Cytokine and apoptosis gene polymorphisms influence the outcome of hepatitis C virus infection
2011-07-03LeilaKsiaaCheikhrouhouImenSfarHajerAounallahSkhiriHoudaAouadiSalwaJendoubiAyedTaiebBenAbdallahKhaledAyedandYousrLakhouaGorgi
Leila Ksiaa Cheikhrouhou, Imen Sfar, Hajer Aounallah-Skhiri, Houda Aouadi, Salwa Jendoubi-Ayed, Taieb Ben Abdallah, Khaled Ayed and Yousr Lakhoua-Gorgi
Tunis, Tunisia
Cytokine and apoptosis gene polymorphisms influence the outcome of hepatitis C virus infection
Leila Ksiaa Cheikhrouhou, Imen Sfar, Hajer Aounallah-Skhiri, Houda Aouadi, Salwa Jendoubi-Ayed, Taieb Ben Abdallah, Khaled Ayed and Yousr Lakhoua-Gorgi
Tunis, Tunisia
BACKGROUND:Hepatitis C virus (HCV) infection is thought to be chronic and the factors leading to viral clearance or persistence are poorly understood. This study was undertaken to investigate the possibility of a significant relationship between the spontaneous clearance or the persistence of hepatitis C virus (HCV) infection and cytokine and apoptosis gene polymorphisms in Tunisian patients on hemodialysis.
METHODS:Polymorphisms of the genes IL-1 (-889 IL-1α, -511 and +3954 IL-1β, IL-1Ra), IL-18 (-137 and -607), IL-12 (-1188) and Apo1/Fas (-670) were determined by PCR-RFLP, PCR-SSP and PCR-VNTR in 100 healthy blood donors and 100 patients infected with HCV and undergoing hemodialysis. The patients were classified into two groups: G1 consisted of 76 active chronic hepatitis patients (positive for HCV RNA) and G2 consisted of 24 hemodialysed patients who spontaneously eliminated the virus (negative for HCV RNA).
RESULTS:The frequency of genotype association [-137GC/ -607CA] IL-18 was higher in G2 (41.7%) than in G1 (15.8%) (P=0.008; OR=0.26; 95% CI, 0.10-0.73). We also found a higher frequency of the AA genotype of the Apo1/Fas gene in G2 (41.6%) than in G1 (17.5%) (P=0.026; OR=3.49; 95% CI, 1.13-10.69). Adjustment for known covariate factors (age, gender and genotype) confirmed these univariate findings and revealed that the genotype association GC-CA of the (-137 and -607) IL-18 gene and the AA genotype of the Apo1/Fas gene were associated with the clearance of HCV (P=0.041 and 0.017, respectively).
CONCLUSION:The two genotypes GC-CA of the (-137 and -607) IL-18 polymorphism and the AA genotype of the Apo1/ Fas gene influence the outcome of HCV infection in Tunisian patients on hemodialysis.
(Hepatobiliary Pancreat Dis Int 2011; 10: 280-288)
hepatitis C virus; spontaneous clearance; cytokine gene polymorphisms; Apo1/Fas gene polymorphisms
Introduction
In 80% of infected subjects, chronic hepatitis C virus (HCV) infection leads to serious consequences including liver cirrhosis and hepatocellular carcinoma (HCC). A minority of patients is able to clear the virus.[1-3]HCV persists despite the presence of specific humoral and cellular immune responses[4]and the factors leading to viral clearance or persistence are poorly understood. But some studies showed that the outcome might already be determined at an early time point after infection.[5]In early HCV infection, a vigorous CD4+T cell response is associated with viral clearance. In contrast, patients developing a chronic infection show a predominant Th2 response. These findings indicate that the ability to mount an efficient cellular immune response is the main mechanism responsible for HCV control, while a defect in this response leads to chronicity.[5-8]The main biological mediators in this immune response are cytokines, lowmolecular weight-peptides produced by a large variety of cells that have a broad range of physiological functions.[9]As the indispensability of cytokines and the cellular immune response in the pathogenesis and eradication of chronic HCV has become clearer, the importance of the host immune response has also been proposed.[10]
The capacity for cytokine production in individuals has a major genetic component. This has been ascribed to polymorphisms within the regulatory regions of cytokine genes. IL-1 is a proinflammatory cytokine that playsa key role in initiating and amplifying the immune response during initial infection.
The most important members of the IL-1 family are IL-1α, IL-1β, and IL-1 receptor antagonist (IL-1Ra). IL-1Ra is a natural competitive inhibitor of IL-1, acting by occupying the cell surface receptor without triggering signal transduction.[11]Genes encoding IL-1 are located in the 450 kb region of chromosome 2q13-14, consisting of three homologous genes, IL-1A, IL-1B, and IL-1RN. Biallelic polymorphisms at positions -889 of the IL-1α gene, and -511 and +3954 of the IL-1β gene have been described, all representing a single nucleotide C/T polymorphism (SNP).[11,12]IL-1Ra contains an 86bp variable number tandem repeat (VNTR) polymorphism in intron 2.[11,13]Being involved in the proinflammatory cytokine network, IL-18 and IL-12 are key cytokines, which exert a synergic effect[14]and are thought to be regulators of cytokine expression, inducing interferon gamma (IFNγ), and enhancing the cytotoxicity of NK cells.[15-18]In the presence of IL-12, IL-18 stimulates IFNγ expression, promoting the Th1-mediated immune response, whereas, without IL-12, IL-18 stimulates Th2 responses. SNPs at position 1188A/C in the 3' untranslated region (UTR) of the IL-12 gene mapped to chromosome 5q31-33[19]and at position -137G/ C and -607C/A in the promoter of the IL-18 gene located on chromosome 11q22.2-q22.3[20]were found to correlate with many diseases.[18,21,22]Recently, apoptosis has been implicated in the pathogenesis of several human diseases.[23]As cytotoxic T lymphocytes play a crucial role in the clearance of viral infection, it has been demonstrated that Fas-based mechanisms account for all T-cell mediated cytotoxicity.[24]Apo1/Fas, one of the mediators of apoptosis, also known as CD95, induces an apoptotic signal in susceptible target cells.[25,26]Gene expression can be regulated by a number of genetic elements located in the 5' upstream region of the gene. A polymorphism located at the -670G/A nucleotide within the enhancer region has been described on the 5' flanking region of the Apo1/Fas gene.[27,28]
In this study, we investigated the possible involvement of functional polymorphisms in IL-1α, IL-1β, IL-1Ra, IL-12, IL-18, and Apo1/Fas in the development of HCV infection in Tunisian patients on hemodialysis.
Methods
Patients and controls
This retrospective study involved 100 HCV-infected individuals with confirmed antibodies to HCV. Patients dialysed during 2004 at different hemodialysis centers (private and public) in various regions of Tunisia were divided into two groups: group 1 (G1) consisting of 76 consecutive patients with persistent infection as assessed by positive HCV RNA for more than one year; and group 2 (G2) consisting of 24 consecutive subjects considered to have spontaneously recovered from HCV infection as suggested by two negative HCV-PCR detections one year apart. Data from each patient included age at diagnosis, gender, and possible risk factors for HCV (such as transfusion and invasive procedures) and were obtained at the hemodialysis centers (Table 1). All patients were negative for hepatitis B surface antigen and human immunodeficiency virus. None of the patients had receivedtreatment for HCV infection before entering the study.
As the control group, we studied 100 ethnically and geographically matched healthy individuals, with evidence of negative tests for HCV antibodies, recruited from blood donors.
All patients and controls gave informed consent to participate in this study, which was approved by the Ethics Committee of Charles Nicolle Hospital in Tunis.
HCV RNA detection
HCV RNA in serum was detected by reverse transcriptase PCR (Inno-Lipa HCV II, Innogenetics, Belgium) according to the manufacturer's instructions. The patients who were HCV-PCR positive on the initial assessment and became consistently HCV-PCR negative were classified HCV-PCR negative.
Typing HCV
HCV genotypes were determined by inverse hybridization using a specific oligonucleotide probe assay (HCV Genotype Assay Lipa Innogenetics).
DNA extraction
Blood samples were collected in EDTA, and DNA was isolated by the Salting-Out method reported by Miller et al.[29]

Table 2. Sequence of primers and PCR conditions
IL-1α, IL-18, and IL-12 genotyping
Polymorphisms were determined using polymerase chain reaction sequence-specific primer (PCR-SSP) methodology with the primers listed in Table 2, and specific allelic variants were identified by PCR amplification. The PCR reaction of all these SNPs was carried out in a final volume of 20 μl with 2 μl 10×PCR buffer, 1.5 mmol MgCl2, 0.2 mmol dNTP, 50 ng/μl genomic DNA, 5 pmol each primer, and 2 U Taq polymerase (Promega, USA). For the two SNPs at positions -137 and -607 of the IL-18 gene, a control forward primer at 2.5 pmol was used covering the polymorphic sites, as an internal positive amplification control. PCR products were visualized by 2% agarose gel electrophoresis stained by ethidium bromide.
IL-1βand Apo1/Fas genotyping
Polymorphisms at positions -511 and +3954 of the IL-1β gene and -670 of the Apo1/Fas gene were genotyped by polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). Each PCR was carried out in a 30 μl reaction mixture containing 100 ng/μl genomic DNA, 0.2 mmol dNTP, 1.5 mmol MgCl2, 10 pmol each primer, and 2 U Taq polymerase. PCR amplification was carried out in a thermal cycler PE 9700 for IL-1β, and 9600 for Apo1/Fas, using the parameters (Table 2). All PCR products were confirmed by 2% agarose gel electrophoresis. Five microliters of reaction mixture from each sample was digested with 4 U AvaI restriction enzyme for IL-1β (-511) at 37 ℃ for 2 hours and 30 minutes, 5 U TaqI restriction enzyme for IL1β (+3954) at 65 ℃ for 5 hours, and 5 U BstoI restriction enzyme for -670 Apo1/Fas at 60 ℃ for 2 hours and 30 minutes. These products were loaded into 3% agarose gels and stained by ethidium bromide.
IL-1Ra genotyping
IL1-Ra intron 2 containing a penta-allelic pairtandem repeat polymorphism was analyzed by PCR. The PCR conditions were the same as for PCR-RFLP. PCR products were sized by electrophoresis on a 2% agarose gel stained with ethidium bromide. The IL-1Ra alleles were coded as follows: allele 1=4 repeats, allele 2=2 repeats, allele 3=5 repeats, allele 4=3 repeats, and allele 5=6 repeats.
Statistical analysis
Genotype and allele frequencies were performed using the Epi-stat program (version 6; Center for Disease Control and Prevention, Atlanta, GA). Statistical comparisons were made between the different groups of patients and controls by the Chi-square test calculated on 2×2 contingency tables. The Fisher's exact test was used when expected cell values were less than 5. A P value less than 0.05 was considered statistically significant. The strength of the association between genotypes or alleles in each group was estimated by calculating the odds ratios (OR) and 95% confidence intervals (CI) using the same software. Logistic regression models were used with clearance of HCV infection (HCV RNA negative) as the response variable to evaluate the relationships with the different factors (including confounders) and to estimate adjusted ORs (Exp(B)).
Results
The clinical and virological characteristics are summarized in Table 1. Our results showed a greater proportion of HCV-PCR positivity in females (55.3%) than in the HCV-PCR negative group (41.7%). However, this difference was not statistically significant. The 1b genotype was predominant in G1 (65.8%) and G2 (72.7%). The risk factors were similar in both groups; none of the patients was an intravenous drug user, 91 patients (91%) had a history indicating risk for exposure to HCV, because of blood transfusion (55%) or surgical or invasive procedures (36%). No significant differences were found between the two groups in distribution according to the region, private or public nature of the hemodialysis center, and the duration of hemodialysis.
All analysed allele frequencies and genotype distributions were in Hardy-Weinberg equilibrium both in the patients and controls.
Frequencies of IL-1 gene cluster
The genotype and allele frequencies at positions -889 of the IL-1α gene, -511 and +3954 of the IL-1β gene, and the IL-1Ra gene are summarized in Table 3. Comparison of genotype and allele frequencies in all HCV-infected patients and controls did not show any significant difference in the IL-1α or the IL-1β gene polymorphisms. Analysis of these frequencies in the two groups of hemodialysed patients showed a decreased frequency of C -889 IL-1α, C -511 IL-1β, and C +3954 IL-1β alleles in G1 (67.8%, 54.6% and 66.4% respectively) compared with G2 (75%, 60.4% and 79.2%, respectively). However, these differences were not statistically significant. Only four of the five kinds of polymorphism of IL-1Ra were found in this study. Allele 1 IL-1Ra was the most frequent in our study groups, followed by allele 2, but no significant differences were found between the patient groups and controls.
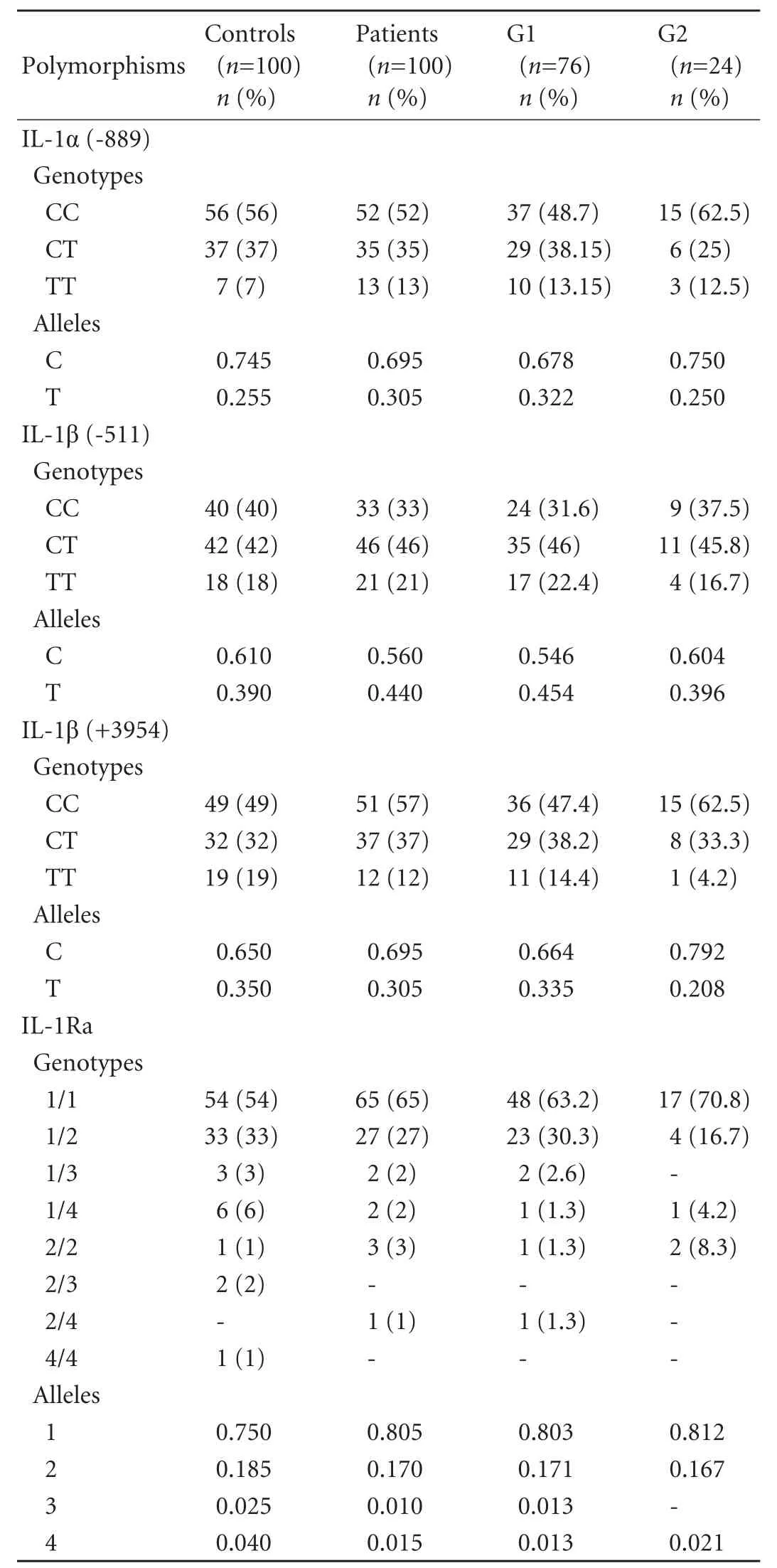
Table 3. Genotype and allele frequencies of IL-1α (-889), IL-1ß (-511 and +3954) and IL-1Ra polymorphisms in HCV-infected patients and controls
Frequencies of IL-12 gene polymorphism
The genotype and allele frequencies at SNP 1188C/A of IL-12 are shown in Table 4. Comparison of genotype and allele frequencies in all HCV-infected patients and controls did not reveal a significant difference. However, comparison of these frequencies accordingto the development of HCV infection revealed a nonsignificant decrease of the A allele frequency in G1 (36.9%) compared with G2 (41.7%).

Table 4. Genotype and allele frequencies of IL-12 1188C/A, IL-18 (-137G/C and -607C/A) and Apo1/Fas gene polymorphisms in HCV-infected patients and controls
Frequencies of IL-18 gene polymorphisms
The genotype and allele frequencies at SNPs -137 GC and -607 CA of IL-18 are shown in Table 4. No significant differences were found in the genotype distribution or in allele frequencies between all HCV-infected patients and controls. When transmission of these genetic variants was analyzed in the two groups of hemodialysed patients, the -137GC genotype was present at a lower frequency in G1 (27.6%) compared with the controls (44%) (P=0.038; OR=2.06; 95% CI, 1.04-4.11). The frequency of this genotype and the CA genotype of IL-18 -607 were also decreased in G1 compared with G2 (27.6% vs. 50%; P=0.043; OR=2.62; 95% CI, 0.92-7.49 and 38.1% vs. 62.5%; P=0.037; OR=2.70; 95% CI, 2.70-7.77, respectively). The transmission of both -137GC and -607CA genotypes in the two groups of patients showed a predominance in G2 (41.7%) compared with G1 (15.8%) (P=0.008; OR=0.26; 95% CI, 0.10-0.73; data not shown). Multivariate analysis showed that the genotypes -137GC and -607CA were significantly associated with spontaneous clearance of HCV after adjustment for age and gender (Exp(B), 3.52; 95% CI, 1.11-11.21; P=0.033) and even when considering the genotype as a confounder (G2, n=11) (Exp(B), 6.02; 95% CI, 1.07-33.78; P=0.041).
Frequencies of Apo1/Fas gene polymorphism
The genotypic distribution and allelic frequencies of Apo1/Fas gene polymorphism (Table 4) in patients and healthy controls did not show any significant difference. However, our results showed a significantly increased frequency of the GA genotype and a decreased frequency of the AA genotype in G1 (59.2% and 17.5%, respectively) compared with G2 (29.2% and 41.6%, respectively) (P=0.019; OR=0.28; 95% CI, 0.09-0.84 and P=0.026; OR=3.49; 95% CI, 1.13-10.69).
Adjustment for known covariate factors (age and gender in a first model and age, gender and genotype in a second model (G2,n=11) confirmed these univariate findings and revealed that the AA genotype of the Apo1/Fas gene was associated with the clearance of HCV ((Exp(B), 4.36; 95% CI, 1.30-14.54; P=0.017) in the first model and (Exp(B), 10.56; 95% CI, 1.39-80.00; P=0.022) in the second model).
Discussion
Our data demonstrate the association between IL-18 and Apo1/Fas gene polymorphisms and the clinical course of HCV infection in a Tunisian population. We chose to examine polymorphisms in the IL-1, IL-12, IL-18, and Apo1/Fas genes, which have demonstrated correlations with cytokine production and have been associated with a range of clinical outcomes in HCV infection, including viral clearance. IL-1 is a multifunctional cytokine with high inflammatory activity. Polymorphisms at positions -889 of the IL-1α gene, -511 and +3954 of the IL-1β gene, and the VNTR of the IL-1Ra gene, have potential functional importance by modulating IL-1 protein production.[11]These loci have been genetically associated with a wide range of diseases. In a Chinese study, the IL-1β -511 TT genotype was associated with susceptibility to gastric cancer,[30]and the IL-1Ra 2/2 genotype was correlated with the same cancer in a Spanish study.[31]In addition, the IL-1Ra 2 allele seems to play a role in protection against HBV infection in a Chinese population.[32]However, little is known about the role of these polymorphisms in HCV infection. A Japanese study reported an association of the IL-1β -511 TT genotype with an increased risk of developing HCV-related HCC.[33]In our study, none of these polymorphisms was associated with clearance, persistence or susceptibility to HCV infection, which is in agreement with other British and Pakistani studies,[34-36]which compared these polymorphisms with spontaneous or treated clearance of HCV infection.
On the other hand, IL-12 is a key regulator of the immune response to Th1 or Th2 categories and plays a role in autoimmune and infectious diseases.[15]IL-12 might therefore have an effect on the pathogenesis of HCV infection by affecting Th1/Th2 balance. It is the major cytokine responsible for differentiation of Th1 cells, which are potent producers of IFNγ. The SNP at position 1188 in the untranslated region of the IL-12 gene is associated with immune-mediated diseases such as type 1 diabetes.[21]A study on the functional characteristics of this polymorphism in an HCV-infected Chinese population shows a decreased frequency of the AA genotype and an increased frequency of the CA genotype in self-limited HCV infection.[16]In a German cohort, subjects who are chronically infected and carriers of the C allele seem to have a better chance of clearing HCV infection after antiviral therapy.[37]Our data clearly exclude a major influence of IL-12 polymorphisms on the natural course of acute HCV infection, because of the non-significant decrease of the A allele frequency between patients with chronic HCV infection and those with spontaneous clearance. In particular, either heterozygosity or homozygosity for the IL-12 1188 CA genotype conferred an increased risk for the development of chronic HCV infection despite recent reports showing impaired IL-12 production in these genotypes.[21,38]Our cohort wassmall and it is possible that it was underpowered.
IL-18, first described as an IFNγ-inducing factor,[17]is a pleiotropic cytokine involved in the regulation of innate and acquired immune responses, playing a key role in autoimmune, inflammatory, and infectious diseases. Potentially, the G/C polymorphism at position -137 could play a major role in the expression of the IL-18 gene.[20]Individuals with the CC genotype at position -137 had higher levels of IL-18 mRNA than other genotypes. Zhang et al[18]reported that the -137C allele is associated with protection against HBV infection in a Chinese cohort. Recently, the same research group suggested that these polymorphisms influence the outcome of HCV infection, with the -607A and -137C alleles and their haplotypes favoring HCV clearance.[39]In our study, the two heterozygous -137GC and -607CA genotypes were correlated with viral clearance. This association was confirmed in multivariate analysis taking age, gender and genotype as confounders. These results suggest that these genotypes exert a synergistic effect on clearance, probably by inducing IFNγ production, which must be necessary during elimination of viral infection. However, the wide confidence intervals reflect the relatively small numbers in the G2 subgroup, with a consequent loss of statistical power. Moreover, lack of analysis of a polymorphic gene effect on protein expression creates additional limits in concluding about the real cause or effect relationship.
A recent report[40]indicated that apoptosis of hepatocytes induced via the Fas system contributes to the elimination of infected cells. In HCV infection, Fas expression in hepatocytes is upregulated in accordance with the severity of liver inflammation.[41]When HCV-specific T cells migrate into hepatocytes and recognize the viral antigen via the T cell receptor, they become activated and express Fas ligand that can transduce the apoptotic death signal to hepatocytes.[23,24]This might be one of the reasons why the A allele or the AA genotype could play a positive role in the immune regulatory system of patients and protect against the development and progression of disease. Our results show that the distribution of genotype and allele frequencies of Apo1/Fas are in agreement with recent Korean[28]and Australian[27]studies, and demonstrate that the AA Apo1/Fas genotype is associated with viral clearance whatever the genotype studied. The association of the GA genotype with HCV persistence supports the previous hypothesis[42-44]that the Apo1/ Fas -670 G allele is likely to affect the progression of HCV infection. Nevertheless, a larger patient cohort is required to reduce the width of the confidence intervals and confirm these observations.
Further functional explorations like the response to HCV antigen stimulation by using peripheral blood mononuclear cells are needed to confirm these results.
We also analyzed the possible role of virus-related factors. The HCV 1b genotype has been shown to play a role in the outcome of HCV-related chronic liver disease and the response to therapy.[2,45]A Japanese study[46]showed a high frequency of this genotype (75%) in persistently infected subjects similar to the increased frequency of the 1b genotype found in our HCVPCR positive group (65.8%). In fact, this genotype is associated with more severe chronic liver disease and a less favorable response to interferon.[1,45]However, clearance of HCV was not found to be associated with genotype 1b. The route of infection could be an important factor in chronic infection. On the other hand, infection frequently occurs through parenteral exposure. In Morocco,[47]blood transfusion is the main risk factor for contamination by HCV (84.6%). In our study, blood transfusion was found in 66.1% of G1 patients and in 77.7% of G2. In fact, our study consisted of hemodialysis subjects who probably had repeated exposure through multiple transfusions that provided larger virus inoculation. However, it is difficult to consider this factor as a cause of HCV persistence, because the number of individuals transfused was the same in both groups of patients. In addition, the high risk of nosocomial HCV transmission in hemodialysed patients suggests a further prospective cohort including patients infected by HCV with normal renal function in order to precisely define the impact of these polymorphisms on the outcome of HCV infection.
In summary, our data suggest that polymorphisms of the IL-18 and the Apo1/Fas genes should be added to the spectrum of immunogenetic factors known to be involved in the outcome of HCV infection in hemodialysed patients. The lack of association between IL-1 and IL-12 gene polymorphisms and the development of HCV infection should be further investigated by large population-based studies. These findings will be useful in the near future to develop appropriate therapeutic strategies for controlling viral hepatitis C and to improve current knowledge about hepatitis C prognosis.
Funding:This study was supported by a grant from the Tunisian Kidney Transplantation Research Fund.
Ethical approval: Not needed.
Contributors: KCL proposed the study and wrote the first draft. ASH analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. LGY is the guarantor.
Competing interest: No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci 2006;3:47-52.
2 Czepiel J, Biesiada G, Mach T. Viral hepatitis C. Pol Arch Med Wewn 2008;118:734-740.
3 Poynard T. Natural History of hepatitis C. Feuillets de biologie 2002;248:19-21. http://cat.inist.fr/?aModele=afficheN&cpsidt= 13935055.
4 Pawlotsky JM. Hepatitis C virus and immune response. Gastroenterol Clin Biol 2001;25:B123-133.
5 Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A 2002;99:15661-15668.
6 Chang KM, Thimme R, Melpolder JJ, Oldach D, Pemberton J, Moorhead-Loudis J, et al. Differential CD4(+) and CD8(+) T-cell responsiveness in hepatitis C virus infection. Hepatology 2001;33:267-276.
7 Zarski JP, Souvignet C. Physiopathology of HCV infection. Gastroenterol Clin Biol 2002;26:B154-162.
8 Cramp ME, Carucci P, Rossol S, Chokshi S, Maertens G, Williams R, et al. Hepatitis C virus (HCV) specific immune responses in anti-HCV positive patients without hepatitis C viraemia. Gut 1999;44:424-429.
9 Dharancy S, Conti F, Desreumaux P, Paris JC, Calmus Y. Cytokine imbalance during hepatitis C virus infection. Consequences in hepatic pathophysiology. Gastroenterol Clin Biol 2001;25:595-604.
10 Fainboim L, Chernavsky A, Paladino N, Flores AC, Arruvito L. Cytokines and chronic liver disease. Cytokine Growth Factor Rev 2007;18:143-157.
11 Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev 2009;20:43-59.
12 Tseng LH, Chen PJ, Lin MT, Singleton K, Martin EG, Yen AH, et al. Simultaneous genotyping of single nucleotide polymorphisms in the IL-1 gene complex by multiplex polymerase chain reaction-restriction fragment length polymorphism. J Immunol Methods 2002;267:151-156.
13 Lennard A, Gorman P, Carrier M, Griffiths S, Scotney H, Sheer D, et al. Cloning and chromosome mapping of the human interleukin-1 receptor antagonist gene. Cytokine 1992;4:83-89.
14 Nakahira M, Ahn HJ, Park WR, Gao P, Tomura M, Park CS, et al. Synergy of IL-12 and IL-18 for IFN-gamma gene expression: IL-12-induced STAT4 contributes to IFN-gamma promoter activation by up-regulating the binding activity of IL-18-induced activator protein 1. J Immunol 2002;168:1146-1153.
15 Sun QL, Ran W. Review of cytokine profiles in patients with hepatitis. World J Gastroenterol 2004;10:1709-1715.
16 Yin LM, Zhu WF, Wei L, Xu XY, Sun DG, Wang YB, et al. Association of interleukin-12 p40 gene 3'-untranslated region polymorphism and outcome of HCV infection. World J Gastroenterol 2004;10:2330-2333.
17 Kohyama M, Saijyo K, Hayasida M, Yasugi T, Kurimoto M, Ohno T. Direct activation of human CD8+ cytotoxic T lymphocytes by interleukin-18. Jpn J Cancer Res 1998;89:1041-1046.
18 Zhang PA, Wu JM, Li Y, Yang XS. Association of polymorphisms of interleukin-18 gene promoter region with chronic hepatitis B in Chinese Han population. World J Gastroenterol 2005;11:1594-1598.
19 Huang D, Cancilla MR, Morahan G. Complete primary structure, chromosomal localisation, and definition of polymorphisms of the gene encoding the human interleukin-12 p40 subunit. Genes Immun 2000;1:515-520.
20 Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol 2001;112:146-152.
21 Davoodi-Semiromi A, Yang JJ, She JX. IL-12p40 is associated with type 1 diabetes in Caucasian-American families. Diabetes 2002;51:2334-2336.
22 Kretowski A, Mironczuk K, Karpinska A, Bojaryn U, Kinalski M, Puchalski Z, et al. Interleukin-18 promoter polymorphisms in type 1 diabetes. Diabetes 2002;51:3347-3349.
23 Wattre P. Apoptosis and virus (Apart from the retrovirus). Revue Française des Laboratoires, mars 1999;311:43-49. doi:10.1016/j.physletb.2003.10.071.
24 Marianneau P, Flamand M, Courageot MP, Deubel V, Desprès P. Cell death by apoptosis in response to infection Dengue virus: consequences in the viral pathogenesis? Annales de Biologie Clinique 1998;56:395-405.http://cat. inist.fr/?aModele=afficheN&cpsidt=2344819.
25 Krammer PH. The CD95(APO-1/Fas)/CD95L system. Toxicol Lett 1998;102-103:131-137.
26 Choi C, Benveniste EN. Fas ligand/Fas system in the brain: regulator of immune and apoptotic responses. Brain Res Brain Res Rev 2004;44:65-81.
27 Huang QR, Morris D, Manolios N. Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol 1997;34: 577-582.
28 Seo JC, Han SW, Yin CS, Koh HK, Kim CH, Kim EH, et al. Evaluation of a Apo-1/Fas promoter polymorphism in Korean stroke patients. Exp Mol Med 2002;34:294-298.
29 Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215.
30 Zeng ZR, Hu PJ, Hu S, Pang RP, Chen MH, Ng M, et al. Association of interleukin 1B gene polymorphism and gastric cancers in high and low prevalence regions in China. Gut 2003;52:1684-1689.
31 Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Natl Cancer Inst 2002;94:1680-1687.
32 Zhang PA, Li Y, Xu P, Wu JM. Polymorphisms of interleukin-1B and interleukin-1 receptor antagonist genes in patients with chronic hepatitis B. World J Gastroenterol 2004;10:1826-1829.
33 Tanaka Y, Furuta T, Suzuki S, Orito E, Yeo AE, Hirashima N, et al. Impact of interleukin-1beta genetic polymorphisms on the development of hepatitis C virus-related hepatocellular carcinoma in Japan. J Infect Dis 2003;187:1822-1825.
34 Constantini PK, Wawrzynowicz-Syczewska M, Clare M, Boron-Kaczmarska A, McFarlane IG, Cramp ME, et al. Interleukin-1, interleukin-10 and tumour necrosis factor-alpha gene polymorphisms in hepatitis C virus infection: an investigation of the relationships with spontaneous viral clearance and response to alpha-interferon therapy. Liver 2002;22:404-412.
35 Minton EJ, Smillie D, Smith P, Shipley S, McKendrick MW, Gleeson DC, et al. Clearance of hepatitis C virus is not associated with single nucleotide polymorphisms in the IL-1, -6, or -10 genes. Hum Immunol 2005;66:127-132.
36 Abbas Z, Moatter T, Hussainy A, Jafri W. Effect of cytokine gene polymorphism on histological activity index, viral load and response to treatment in patients with chronic hepatitis C genotype 3. World J Gastroenterol 2005;11:6656-6661.
37 Mueller T, Mas-Marques A, Sarrazin C, Wiese M, Halangk J, Witt H, et al. Influence of interleukin 12B (IL12B) polymorphisms on spontaneous and treatment-induced recovery from hepatitis C virus infection. J Hepatol 2004;41: 652-658.
38 Morahan G, Huang D, Wu M, Holt BJ, White GP, Kendall GE, et al. Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet 2002;360:455-459.
39 An P, Thio CL, Kirk GD, Donfield S, Goedert JJ, Winkler CA. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J Infect Dis 2008;198:1159-1165.
40 Oksuz M, Akkiz H, Isiksal YF, Saydaogluc G, Serin M, Kayaselcuk F, et al. Expression of Fas antigen in liver tissue of patients with chronic hepatitis B and C. Eur J Gastroenterol Hepatol 2004;16:341-345.
41 Neuman MG, Benhamou JP, Marcellin P, Valla D, Malkiewicz IM, Katz GG, et al. Cytokine--chemokine and apoptotic signatures in patients with hepatitis C. Transl Res 2007;149:126-136.
42 Bel Hadj Jrad B, Mahfouth W, Bouaouina N, Gabbouj S, Ahmed SB, Ltaief M, et al. A polymorphism in FAS gene promoter associated with increased risk of nasopharyngeal carcinoma and correlated with anti-nuclear autoantibodies induction. Cancer Lett 2006;233:21-27.
43 Zhu Q, Wang T, Ren J, Hu K, Liu W, Wu G. FAS-670A/ G polymorphism: A biomarker for the metastasis of nasopharyngeal carcinoma in a Chinese population. Clin Chim Acta 2010;411:179-183.
44 Ben Aleya W, Sfar I, Mouelhi L, Aouadi H, Makhlouf M, Ayed-Jendoubi S, et al. Association of Fas/Apo1 gene promoter (-670 A/G) polymorphism in Tunisian patients with IBD. World J Gastroenterol 2009;15:3643-3648.
45 Pawlotsky JM. Hepatitis C virus resistance to antiviral therapy. Hepatology 2000;32:889-896.
46 Kuzushita N, Hayashi N, Moribe T, Katayama K, Kanto T, Nakatani S, et al. Influence of HLA haplotypes on the clinical courses of individuals infected with hepatitis C virus. Hepatology 1998;27:240-244.
47 Benammar L, Rhou H, Ezzaitouni F, Kouider N, OuzeddounN, Bayahya R, et al. Hepatite C virus in hemodialysis patients CHU RABAT, prevalence and risk factors. Médecine du Maghreb 2001;89:17-20. http://www.santetropicale.com/ resume/8904.pdf.
Received September 28, 2010
Accepted after revision December 21, 2010
Author Affiliations: Immunology Research Laboratory of Kidney Transplantation and Immunopathology (Laboratoire de Recherche: LR03SP01), Charles Nicolle Hospital, Thunis, Tunisia (Ksiaa Cheikhrouhou L, Sfar I, Aouadi H, Jendoubi-Ayed S, Ben Abdallah T, Ayed K and Lakhoua-Gorgi Y), and National Health Institute, Tunis, Tunisia (Aounallah-Skhiri H)
Yousr Lakhoua-Gorgi, Professor, Laboratory of Immunology, Charles Nicolle Hospital, Boulevard 9 Avril, 1006 Tunis, Tunisia (Tel: 0021671578055; Fax: 0021671561156; Email: yousr.gorgi@rns.tn)
© 2011, Hepatobiliary Pancreat Dis Int. All rights reserved.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Hepatobiliary & Pancreatic Diseases International (HBPD INT)
- Iron overload and HFE gene mutations in Polish patients with liver cirrhosis
- Predictive value and main determinants of abnormal features of intraoperative cholangiography during cholecystectomy
- Emergency re-routing of anterior sector venous outflow for right lobe living donor liver transplantation including the middle hepatic vein
- Long-term survival of a patient after resection of a gastrointestinal stromal tumor arising from the pancreas
- Persistent port-site sinus in a patient after laparoscopic cholecystectomy: watch out for gallbladder tuberculosis
