Simultaneous recovery of dual pathways for ammonia metabolism do not improve further detoxif i cation of ammonia in HepG2 cells
2013-06-01
Fuzhou, China
Simultaneous recovery of dual pathways for ammonia metabolism do not improve further detoxif i cation of ammonia in HepG2 cells
Fei-Yuan Zhang, Nan-Hong Tang, Xiao-Qian Wang, Xiu-Jin Li and Yan-Ling Chen
Fuzhou, China
BACKGROUND:Key enzyme def i ciency in the dual-pathway of ammonia metabolism leads to low detoxif i cation capacity of HepG2 cells. Previously, we established a HepG2/AFhGS cell line with overexpression of human glutamine synthetase (hGS) in pathway 1 and a HepG2/(hArgI+hOTC)4 cell line with overexpression of human arginase I (hArgI) and human ornithine transcarbamylase (hOTC) in pathway 2. The present study aimed to investigate whether simultaneous recovery of the two pathways contributes to the further improvement of ammonia detoxif i cation in HepG2 cells.
METHODS:We adopted a recombinant retrovirus carrying the hGS gene to infect HepG2/(hArgI+hOTC)4 cells and selected a new recombinant HepG2 cell line. The capacities of ammonia tolerance and detoxif i cation in cells were detected by biochemical methods. Cell cycle PCR chip was used to assess the changes of gene expression.
RESULTS:Introducing hGS into HepG2/(hArgI+hOTC)4 cells did not lead to hGS overexpression, but inhibited hArgI expression. The levels of synthetic glutamine and urea in HepG2/(hArgI+hOTC+AFhGS)1 cells were signif i cantly lower than those in HepG2/(hArgI+hOTC)4 cells when cultured in the medium with 10 and 15 mmol/L glutamate (Glu) and with 60 and 180 mmol/L NH4Cl, respectively. In addition, the comparison of different cell growth showed that HepG2/AFhGS cells signif i cantly lagged behind the other cells by the 5th and 7th day, indicating that introduction of hGS impedes HepG2 cell proliferation. Analysis of the mechanism suggested that the decreased expression of BCL2 played an important role.
CONCLUSIONS:This study demonstrated that the recovery of two ammonia metabolic pathways in HepG2 cells is not helpfulin increasing ammonia metabolism. The reinforcement of the pathway of urea metabolism is more important and valuable in improving the ammonia metabolism capacity in HepG2 cells.
(Hepatobiliary Pancreat Dis Int 2013;12:525-532)
glutamine synthetase;urea cycle; ammonia metabolism; liver cell
Introduction
Patients with acute hepatic failure waiting for liver recovery or transplantation require an effective bioartif i cial liver (BAL) system to serve as a temporary substitute for basic detoxif i cation and excretory functions.[1]The most important or central part of the system is composed of bioactive materials called hepatocytes.[2]
Primary human hepatocytes in culture may maintain liver-specif i c functions. However, they may quickly lose their abilities of proliferation. Although pig hepatocytes have physiological characteristics and metabolic functions similar to those of human hepatocytes, the presence of several defects cannot be ignored such as the antigenicity of secreted proteins and the possibility of porcine retrovirus infection.[3,4]The human hepatoblastomaderived cell line HepG2 may reveal a variety of typical hepatic cell-like features and possess certain synthesis and detoxif i cation capabilities. Therefore, HepG2 may become a promising cell source for the BAL system. HepG2 retains its functions and capability of inf i nite proliferation.[5]However, the diminished capability of ammonia metabolism limits the application of HepG2 cells. The decreased expressions of glutamine synthetase (GS) in pathway 1 and key enzyme arginase I (ArgI) and ornithine transcarbamylase (OTC) in pathway 2 root the metabolic def i ciency of ammonia. Enosawa et al[6,7]transferred the mouse GS (mGS) gene into HepG2 cellsand found that the growth of HepG2/mGS cells in the bioreactor signif i cantly extends the survival time of pig with ischemic liver failure. Mavri-Damelin et al[8]adopted instantaneous double-gene transfection and enabled the urea circulatory function of HepG2 cells to be partially recovered. We also established the HepG2/ AFhGS cell line[9](with a successful and stable expression of human GS) and HepG2/(hArgI+hOTC)4 cell line[10](with overexpressions of human ArgI and OTC), and found that the ammonia-reducing capacities of these cells are greatly increased compared with HepG2 cells.
Two metabolic pathways of ammonia coexist in hepatocytes, indicating that under normal circumstances, hepatocytes can automatically adjust the direction of metabolism according to the concentration of ammonia in the environment. They can come into a mutual relationship via their respective products as well. Nakamura et al[11]found that L-glutamine (Gln) has the function to protect gastric epithelial cellsin vitroin the case of high concentration of ammonia. The mechanism might involve the activation of ArgI which promotes the transformation from ammonia into urea. These studies showed that Gln, as a product of the metabolic pathway 1 of ammonia in the cells, might further improve the metabolic pathway 2 of ammonia. Based on these preliminary works, we hypothesized that the two pathways of ammonia metabolism in hepatocytes are not isolated. Unifying these two pathways might augment the metabolism of ammonia in HepG2 cells. Therefore, we infected HepG2/(hArgI+hOTC)4 cells with recombinant retrovirus carrying the hGS gene (including AFP promoter) and thus, HepG2 cells hold two metabolic pathways of ammonia. We evaluated the impact of the dual-pathway recovery of ammonia metabolism on the biological function of HepG2 cells.
Methods
Cell culture
The HepG2 cells of human hepatoblastoma-derived cell line were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in a high glucose DMEM medium supplemented with 10% fetal bovine serum in a 5% CO2incubator at 37 ℃. The PA317 cell line was maintained in our laboratory. The HepG2/con, HepG2/AFhGS, HepG2/ (hArgI+hOTC)4 cells, and the retrovirus vector pLNCX and pLNAFhGS were established previously.[9,10]
Establishment of recombinant HepG2 cells with simultaneous recovery of two metabolic pathways of ammonia
pLNCX and pLNAFhGS were transducted into the PA317 cell line by using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). The PA317/X and PA317/AFhGS cell clones were selected by G418 (200 μg/mL, Life Technologies, Carlsbad, CA, USA) and subcultured for amplif i cation. Furthermore, the supernatants of these cells containing different recombinant retroviruses were collected to infect HepG2/con and HepG2/(hArgI+ hOTC)4 cells respectively. By screening with G418 (1 mg/mL) for 15 days, the resistant cell clones including HepG2/Mock, HepG2/(hArgI+hOTC+AFhGS)1 and HepG2/(hArgI+hOTC+AFhGS)2 were obtained.
Measurement of hGS mRNA expression
Total RNA was extracted from HepG2, HepG2/ Mock, HepG2/AFhGS, HepG2/(hArgI+hOTC)4 and HepG2/(hArgI+hOTC+AFhGS)1 cells. The fragments of hGS (1139 bp) and AFhGS (2189 bp) were amplif i ed by PCR. The primer sequences were as follows: hGS, sense 5'-CCC AAG CTT ATG ACC ACC TCA GCA AGT T-3' and antisense (AS) 5'-CGG TTA ACT TAA TTT TTG TAC TGG AAG GGC-3'; AFhGS, sense 5'-CGG GAT CCG CCT GTC ATA CAG CTA ATA-3' and AS 5'-CGG TTA ACT TAA TTT TTG TAC TGG AAG GGC-3'; β-actin sense, 5'-TGC TGT CCC TGT ATG CCT CT-3' and AS 5'-GGT CTT TAC GGA TGT CAA CG-3'. The PCR product underwent electrophoresis and relative quantity intensity analysis, taking the ratio of hGS to β-actin.
Immunoblotting
Whole cell extracts were used for immunoblotting as described elsewhere[10]and they were visualized by antibodies against hGS (Abcam, Cambridge, UK), hArgI, hOTC, human carbamoyl phosphate synthetase I (hCPSI), human argininosuccinate synthetase (hASS), human argininosuccinate lyase (hASL), human arginase II (hArgII) and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) followed by HRP-labeled IgG and detected by enhanced chemiluminescence. The relative amount of each protein band was quantif i ed as a ratio to the β-actin band indicated underneath each gel using the densitometric scanning software Quantity One (BIO-RAD, Hercules, CA, USA).
Cell proliferation
In total, 5×103cells were cultured for 1, 3, 5 and 7 days in 96-well plates. Then, 100 μL of 1:10 (v/v) CCK-8 solution (Dōjindo Molecular Technologies, Kumamoto, Japan) was added to wells after incubation. Two hours later, the formazan formed were aspirated into another 96-well plate for absorbance measurement at 450 nm in a microplate reader.
Measurement of cell tolerance to ammonia poisoning
In total, 1×105cells were cultured in 96-well plates. After cell adherence, the control group (HepG2) was given the experimental medium, whereas the experimental groups received 30-180 mmol/L NH4Cl prepared with standard (STD) buffer containing 147 mmol/L Na+, 5 mmol/L K+, 131 mmol/L Cl-, 1.3 mmol/L Mg2+, 1.3 mmol/L, 2 mmol/L Ca2+, 25 mmol/L, 15 mmol/L 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES), and 20 mmol/L D-glucose (pH 7.4) at 37 ℃ for 12 hours. The supernatants in the 96-well plates were discarded and replaced with 100 μL of CCK-8 solution. OD450was measured for every well in a microplate reader. The median lethal dose (LD50) of NH4Cl was used to determine the ammonia tolerance capacity of the cells. For detecting the effects of Gln and glutamate (Glu) on cell tolerance to ammonia, different concentrations of Gln or Glu solution (0, 5, 10, 15, 20, and 25 mmol/L) were prepared using STD buffer with 120 mmol/L NH4Cl. The other procedures applied were the same as those described above.
Measurement of Gln and urea production, hArgI and hOTC enzymatic activity in cells
Gln and urea production, hArgI and hOTC enzymatic activities in cells were measured as reported previously.[10]
Cell cycle PCR chip microarray
We used a human cell cycle RT2Prof i ler PCR array (Qiagen, Duesseldorf, Germany) to simultaneously analyze the mRNA levels of 84 genes involved in regulation of cell cycle (Human Cell Cycle array, PAHS-020A) in HepG2, HepG2/AFhGS and HepG2/ (hArgI+hOTC)4 cells. A total of 107HepG2/AFhGS (test1), HepG2/(hArgI+hOTC)4 (test2) and HepG2 cells were harvested at day 3 ofin vitroculture for total RNA extraction. RNA quality was assessed using a RNeasy® MinElute™ kit (Qiagen) before cDNA synthesis using 1.5 μg total RNA per sample. The resulting cDNA was used as template to perform quantitative polymerase chain reaction (qPCR) analysis. Data were analyzed by the ∆∆cycle threshold method to determine the fold changes in expression. 3.0-fold change or more were considered differentially expressed between the test and control groups.
Among the differential expression genes screened by cDNA microarrays, CDC16, HERC5 and BCL2 genes were selected to be further conf i rmed by reverse transcriptase-polymerase chain reaction (RT-PCR). The fragments of CDC16 (458 bp), HERC5 (349 bp), BCL2 (339 bp) and β-actin (462 bp) cDNA were amplif i ed by PCR. The primer sequences were as follows: CDC16, sense 5'-GCA ATT GGG AAC GAG GTA AC-3' and AS 5'-GAG GTT TCC AAT GGC GTA AG-3'; HERC5, sense 5'-GGA GTA CCC TTG GCT CAG AT-3' and AS 5'-GCC ACC TTC CAC ATG CTA T-3'; BCL2, sense 5'-TTC GCC GAG ATG TCC AG-3' and AS 5'-GGC CAA ACT GAG CAG AGT CT-3'. The results were normalized with respect to β-actin.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 for Windows. The data were expressed as mean±SD. Differences between the groups were evaluated with ANOVA or factorial design ANOVA and considered statistically signif i cant whenP<0.05.
Results
Comparison of the fi ve cell lines for hGS expression
The hGS mRNA expression in HepG2/(hArgI+hOTC+ AFhGS)1 cells was relatively intensif i ed compared with that in HepG2/(hArgI+hOTC+AFhGS)2 (data not shown). As shown in Fig. 1A, the expression of hGS mRNA in HepG2/AFhGS and HepG2/(hArgI+hOTC+ AFhGS)1 cells is the most prominent (1139 bp), and the AFhGS mRNA with AFP promoter and enhancer is merely expressed in HepG2/AFhGS and HepG2/(hArgI+ hOTC+AFhGS)1 cells (2189 bp). In addition, the hGS expression in HepG2/AFhGS, HepG2/(hArgI+hOTC)4 and HepG2/(hArgI+hOTC+AFhGS)1 cells is relatively higher than that in HepG2 and HepG2/Mock cells (Fig. 1B).
Comparison of cell lines for ammonia tolerance and detoxif i cation
As shown in Fig. 2A, the LD50of NH4Cl for HepG2 and HepG2/Mock cells was about 60 mmol/L. LD50was between 120 mmol/L and 150 mmol/L for HepG2/AFhGS cells, 120 mmol/L for HepG2/(hArgI+hOTC+AFhGS)1 cells, and nearly 180 mmol/L for HepG2/(hArgI+hOTC)4 cells, respectively. The Gln production in HepG2, HepG2/ Mock, and HepG2/(hArgI+hOTC+AFhGS)1 cells in the presence of 10 and 15 mmol/L Glu was lower than that in HepG2/AFhGS and HepG2/(hArgI+hOTC)4 cells (Fig. 2B). The urea production in HepG2, HepG2/Mock, HepG2/AFhGS, and HepG2/(hArgI+hOTC+AFhGS)1 cells in the presence of 60-180 mmol/L NH4Cl was signif i cantly lower than that in HepG2/(hArgI+hOTC)4 cells with the same concentration of NH4Cl (Fig. 2C). These results indicated that the HepG2/(hArgI+hOTC)4 cells have the highest capacity for ammonia tolerance and detoxif i cation.
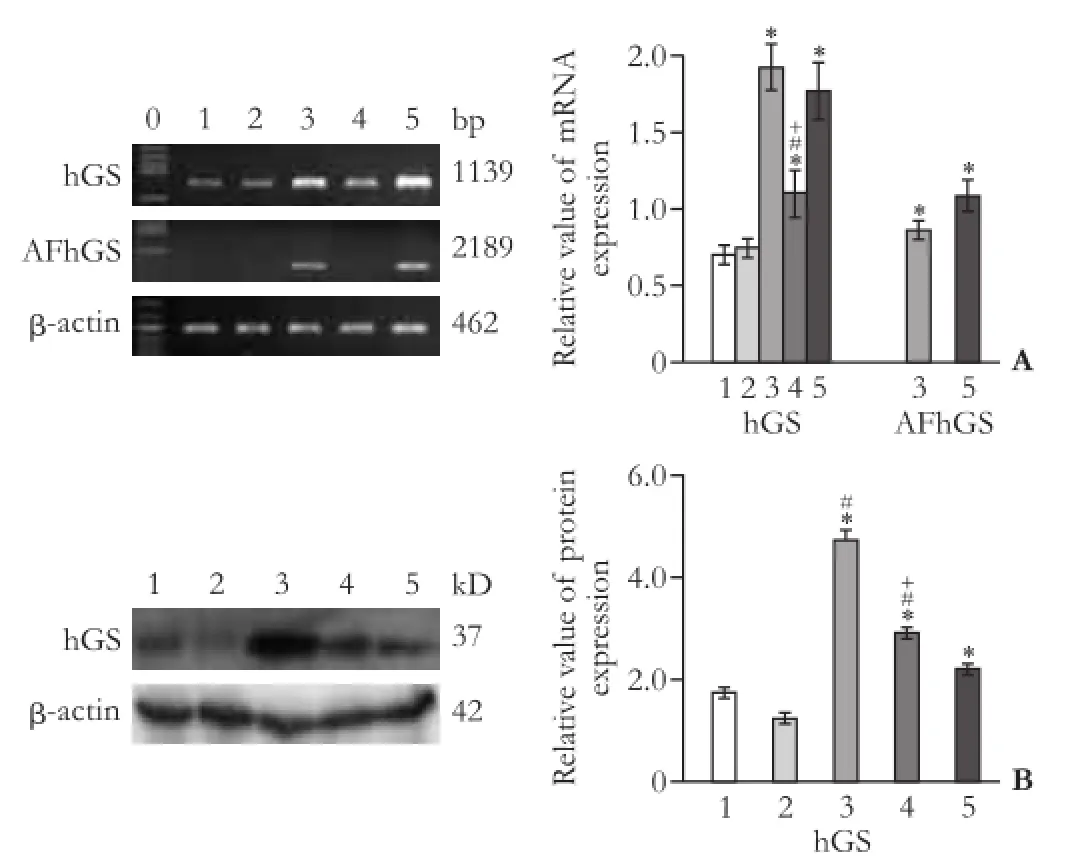
Fig. 1.Comparison of the five cell lines for hGS expression.A: Representative gel images from three independent studies for mRNA expression of hGS and AFhGS in fi ve cell lines detected by RT-PCR.B: Representative immunoblots from three independent studies for hGS and β-actin protein expressions in fi ve cell lines. The expression of HepG2/AFhGS cells is the most prominent. *:P<0.05, vs HepG2 and HepG2/Mock; #:P<0.05, vs HepG2/(hArgI+ hOTC+AFhGS)1; +:P<0.05, vs HepG2/AFhGS. 0: Marker; 1: HepG2; 2: HepG2/Mock; 3: HepG2/AFhGS; 4: HepG2/(hArgI+ hOTC)4; 5: HepG2/(hArgI+hOTC+AFhGS)1.
Impact of introduction of hGS on expression of urea metabolism-related enzymes
To clarify whether the effects of hGS transduction on the ammonia metabolism of HepG2 and HepG2/ (hArgI+hOTC)4 cells are related to the changes in urea metabolism, we compared the expressions of enzymes in urea cycle and ArgII proteins among the fi ve cell lines. The hGS expression in HepG2/AFhGS, HepG2/ (hArgI+hOTC)4, and HepG2/(hArgI+hOTC+AFhGS)1 cells was relatively higher than that in HepG2 and HepG2/Mock cells; the expression of hGS in HepG2/AFhGS cells was superior to that in other cells. Introducing hGS into HepG2 cells signif i cantly increased the expressions of hASL and hArgII, whereas its introduction into HepG2/(hArgI+hOTC)4 cells decreased the expressions of hArgI and hASL. The hArgI expression in HepG2/(hArgI+hOTC) 4 cells was higher than that in other four cell lines. The hOTC expression in HepG2/(hArgI+hOTC)4 and HepG2/ (hArgI+hOTC+AFhGS)1 cells was higher than that in HepG2, HepG2/Mock, and HepG2/AFhGS (Fig. 3A). Furthermore, we investigated whether introducing hGS has an impact on hArgI and hOTC functions and on the enzyme activity. The absorbance increased with a progressive increase in extracted protein concentration. The hArgI enzyme activity (protein concentration >16 μg/μL) in HepG2/(hArgI+hOTC)4 cells was higher than that in the other cells. When the protein concentration was >0.2 μg/μL, the activity of hOTC in HepG2/ (hArgI+hOTC)4 and HepG2/(hArgI+hOTC+AFhGS)1 cells was higher than that in HepG2, HepG2/Mock,and HepG2/AFhGS cells (Fig. 3B and C). These results suggested that introducing hGS does not promote the expressions of enzymes in the urea metabolic pathway and may not be conducive to ammonia metabolism.
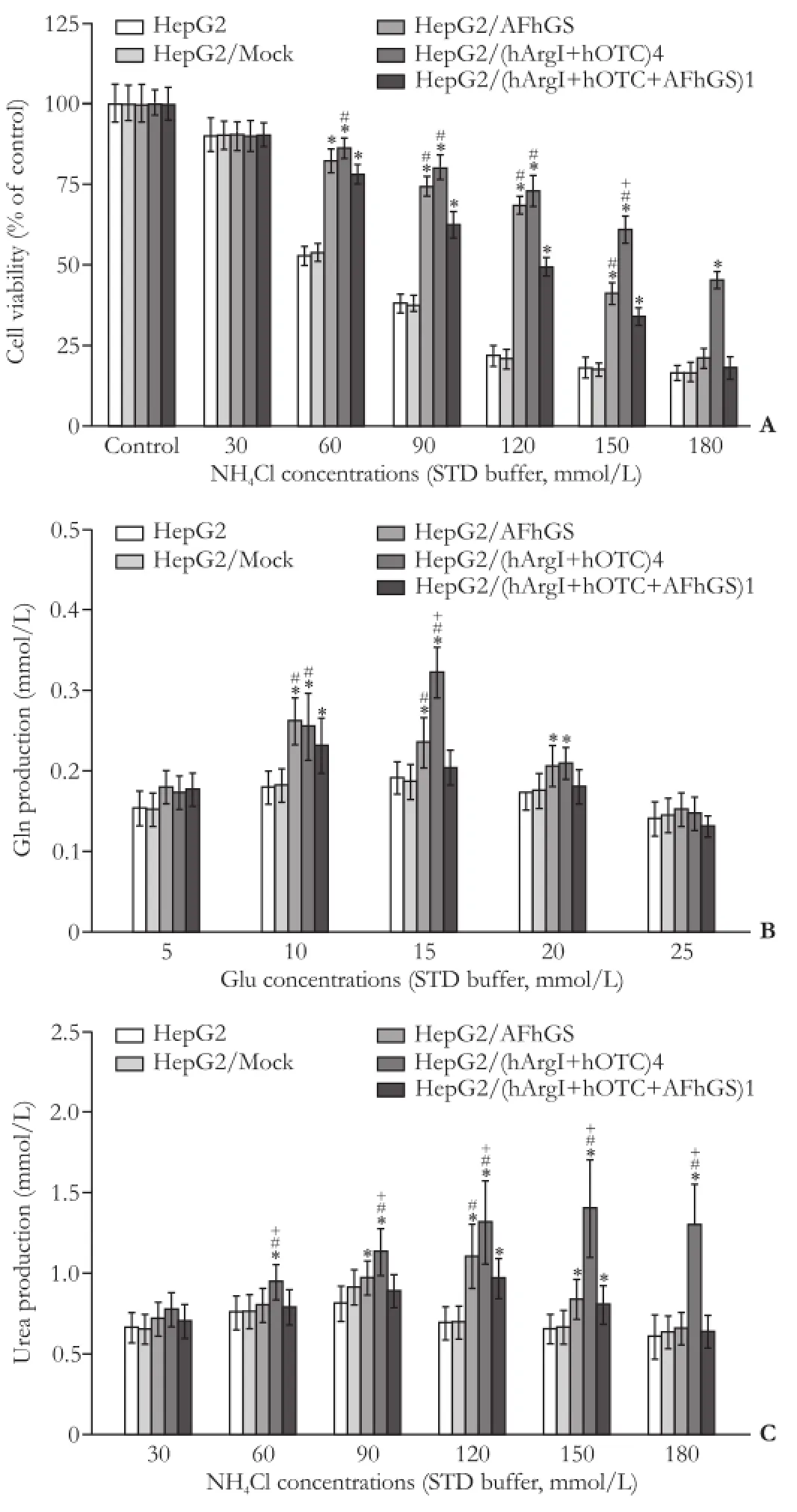
Fig. 2.Comparison of cell lines for ammonia tolerance and detoxif ication.A: Cell viability with NH4Cl at different concentrations. Data represent the mean±SD (n=4) of cell viability.B: Comparison of synthetic amounts of Gln produced by the different cell lines. Data represent the mean±SD (n=3) of Gln production.C: Comparison of urea production. Data represent the mean±SD (n=3) of urea production. *:P<0.05, vs HepG2 and HepG2/Mock; #:P<0.05, vs HepG2/(hArgI+hOTC+AFhGS)1; +:P<0.05, vs HepG2/AFhGS.
Proliferation capacities of cell lines in different culture conditions
The transduction of gene into cells may lead to changes in cell proliferation. Therefore, we compared the growth of the fi ve cells in different culture conditions. The viability of all cells rose with increasing Gln concentration in the presence of 120 mmol/L NH4Cl and peaked when the Gln concentration was 25 mmol/L (Fig. 4A). Meanwhile, the cell viability peaked in thepresence of 120 mmol/L NH4Cl and 10 or 15 mmol/L Glu (Fig. 4B). However, on day 3 after culture under the normal condition, the proliferation of HepG2/AFhGS cells began to slow down; on days 5 and 7, the growth of HepG2/AFhGS cells was markedly lower than that of the other cells (P<0.05) (Fig. 4C). These results indicate that hGS overexpression can protect cells in an environment with a high concentration of ammonia by producing more Gln but suppresses cell viability under normal culture conditions with prolonged incubation time.
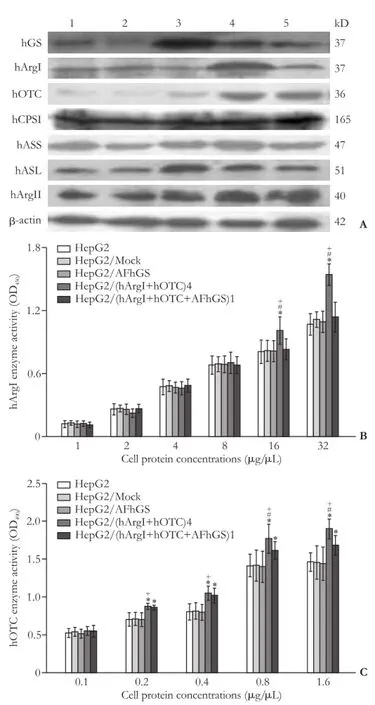
Fig. 3.Impact of the introduction of hGS on the expression of urea metabolism-related enzymes.A: Representative immunoblots from three independent studies for hGS, hArgI, hOTC, hCPSI, hASS, hASL and hArgII. 1: HepG2; 2: HepG2/Mock; 3: HepG2/AFhGS; 4: HepG2/(hArgI+hOTC)4; 5: HepG2/(hArgI+ hOTC+AFhGS)1.B,C: Data represent the mean±SD (n=3) of hArgI and hOTC enzyme activities. *:P<0.05, vs HepG2 and HepG2/Mock; #:P<0.05, vs HepG2/(hArgI+hOTC+AFhGS)1; +:P<0.05, vs HepG2/AFhGS.

Fig. 4.Cell proliferation curves.A: Data represent the mean ±SD (n=4) of cell viability. *:P<0.05, vs 0 mmol/L Gln.B: Data represent the mean±SD (n=4) of cell viability. *:P<0.05, vs 0 mmol/L Glu.C: Data represent the mean±SD (n=4) of cell proliferation. *:P<0.05, vs HepG2, HepG2/Mock, HepG2/(hArgI+hOTC)4, and HepG2/ (hArgI+hOTC+AFhGS)1 cells.

Fig. 5.Analysis of cell cycle PCR chip and its validation.A: Genes with which the ratio of the change multiple of HepG2/AFhGS cells to that of HepG2 cells significantly increased were HERC5 (+3.18) and CDC16 (+3.46), whereas that with which the ratio decreased was BCL2 (-26.18).B: The gene with which the ratio of the change multiple of HepG2/(hArgI+hOTC)4 cells to that of HepG2 cells increased signif i cantly was CDC16 (+4.59).C: mRNA expression levels and relative expression intensities of BCL2, HERC5, and CDC16 in the HepG2, HepG2/Mock, HepG2/AFhGS, HepG2/(hArgI+hOTC)4, and HepG2/(hArgI+hOTC+AFhGS)1 cells. Data represent the mean±SD (n=3) of relative mRNA expression. *:P<0.05, vs HepG2 and HepG2/Mock; #:P<0.05, vs HepG2/(hArgI+hOTC+AFhGS)1; +:P<0.05, vs HepG2/AFhGS. 0: Marker; 1: HepG2; 2: HepG2/Mock; 3: HepG2/AFhGS; 4: HepG2/ (hArgI+hOTC)4; 5: HepG2/(hArgI+hOTC+AFhGS)1.
Analysis of cell cycle PCR chip and its validation
Based on the above fi ndings, we adopted cell cycle PCR chip to analyze the changes in cell cycle-related genes in HepG2, HepG2/AFhGS, and HepG2/(hArgI+ hOTC)4 cells. The ratio of the change multiple of relevant genes in HepG2/AFhGS to HepG2 and that in HepG2/(hArgI+hOTC)4 to HepG2 are shown in panels A and B of Fig. 5, respectively. The gene expressions of BCL2, HERC5, and CDC16 changed signif i cantly. We further verify the mRNA expressions of these genes in the HepG2, HepG2/Mock, HepG2/AFhGS, HepG2/ (hArgI+hOTC)4, and HepG2/(hArgI+hOTC+AFhGS)1 cells (Fig. 5C). The results showed that decreased proliferation of HepG2/AFhGS cells is related to BCL2, HERC5, or CDC16.
Discussion
The liver is the main organ for the ammonia metabolism. Key enzymes involved in the two metabolic pathways of ammonia include GS expressed in the cytoplasm of liver cells around the hepatic vein and CPSI, OTC, ASS, ASL, and ArgI, which participate in the ornithine cycle of hepatocytes around the portal vein.[12]In this study, we tried to improve the metabolism of ammonia in HepG2 cells by co-overexpression of the metabolic enzymes of ammonia. The introduction of GS played varied roles in the pathway enzymes of urea metabolism but did not improve any metabolic pathways of urea.
Although HepG2/AFhGS and HepG2/(hArgI+hOTC+ AFhGS)1 cells showed higher ammonia tolerances and better ammonia-reducing capacities than HepG2 (parent cell), they were not as effective as HepG2/(hArgI+ hOTC)4 cells. We speculated that these fi ndings might be due to the following reasons. 1) Differences in hGS expression. Overexpression of hGS mRNA and protein was found in HepG2/AFhGS cells, which is signif i cantly increased compared with HepG2 cells. The hGS expression in HepG2/(hArgI+hOTC)4 cells was higher than that in HepG2 cells.[10]However, the introduction of AFhGS [HepG2/(hArgI+hOTC+AFhGS)1 cells] did not increase hGS protein expression although hGS mRNA expression was augmented. This illustrated that the introduction of hArgI and hOTC into HepG2cells had brought the two metabolic pathways of ammonia into equilibrium, and the re-transcripted hGS gene might be affected by the translational level and could not achieve better results; 2) The introduction of hGS had negative impact on the pathway of urea metabolism. The hArgI expression in HepG2/AFhGS and HepG2/(hArgI+hOTC+AFhGS)1 cells was decreased more signif i cantly than HepG2/ (hArgI+hOTC)4 cells. However, the hOTC expression in HepG2/(hArgI+hOTC+AFhGS)1 cells, as well as the hASL expression in HepG2/AFhGS and HepG2/ (hArgI+hOTC+AFhGS)1 cells, was increased. Thus, the impact of the introduction of hGS into HepG2 and HepG2/(hArgI+hOTC)4 cells on the key enzymes of the urea metabolic pathway was the sacrif i ce of hArgI; 3) The introduction of hGS promoted hArgII expression. As another important reaction of urea synthesis in cells, hArgII was expressed in the mitochondria of a variety of tissues,[13,14]and directly catalyzed arginine and produced urea, instead of being directly involved in the metabolism of ammonia. In any environment with high ammonia concentration, the resultant arginine may be transformed into urea through hArgII for HepG2/(hArgI+hOTC)4 cells with relatively sound urea pathways, achieving more complete metabolism; 4) In the two metabolic pathways of ammonia, GS is characterized by high aff i nity. However, in any environment with low ammonia concentration, GS prioritizes Gln synthesis.[15]In any environment with a high ammonia concentration, the key enzymes of urea synthesis are speculated to play a major role and to prioritize urea synthesis.
In this study, we also investigated the protective effect of Gln on hepatocytes in an environment with a high concentration of ammonia and compared the effect of Gln and Glu on the fi ve cell lines. In any environment with a high concentration of ammonia, cellular activity was increased with the elevation of Gln concentration, which conformed the protective effect of Gln on cells.[11]However, cellular activity increased with certain Glu concentration, higher Glu concentration inhibited cellular activity. Glu possibly reacts with ammonia and generates Gln at a certain concentration, which promotes the survival of cells. However, Gln generated at the cellular level is very limited and plays an insignif i cant role in promoting cell survival. The increase of Glu concentration might inhibit the absorption of cystine and reduce the level of intracellular glutathione and thus, activate the Ca2+pathway and eventually lead to apoptosis.[16]In addition, although Gln plays an important role in cell growth, some studies have shown that it is closely associated with the incidence of hepatic encephalopathy, and therefore, Gln is a pathological factor.[17]The overexpression of hGS in liver failure may not be a favorable factor.
Cell proliferation assays showed that HepG2/AFhGS cell growth slowed down on the 3rd day and signif i cantly lagged behind the other cells by the 5th and 7th day. Presumably, hGS introduction induces apoptosis. This hypothesis was conf i rmed by the results of cell cycle PCR chip. HepG2/AFhGS cells have three kinds of gene abnormalities. The expression levels of CDC16[18,19]and HERC5,[20]which are involved in cell division, increased to a certain extent. However, BCL2[21], an apoptosis inhibitor, signif i cantly decreased. In addition, because hGS is an ATP-dependent enzyme, hGS overexpression consumes excessive amounts of ATP,[22]which is also another cause of apoptosis.
In summary, a series of experiments in this study proved that improving the overall ammonia-reducing capacity of HepG2 cells through the recovery of the two metabolic pathways of ammonia is not practical. We believe the recovery of the urea metabolic pathway is more important and more valuable in improving the ammonia-reducing capacity in HepG2 cells. In future animal experiments, we will also evaluate the practical value of recombinant HepG2/(hArgI+hOTC)4 cells and choose the best material of cells for the development of a BAL liver system.
Contributors:TNH and CYL contributed to the conception and design of research. ZFY, WXQ and LXJ performed experiments. ZFY and WXQ analyzed data and prepared fi gures. ZFY and TNH interpreted results of experiments and drafted manuscript. TNH is the guarantor.
Funding:This study was supported by grants from the National Natural Science Foundation of China (30972926), the Professor's Academic Development Foundation of Fujian Medical University (JS11004), and the Natural Science Foundation of Fujian Province (2013J01309).
Ethical approval:Not needed.
Competing interest:No benef i ts in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Gerlach JC, Zeilinger K, Patzer Ii JF. Bioartif i cial liver systems: why, what, whither? Regen Med 2008;3:575-595.
2 Nibourg GA, Huisman MT, van der Hoeven TV, van Gulik TM, Chamuleau RA, Hoekstra R. Stable overexpression of pregnane X receptor in HepG2 cells increases its potential for bioartif i cial liver application. Liver Transpl 2010;16:1075-1085.
3 Frühauf JH, Mertsching H, Giri S, Frühauf NR, Bader A. Porcine endogenous retrovirus released by a bioartif i cial liver infects primary human cells. Liver Int 2009;29:1553-1561.
4 Nyberg SL, Hibbs JR, Hardin JA, Germer JJ, Platt JL, Paya CV, et al. Inf l uence of human fulminant hepatic failure sera on endogenous retroviral expression in pig hepatocytes. Liver Transpl 2000;6:76-84.
5 Tsiaoussis J, Newsome PN, Nelson LJ, Hayes PC, Plevris JN. Which hepatocyte will it be? Hepatocyte choice for bioartif i cial liver support systems. Liver Transpl 2001;7:2-10.
6 Enosawa S, Miyashita T, Suzuki S, Li XK, Tsunoda M, Amemiya H, et al. Long-term culture of glutamine synthetase-transfected HepG2 cells in circulatory fl ow bioreactor for development of a bioartif i cial liver. Cell Transplant 2000;9:711-715.
7 Enosawa S, Miyashita T, Saito T, Omasa T, Matsumura T. The signif i cant improvement of survival times and pathological parameters by bioartif i cial liver with recombinant HepG2 in porcine liver failure model. Cell Transplant 2006;15:873-880.
8 Mavri-Damelin D, Eaton S, Damelin LH, Rees M, Hodgson HJ, Selden C. Ornithine transcarbamylase and arginase I def i ciency are responsible for diminished urea cycle function in the human hepatoblastoma cell line HepG2. Int J Biochem Cell Biol 2007;39:555-564.
9 Tang NH, Wang XQ, Li XJ, Chen YL. Ammonia metabolism capacity of HepG2 cells with high expression of human glutamine synthetase. Hepatobiliary Pancreat Dis Int 2008;7: 621-627.
10 Tang N, Wang Y, Wang X, Zhou L, Zhang F, Li X, et al. Stable overexpression of arginase I and ornithine transcarbamylase in HepG2 cells improves its ammonia detoxif i cation. J Cell Biochem 2012;113:518-527.
11 Nakamura E, Hagen SJ. Role of glutamine and arginase in protection against ammonia-induced cell death in gastric epithelial cells. Am J Physiol Gastrointest Liver Physiol 2002; 283:G1264-1275.
12 Häussinger D, Schliess F. Glutamine metabolism and signaling in the liver. Front Biosci 2007;12:371-391.
13 Yu H, Iyer RK, Kern RM, Rodriguez WI, Grody WW, Cederbaum SD. Expression of arginase isozymes in mouse brain. J Neurosci Res 2001;66:406-422.
14 Mavri-Damelin D, Damelin LH, Eaton S, Rees M, Selden C, Hodgson HJ. Cells for bioartif i cial liver devices: the human hepatoma-derived cell line C3A produces urea but does not detoxify ammonia. Biotechnol Bioeng 2008;99:644-651.
15 Häussinger D, Sies H, Gerok W. Functional hepatocyte heterogeneity in ammonia metabolism. The intercellular glutamine cycle. J Hepatol 1985;1:3-14.
16 Li Y, Maher P, Schubert D. Phosphatidylcholine-specif i c phospholipase C regulates glutamate-induced nerve cell death. Proc Natl Acad Sci U S A 1998;95:7748-7753.
17 Albrecht J, Dolińska M. Glutamine as a pathogenic factor in hepatic encephalopathy. J Neurosci Res 2001;65:1-5.
18 Wang Q, Moyret-Lalle C, Couzon F, Surbiguet-Clippe C, Saurin JC, Lorca T, et al. Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene 2003;22:1486-1490.
19 Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, et al. TFDP1, CUL4A, and CDC16 identif i ed as targets for amplif i cation at 13q34 in hepatocellular carcinomas. Hepatology 2002;35:1476-1484.
20 Kroismayr R, Baranyi U, Stehlik C, Dorf l eutner A, Binder BR, Lipp J. HERC5, a HECT E3 ubiquitin ligase tightly regulated in LPS activated endothelial cells. J Cell Sci 2004;117:4749-4756.
21 Ruvolo PP, Deng X, May WS. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 2001;15:515-522.
22 Kubota Y, Kato K, Dairaku N, Koike T, Iijima K, Imatani A, et al. Contribution of glutamine synthetase to ammoniainduced apoptosis in gastric mucosal cells. Digestion 2004;69: 140-148.
Received December 12, 2012
Accepted after revision June 5, 2013
AuthorAff i liations:Fujian Institute of Hepatobiliary Surgery, Fujian Medical University Union Hospital, Fuzhou 350001, China (Zhang FY, Tang NH, Wang XQ, Li XJ and Chen YL)
Nan-Hong Tang, PhD, Fujian Institute of Hepatobiliary Surgery, Fujian Medical University Union Hospital, Fuzhou 350001, China (Fax: 86-591-83357896; Email: fztnh@sina.com)
© 2013, Hepatobiliary Pancreat Dis Int. All rights reserved.
10.1016/S1499-3872(13)60083-1
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Diagnostic accuracy of enhanced liver fi brosis test to assess liver fi brosis in patients with chronic hepatitis C
- Clinical and economic consequences of pancreatic fi stula after elective pancreatic resection
- Retrohepatic vena cava deroof i ng in living donor liver transplantation for caudate hepatocellular carcinoma
- Mattress sutures for the modif i cation of end-toend dunking pancreaticojejunostomy
- Risk factors and clinical characteristics of portal vein thrombosis after splenectomy in patients with liver cirrhosis
- Optimal central venous pressure during partial hepatectomy for hepatocellular carcinoma
