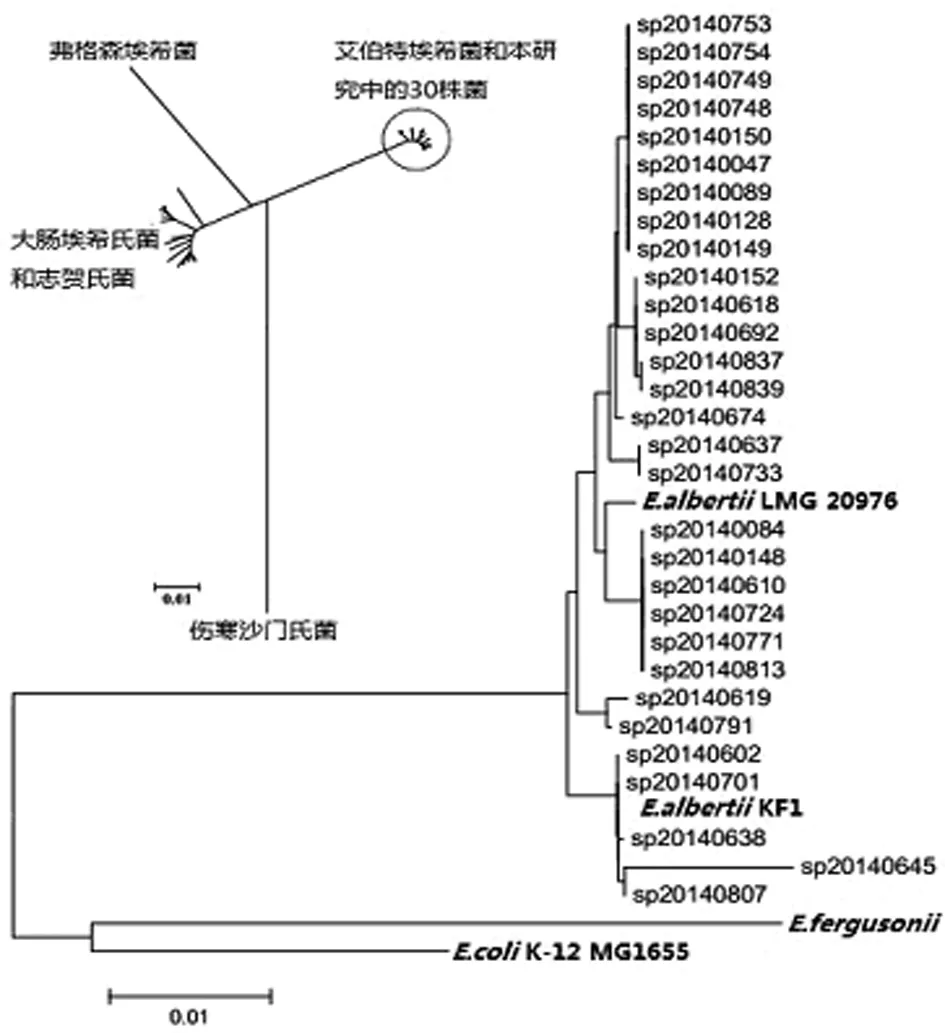
图1 30株菌与大肠埃希菌、弗格森埃希菌、艾伯特埃希菌、志贺氏菌、伤寒沙门氏菌系统发育关系分析结果
Fig.1 Phylogenetic relationships of 30 strains withE.coli,E.fergusonii,E.albertii,Shigellaspp. andSalmonellaentericaserovar Typhi
2.3 GenBank核酸序列索取号 本研究中30株艾伯特埃希菌的7个管家基因已全部上传至GenBank数据库,登陆号为:KP015856-KP016011、KP064411-KP064472。
3 讨 论
艾伯特埃希菌是一种新发现的肠道致病菌。起初通过常规检测方法被鉴定为致病性大肠杆菌(EPEC)或者出血性大肠杆菌(EHEC),对该菌的误诊漏诊容易造成公众健康潜在的危险,也会影响临床治疗效果。由于目前对其研究较少,仍没有标准化检测方法和商品化鉴定系统,利用分子生物学技术在基因水平上研究遗传进化关系,则成为菌种鉴定的关键。
利用MLST对30株疑似艾伯特埃希菌分型过程中,存在个别菌株的管家基因序列在大肠杆菌MLST数据库中未找到完全匹配的等位基因型,可能属于新型别。采用双向测序,将其完好的序列和峰图文件上传至MLST数据库以待确认,上传前需核查PCR产物序列和峰图文件,避免误判等位基因型及新型别;此外,在本实验样品7个管家基因PCR扩增时,除purA外的其余6个管家基因产物测序峰图基本为单峰,个别出现重峰、杂峰的通过多次扩增和降低模板浓度得到改善;而全部样品purA基因测序产物重峰、杂峰情况严重,存在非特异产物干扰,结果无法用于比对分析。利用提高退火温度,降低模板浓度,多次扩增后测序效果并不理想,最终改用数据库中purA其它引物进行扩增后,产物特异性明显提高,正反向测序峰图完好。这也提示我们发现测序峰图不完好时,可通过多次PCR扩增测序、优化扩增条件和选取不同引物进行解决。
传统检测方法在新病原菌鉴定中存在局限,而MLST可通过研究管家基因图谱与参考菌株相应序列构建系统进化树,根据聚类分析结果判断菌种归属,在新病原菌的鉴定和常见细菌新的变异方面发挥积极的作用。本研究MLST聚类分析结果显示30株菌为艾伯特埃希菌,属国内首次发现并证实艾伯特埃希菌的存在。然后MLST针对的7个管家基因是大肠埃希菌的保守基因,如若标本中混有大肠杆菌,PCR扩增为混合产物无法区分,故不能直接作为该细菌PCR快速诊断。目前拟针对艾伯特埃希菌的特异基因建立PCR快速诊断方法。
综上所述,MLST为最终确认艾伯特埃希菌提供依据。同时也适用于其他新病原菌的诊断以及后续的分子流行病学研究、菌群遗传多态性、溯源等,为制定有效的监测体系和预防疾病的流行与暴发提供有力的技术支持。
[1]Albert MJ, Alam K, Islam M, et al.Hafniaalvei, a probable cause of diarrhea in humans[J]. Infect Immun, 1991, 59: 1507-1513.
[2]Konno T, Yatsuyanagi J, Takahashi S, et al. Isolation and identification ofEscherichiaalbertiifrom a patient in an outbreak of gastroenteritis[J]. Jap J Infect Dis, 2012, 65: 203-207. DOI: 10.7883/yoken.65.203
[3]Ooka T, Seto K, Kawano K, et al. Clinical significance ofEscherichiaalbertii[J]. Emerg Infect Dis, 2012, 18: 488-492. DOI: 10.3201/eid1803.111401
[4]Asoshima N, Matsuda M, Shigemura K, et al. Identification ofEscherichiaalbertiias a causative agent of a food-borne outbreak occurred in 2003[J]. Jap J Infect Dis, 2014, 67: 139-140.
[5]Murakami K, Etoh Y, Tanaka E, et al. Shiga toxin 2f-producingEscherichiaalbertiifrom a symptomatic human[J]. Jap J Infect Dis, 2014, 67: 204-208.
[6]Oaks JL, Besser TE, Walk ST, et al.Escherichiaalbertiiin wild and domestic birds[J]. Emerg Infect Dis, 2010, 16: 638-646. DOI: 10.3201/eid1604.090695
[7]Ooka T, Tokuoka E, Furukawa M, et al. Human gastroenteritis outbreak associated withEscherichiaalbertii, Japan[J]. Emerg Infect Dis, 2013, 19: 144-146. DOI: 10.3201/eid1901.120646
[8]Huys G, Cnockaert M, Janda JM, et al.Escherichiaalbertiisp. nov., a diarrhoeagenic species isolated from stool specimens ofBangladeshichildren[J]. Int J Systemat Evolutionary Microbiol, 2003, 53: 807-810.
[9]Ji XW, Liao YL, Mao XH, et al. The research progress of MLST analysis in the application of Pathogenic microorganisms genotyping[J]. Int J Lab Med, 2011, 32(2): 246-247. DOI: 10.3969/j.issn.1673-4130.2011.02.051 (in Chinese) 姬小薇,廖亚玲,毛旭虎,等. MLST分析在病原微生物基因分型应用中的研究进展[J].国际检验医学杂志,2011,32(2):246-247.
[10]Zuo TT, Li YW, Han XL, et al. Two new sequence type isolates ofBacillusanthracisby multilocus sequence typing[J]. Acta Microbiologica Sinica, 2012, 52(1): 120-123. (in Chinese) 左庭婷,李岩伟,韩雪莲,等. 两株炭疽芽胞杆菌 MLST 新序列型(ST) [J].微生物学报,2012,52(1):120-123.
[11]Deng XL, Guan DW, Li W, et al. MLST technique applied to molecular epidemiological research on neisseria meningitides isolates[J]. South China J Prev Med, 2008, 34(3): 10-14. (in Chinese) 邓小玲,管大伟,黎薇,等. MLST分型技术应用于脑膜炎奈瑟菌的分子流行病学研究[J]. 华南预防医学,2008,34(3):10-14.
[12]Bai XN, Zhao AL, Xia SL, et al. Multilocus sequence typing of non-O157 Shiga toxin-producingEscherichiacoliisolates[J]. Chin J Zoonoses, 2012, 28(6): 544-548. (in Chinese) 白向宁,赵爱兰,夏胜利,等. 非O157产志贺毒素大肠杆菌分离株的多位点序列分型研究[J]. 中国人兽共患病学报,2012,28(6): 544-548.
[13]Bai XN, Wang H. The research progress onEscherichiaalbertii[J]. Chin J Zoonoses, 2014, 34(2): 154-158. DOI: 10.3760/cma.j.issn.0254-5101.2014.02.016 (in Chinese) 白向宁,王红. 艾伯特埃希菌研究进展[J]. 中华微生物学和免疫学杂志,2014,34(2): 154-158.
DOI:10.3969/j.issn.1002-2694.2015.11.010
Abstract:MicroRNAs (miRNAs) are key regulators of gene expression. The miR-36 family has only been authentically found in helminth includingAscarisspp.,Caenorhabditiselegans,Brugiamalayi, andSchistosomamansoni. And the family member named miR-36f was identified to exist only in the egg and larval stage ofAscarissuum(A.suum) in our previous study.We herein preliminarily evaluated the function of miR-36f via its mimics and inhibitors by usingA.suumas model organism. The miR-36f mimics, inhibitors, mimics negative controls and inhibitor negative controls were absorbed by the different groups larvae via soaking method. Three days later, the mice were infected with the larvae above. And the larvae were recovered from the liver and lung of the mice respectively. The larvae of each group were counted and their length and width were measured, and the two target genes of miRNA were analyzed by qRT-PCR.The results showed that development and infective numbers of theA.suuminfective larvae were significantly influenced by miR-36f inhibitors (P<0.05), when compared with the mimics (P>0.05).The expression of the two key target genes of miR-36f named OST48 and cytochrome b were similar to that of inhibitor and mimics. When them iR-36f was influenced by its inhibitor, the development and infective ability ofA.suumlarvae were reduced, which indicated that the miR-36f might be associated with development and infectivity of theA.suumlarvae. Furthermore, miR-36f inhibitors seemed more effective than its mimics. The results would be helpful for better understanding and novel control strategy for parasitic helminths.
Keywords:Ascarissuum; asu-miR-36f; infectivity; development
Supported by the Science Fundfor Creative Research Groups of Gansu Province (Grant No.1210RJIA006)
Corresponding author: Xu Min-jun, Email:xuminjun@caas.cn
MicroRNAs (miRNAs) are small non-coding RNAs with increasingly recognized regulating roles played in gene expression in animals, plants and parasites by binding to mRNAs in 3′-untranslated regions (3′-UTR), and also in coding domain sequences or 5′-UTR[1-4]. And up to now, members of the miR-36 family have been only authenticated in helminths, includingAscarisspp.[5],Caenorhabditiselegans[6],Brugiamalayi[7],Schmidteamediterranea[8], andSchistosomamansoni[9]. The miR-36 family members were highly conserved, stayed in cluster in different organisms, and adult- and larva-specifically expressed; knocking down the miR-36 cluster will lead to embryonic and larval deadliness[10-11]. These reports indicated that the miR-36 members play vital roles in worms. Besides, the main control measure for the epidemic of worms at present is anthelmintic drugs, which easily results in the drug resistance of the parasite and drug residues in animal products[12-14]. Therefore, alternative controlling measure is urgently needed for the replacement of anthelmintic drugs for animal welfare and food safety, and changing miRNA expression abundance with convenient way is likely to be a new way to prevent and control the parasites.
A.suumis a globally parasitic nematode which causes infection in pigs and human with high prevalence rates, especially in developing countries[15-17], and the identification of its genome, proteome, and transcriptome made the parasite be a fine model[18-20]. We herein usedA.suumas a model to evaluate the roles of one miR-36 member named miR-36f, which is specifically expressed in the egg and larval stage ofA.suum. Furthermore, two key target genes of miR-36f, named OST48 (gi|171279896) and cytochrome b (gi|76250725), were detected with qRT-PCR after the over- and down-regulation of miR-36f with the corresponding mimics and inhibitors.
Materials and methods
Larvae preparation
A.suumembryos were collected from the uteri of live adult females, which were obtained from slaughtered pigs in Lanzhou, Gansu Province of China. The embryos were incubated in a culture dish at 28 ℃ for 30 days to allow embryonic development. After that, eggshells were removed with 7.5% v/v sodium hypochlorite overnight at 37 ℃ and shaking with glass beads[21]. A total of 1.0×105larvae were obtained and cultured in DMEM (HyClone) containing 50% calf serum, penicillin G potassium (1 000 units/mL), amphotericin B (10 μg/mL), and streptomycin sulfate (1 mg/mL). The mixture was then evenly spread into a dish of 5 holes, and the miRNA mimics, inhibitors, mimics negative controls and inhibitor negative controls were added to the 4 holes respectively, with the last one left as blank control. The experimental nematodes were cultured at 37 ℃ with 5% CO2for 72 h following gently hand shook 2-3 times each day.
Animals
Fifty 8-week-old BALB/c mice of specific-pathogen-free (SPF) grade were purchased from Lanzhou Veterinary Research Institute Animal Center. The mice were equally divided into five groups, housed in sterile cages, and fed with pelleted food and wateradlibitum. And the mice were acclimatized to these conditions for one week before the experiment. Animal experiments were performed strictly according to the institutional guidelines for animal ethics of China.
Preparation of miRNA inhibitors, mimics and negative controls
The miRNA inhibitors, mimics, and their negative controls (Table 1) were obtained from Invitrogen (Shanghai) and prepared a final concentration of 20 μM by following the instructions respectively. Nucleotides of miRNA inhibitor and its control were modified with 2’-O-methyl.
Challenge infection and measurement of larvae
After soaking for 72 h, larvae of the 5 groups were collected separately. And the 5 groups BALB/c
Tab.1 Sequences of miRNA inhibitor, mimic and their negative controls
Note:aN.C indicates negative control.
mice were subjected to be orally infected with about 5 000 larvae for each. Baermann method was adopted to recover the larvae that migrated to the livers and lungs 4 days later[22]. In a nutshell, livers and lungs of the experimental and the control mice were collected and carefully ground with a mortar, and transferred to a small gauze bag containing 30 mL sterile physiological saline with kanamycin (100 mg/L) and ampicillin (100 mg/L) added, and incubated at 37 ℃ for 24 hours. The larvae recovered were counted and measured with a microscope having micrometer (Olympus) with 20 worms detected for each.
qRT-PCR of target genes after miRNA interventions
Two target genes named OST48 and cytochrome b were then quantified by qRT-PCR following the over-and/or down-regulation of miR-36f. Briefly, total RNA from the transfectedA.suumlarvae were extracted with Trizol reagent (Invitrogen) in accordance with the manufacture’s protocol. The purity of total RNA was confirmed by BioPhotometer plus (Eppendorf). Reverse transcription to obtain cDNA by using EasyScript First-Strand cDNA Synthesis SuperMix (Transgene, Beijing). Real-time quantitative PCR was conducted using an ABI PRISM®7500 Sequence Detection System. All of the primers (Table 2) were synthesized by Shenggong Co, Ltd., China. The actin gene (EU109284) was used as the endogenous control. The amplification cycle conditions were performed as follows: 95 ℃ for 5 min, followed by 40 cycles of 95 ℃ for 15 s, 65 ℃ for 15 s, and 72 ℃ for 32 s. The relative quantification of each miRNA and its target gene was calculated using the equation: N = 2-ΔCt, ΔCt = CtmiRNA-Ctactin[23]. All reactions were conductedin triplicate.
Tab.2 Sequences of primers used in the amplification of target and endogenous control genes
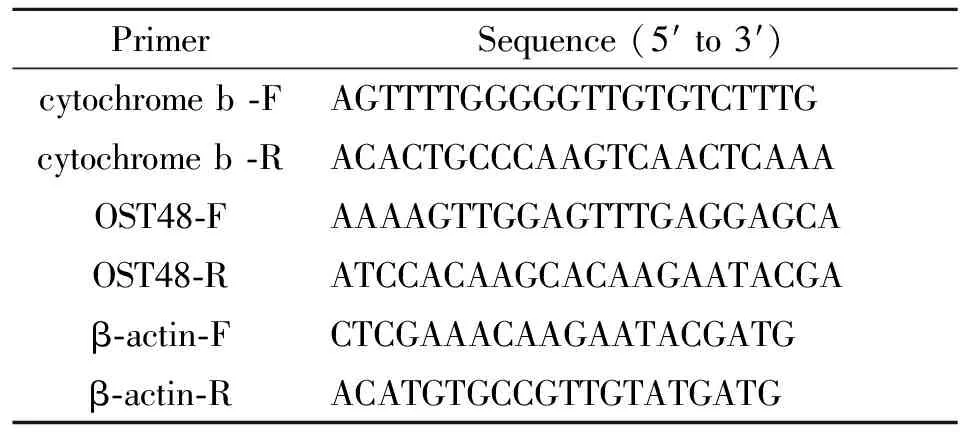
PrimerSequence(5′to3′)cytochromeb⁃FAGTTTTGGGGGTTGTGTCTTTGcytochromeb⁃RACACTGCCCAAGTCAACTCAAAOST48⁃FAAAAGTTGGAGTTTGAGGAGCAOST48⁃RATCCACAAGCACAAGAATACGAβ⁃actin⁃FCTCGAAACAAGAATACGATGβ⁃actin⁃RACATGTGCCGTTGTATGATG
Results and discussion
There was a significant difference between the inhibitor treated group and the control, including the larvae recovered number, body length and body width (P<0.05). For larvae recovered number, there was 42.5±23.72 larvae recovered from the inhibitor treated groups (P<0.05), while it was 69.44±34.86, 55.77±27.30, 67.86±27.82, 75.00±33.07 recovered from the mimics groups and other 3 control groups, respectively (Figure 1). For body length, it measures 367±133.29 μm in average in the inhibitor group, while that of controls is 460.82±165.12 μm in average (P<0.05) (Figure 2A). For the body width, it is 17.04±6.37 μm and 23.44±6.90 μm respectively for the inhibitor treated groups and controls (P<0.05)(Figure 2B). For the immune protection of the
host, most of the invasion larvae were killed with only those worms strongly enough to survive. Therefore, although 5 000 larvae were orally infected, only a small number of larvae were successfully recovered, similar to the report by Islam et al[24]. No significant differences were found (P>0.05) in mimics, which was similar to the study reported by Puthiyakunnon[25].This phenomenon might suggest that, for its specific and crucial regulating functions for the development and infectivity of larvae, miR-36f was also critically controlled by organism, and any changes over the saturation level will not obtain functional enhancement.
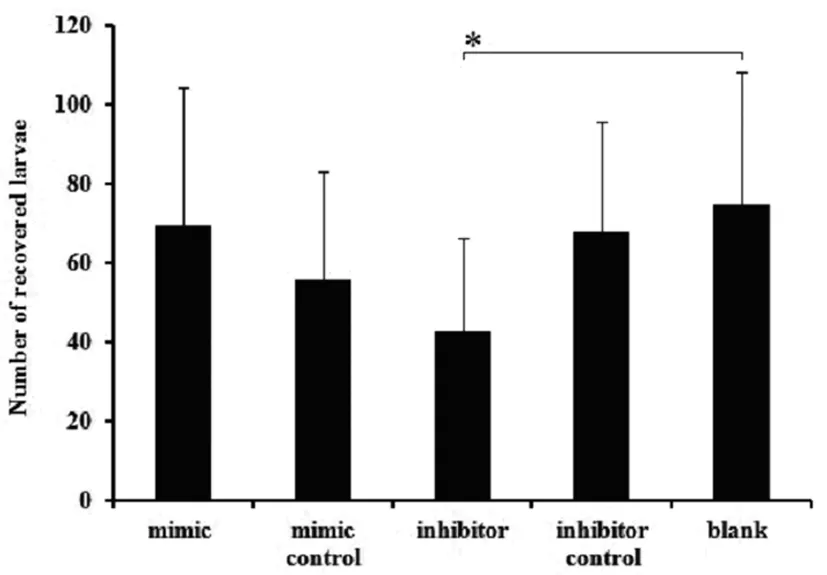
Data are expressed as the mean values±standard deviations in each group. The symbol"*" indicatesP<0.05.
Fig.1 Number of recoveredA.suumlarvae from the five experimental groups
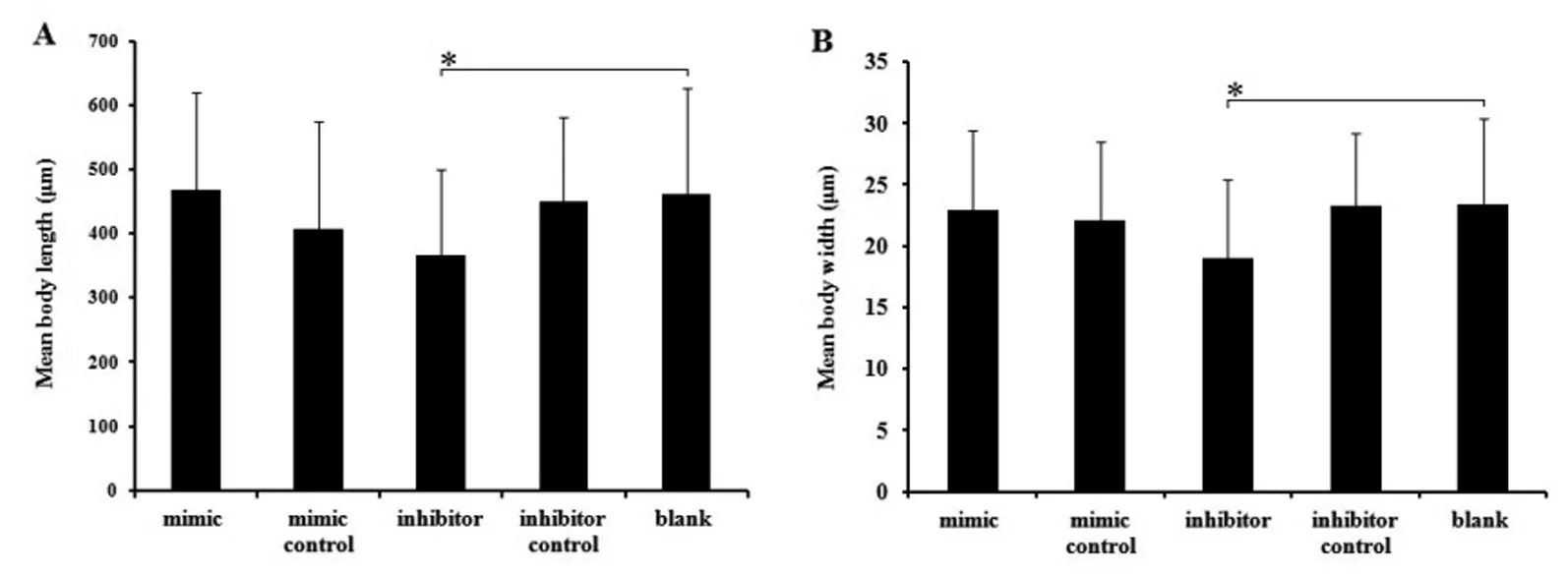
Data are expressed as the mean values±standard deviations in each group. The symbol"*" indicates P<0.05.Fig.2 Mean body length (A) and width (B) of recovered larvae from the five experimental groups
The expression of OST48 genes was significantly up-regulated after inhibitor treatment(P<0.05), and only slightly decreased after mimics treated (Figure 3A). In contrast to cytochrome b, OST48 was significantly reduced with the mimics (P<0.01), and only slightly increased after inhibitors treatment (P>0.05) (Figure 3B). The two target genes were obtained via enrichment analysis from a total of 216 target genes of miR-36f in our previous study, indicating essential roles of the two genes in the regulating network of miR-36f for the organisms.OST48 is an important part of the oligosaccharyl transferase which plays an important role in N-linked glycosylation[26-27]. And cytochrome b is involved in the oxidative phosphorylation, and is important for cellular metabolism[28-30]. When treated with mimics or inhibitors,miR-36f of the organism was influenced, leading to a changed regulation and metabolism balance, and the expression changes of targets. The phenomenon indicated that miR-36f did have functions of gene regulation in the worms. Besides, it was found that the two targets were not changed as similar trends, which confirmed the previous conclusion that cellular pathways might be regulated with one or more miRNAs[31].

A: Quantification of OST48. B: Quantification of cytochrome b. The symbol"*" indicates P<0.05; "**" indicates P<0.01.
Fig.3 Target gene relative expression analysis of groups interfered with mimics, inhibitors, negative control, inhibitor negative control and normal saline by qRT-PCR
Conclusions
By usingA.suumas model organism, we identified that the egg and larva stage-specific miRNA, miR-36f, involves in the infectivity and development of the parasite, which would help for better understanding and novel control strategy development of parasitic helminths.
References
[1]Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function[J]. Cell, 2004, 116(2): 281-297.
[2]Lai EC. MicroRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation[J]. Nat Genet, 2002, 30(4): 363-364.
[3]Ivashchenko AT, IssabekovaAS, Berillo OA. MiR-1279, miR-548j, miR-548m, and miR-548d-5p binding sites in CDSs of paralogous and orthologous PTPN12, MSH6, and ZEB1 genes[J]. Biomed Res Int, 2013, 2013: 902467. DOI: 10.1155/2013/902467
[4]da Sacco L, Masotti A. Recent insights and novel bioinformatics tools to understand the role of MicroRNAs binding to 5’untranslated region[J]. Int J Mol Sci, 2012, 14(1): 480-495. DOI: 10.3390/ijms14010480
[5]Wang J, Czech B, Crunk A, et al. Deep small RNA sequencing from the nematodeAscarisreveals conservation, functional diversification, and novel developmental profiles[J]. Genome Res, 2011, 21(9): 1462-1477. DOI: 10.1101/gr.121426.111
[6]de Wit E, Linsen SE, Cuppen E, et al. Repertoire and evolution of miRNA genes in four divergent nematode species[J].Genome Res, 2009, 19(11):2064-2074. DOI: 10.1101/gr.093781.109
[7]Poole CB, Davis PJ, Jin J, et al. Cloning and bioinformatic identification of small RNAs in the filarial nematode,Brugiamalayi[J]. Mol Biochem Parasitol, 2010, 169(2): 87-94. DOI: 10.1016/j.molbiopara.2009.10.004
[8]Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the PlanarianSchmidteamediterranea: a model system for stem cell biology[J]. RNA, 2006, 12(9): 1640-1649.
[9]Marco A, Kozomara A, Hui JH, et al. Sex-biased expression of microRNAs inSchistosomamansoni[J]. PLoS Negl Trop Dis, 2013, 7(9): e2402. DOI: 10.1371/journal.pntd.0002402
[10]Miska EA, Alvarez-Saavedra E, Abbott AL, et al. MostCaenorhabditiselegansmicroRNAs are individually not essential for development or viability[J]. PLoS Genet, 2007, 3(12): e215.
[11]Alvarez-Saavedra E, Horvitz HR. Many families ofC.elegansmicroRNAs are not essential for development or viability[J]. CurrBiol, 2010, 20(4): 367-373. DOI: 10.1016/j.cub.2009.12.051
[12]Holm SA, Sorensen CR, Thamsborg SM, et al. Gastrointestinal nematodes and anthelmintic resistancein Danish goat herds[J]. Parasite, 2014, 21: 37. DOI: 10.1051/parasite/2014038
[13]Martinez-Valladares M, Cordero-Perez C, Rojo-Vazquez FA. Efficacy of an antihelmintic combination in sheep infected withFasciolahepaticaresistant to albendazole and clorsulon[J]. ExpParasitol, 2014, 136: 59-62. DOI: 10.1016/j.exppara.2013.10.010
[14]Urban JF Jr, Hu Y, Miller MM, et al.Bacillusthuringiensis-derived Cry5B has potent anthelmintic activity againstAscarissuum[J]. PLoS Negl Trop Dis,2013, 7(6): e2263. DOI:10.1371/journal.pntd.0002263
[15]Buxton SK, Robertson AP, Martin RJ. Diethylcarbamazine increases activation of voltage-activated potassium (SLO-1) currents inAscarissuumand potentiates effects of emodepside[J]. PLoS Negl Trop Dis, 2014, 8(11): e3276. DOI: 10.1371/journal.pntd.0003276
[16]Nejsum P, Parker ED Jr, Frydenberg J, et al. Ascariasis is a zoonosis in Denmark[J]. J ClinMicrobiol, 2005, 43(3):1142-1148.
[17]Morimoto M, Zarlenga D, Beard H, et al.Ascarissuum: cDNA microarray analysis of 4th stage larvae (L4) during self-cure from the intestine[J]. Exp Parasitol, 2003, 104(3-4): 113-121.
[18]Jex AR, Liu S, Li B, et al.Ascarissuumdraft genome[J]. Nature, 2011, 479(7374): 529-533. DOI: 10.1038/nature10553
[19]Chehayeb JF, Robertson AP, Martin RJ, et al. Proteomic analysis of adultAscarissuumfluid compartments and secretory products[J]. PLoS Negl Trop Dis, 2014, 8(6): e2939. DOI: 10.1371/journal.pntd.0002939
[20]Ma X, Zhu Y, Li C, et al. Comparative transcriptome sequencing of germline and somatic tissues of theAscarissuumgonad[J]. BMC Genomics, 2011, 12:481. DOI: 10.1186/1471-2164-12-481
[21]Xu MJ, Chen N, Song HQ, et al.RNAi-mediated silencing of a novelAscarissuumgene expression in infective larvae[J]. Parasitol Res, 2010, 107(6): 1499-1503. DOI: 10.1007/s00436-010-2027-3
[22]Chen N, Xu MJ, Nisbet AJ, et al.Ascarissuum: RNAi mediated silencing of enolase gene expression in infective larvae[J]. Exp Parasitol, 2011, 127(1): 142-146. DOI: 10.1016/j.exppara.2010.07.019
[23]Sanchez-Freire V, Ebert AD, Kalisky T, et al. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns[J]. Nat Protoc, 2012, 7(5): 829-838. DOI:10.1038/nprot.2012.021
[24]Islam MK, Miyoshi T, Yamada M, et al. Pyrophosphatase of the roundwormAscarissuumplays an essential role in the worm’s molting and development[J]. Infect Immun. 2005, 73(4): 1995-2004.
[25]Puthiyakunnon S, Yao Y, Li Y, et al. Functional characterization of three microRNAs of the Asian tiger mosquito,Aedesalbopictus[J]. Parasit Vectors, 2013, 6(1): 230. DOI: 10.1186/1756-3305-6-230
[26]Mohorko E, Glockshuber R, Aebi M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation[J]. J Inherit Metab Dis, 2011, 34(4): 869-878. DOI:10.1007/s10545-011-9337-1
[27]Roboti P, High S. The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation[J]. J Cell Sci,2012, 125(Pt 14): 3474-3484. DOI: 10.1242/jcs.103952
[28]Barton V, Fisher N, Biagini GA, et al. InhibitingPlasmodiumcytochrome bc 1: a complex issue[J]. Curr Opin Chem Biol, 2010, 14(4): 440-446. DOI: 10.1016/j.cbpa.2010.05.005
[29]Tucker EJ, Wanschers BF, Szklarczyk R, et al. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression[J]. PLoS Genet, 2013, 9(12): e1004034. DOI: 10.1371/journal.pgen.1004034
[30]Hildenbeutel M, Hegg EL, Stephan K, et al. Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation[J]. J Cell Biol, 2014, 205(4): 511-524. DOI: 10.1083/jcb.201401009
[31]Mo YY. MicroRNA regulatory networks and human disease[J]. Cell Mol Life Sci, 2012, 69(21): 3529-3531. DOI: 10.1007/s00018-012-1123-1
Received:2015-09-03;Revision accepted:2015-10-26
Application of multi-locus sequence typing (MLST)in the identification ofEscherichiaalbertii
LIU Xiang1,XU Yan-mei2,WANG Bin1,DENG Jian-ping1,XIAO Bo1,SUN Song-song1,ZHOU Yang1,XIONG Yan-wen2,WANG Hong1
(1.ZigongCenterforDiseaseControlandPrevention,Zigong643000,China;2.StateKeyLaboratoryforInfectiousDiseasePreventionandControl,NationalInstituteforCommunicableDiseaseControlandPrevention,ChineseCenterforDiseaseControlandPrevention,Beijing102206,China)
We studied the application of multilocus sequence typing (MLST) in the discovery and identification of a new pathogenEscherichiaalbertii. MLST was performed according to theE.coliMLST database using seven housekeeping genes. The 7 housekeeping genes of 30 suspectedEscherichiaalbertiistrains isolated from retail raw meats were amplified, sequenced and analysed by DNA star software. The nucleotide sequences were uploaded toE.coliMLST database to confirm the allele number and the sequence type (ST type). To analyse the phylogenetic relationships between the 30 strains and the reference strains ofE.coli,E.fergusonii,E.albertii,Shigellaspp. andSalmonellaentericaserovar Typhi, the phylogenetic tree was constructed with the neighbor-joining method of MEGA 6. Results showed that 30 strains were classified into 7 new STs (16 strains) and 4 known STs (14 strains), the analysis of N-J showed high homology withE.albertiistrain, while were highly divergent fromE.coli,Shigellasp.,E.fergusoniiandSalmonellaentericaserotype Typhi stains. MLST accurately determine the relationship between species ofE.albertiiat the level of housekeeping genes. And it is the first time of discovering and confirming the existence ofE.albertiiin China.
multilocus sequence typing (MLST);E.albertii; new pathogens
s: Wang Hong, Email: 460973389@qq.com; Xiong Yan-wen, Email: xiongyanwen@icdc.cn
Effects of miR-36f on larval infection and development ofAscarissuum
FENG Sheng-yong1,FU Jing-hua1,2,SHAO Chang-chun1,3,ZHU Xing-quan1,XU Min-jun1,2
(1.StateKeyLaboratoryofVeterinaryEtiologicalBiology,KeyLaboratoryofVeterinaryParasitologyofGansuProvince,LanzhouVeterinaryResearchInstitute,ChineseAcademyofAgriculturalSciences,Lanzhou730046,China;2.CollegeofAnimalScience,SouthChinaAgriculturalUniversity,Guangzhou510642,China;3.CollegeofVeterinaryMedicine,YangzhouUniversity,Yangzhou225009,China)
自贡市重点科技计划项目(No.2013S06)和四川省卫生和计划生育委员会科研课题(No.150259)
王 红,Email: 460973389@qq.com; 熊衍文,Email: xiongyanwen@icdc.cn
1.四川省自贡市疾病预防控制中心,自贡 643000; 2.中国疾病预防控制中心传染病预防控制所,传染病预防控制国家重点实验室,北京 102206
10.3969/j.issn.1002-2694.2015.11.009
R378
A
1002-2694(2015)11-1033-04
2015-02-05;
2015-07-26

Supported by the Zigong Key Science and Technology Program (No. 2013S06) and Grant from the Health and Family Planning Commission of Sichuan Province (No. 150259)
