High Expression of ERCC1 Is a Poor Prognostic Factor in Chinese Patients with Non-small Cell Lung Cancer Receiving Cisplatin-based Therapy
2010-01-08QingzhiGuoJieWangHuaBaiTongtongAnMeinaWuJunZhaoLuYangJianchunDuanYuyanWangZhijieWang
Qing-zhi Guo,Jie Wang,Hua Bai,Tong-tong An,Meina Wu,Jun Zhao,Lu Yang Jian-chun Duan,Yu-yan Wang,Zhi-jie Wang
Key laboratory of Carcinogenesis and Translational Research (Ministry of Education),Department of Medical Oncology,Peking University Cancer Hospital & Institute,Beijing 100142,China
INTRODUCTION
Lung cancer is a most common malignant tumor worldwide.According to the World Health Organization (WHO)in 2002,there are 1.35 million new cases of lung cancer,and 1.18 million deaths,accounting for 20.0% incidence of total cancer incidence,23.8% of cancer deaths in the world[1].In China the incidence and mortality of lung cancer are also ranks first in cancer,and continue upward trend.It is predicted that by 2010,the number of new cases of lung cancer in China will reach 441,000 with 378,000 deaths,being the leading cause of deaths from cancer[2].For advanced non-small cell lung cancer(NSCLC).platinum-based regimen chemotherapy is still the main treatment,and platinum-based regimen are recommended as standard first-line setting,but the current efficiency of chemotherapy is only 20%-40%,with median survival time of 8-10 months[3].Most of patients had the side effects of chemotherapy,but no effect.The possible reason of the significant differences in efficacy in the chemotherapy of non-small cell lung cancer lies in the difference of pharmacogenomics which results in different drug efficacy.Thus,according to the different conditions of each patient,to develop specific treatment programs to give the most appropriate and effective individualized therapy is the trend of the lung cancer treatment.
Cisplatin can cause monoadducts and intrastrand or interstrand cross-links in DNA[4],thereby preventing its replication and transcription,leading to DNA damage and cell death.The DNA lesions can be repaired by DNA repaired mechanisms and activities of molecules may impact treatment activity of platinum.The excision repair cross- complementtation 1 (ERCC1),a structure-specific DNA repair endonuclease responsible for the 5-prime incision,is one of the key enzymes of nucleotide excision repair(NER)pathway[5].A number of studies have shown that the lower expression levels of ERCC1 mRNA,the longer survival in patients,suggesting that it may be a predictor of efficacy,prognostic and survival in NSCLC patients with cisplatin-containing chemotherapy[6-12].
Ribonucleotide reductase M1 (RRM1)is a key enzyme involved in DNA synthesis,catalyzing the biosynthesis of deoxyribonucleotides from the corresponding ribonucleotides,thus plays an important role in the DNA synthesis and repair.Data indicate that higher levels of RRM1 are associated with chemoresistance to gemcitabine-based therapies.Up-regulation of RRM1 has been previously observed pre-clinically in different gemcitabine-resistant human cell lines[13,14].The patients with high expression of RRM1 have a longer survival than that with low expression of RRM[15-17].
The aim of our work is to validate,in a retrospective study,the prognostic and predict relevance of ERCC1 and RRM1 mRNA expression levels in Chinese advanced NSCLC patients treated with cisplatin-based therapies.
MATERIALS AND METHODS
Patients and Samples
A total of 160 formalin-fixed,paraffin-embedded(FFPE)bronchoscope or fine needle aspiration (FNA)biopsy tumor samples were analyzed to determine ERCC1 and RRM1 mRNA expression levels.They were obtained from the unresectable local advanced and metastaric NSCLC patients who were treated in Beijing Cancer Hospital (Thoracic Oncology Unit)between September 2001 and October 2008.
All enrolled patients had clinical and pathological information including age,gender,date of initial diagnosis and date of death or last follow-up.In addition,the patients enrolled in this study met the following criteria: received two cycles of first-line cisplatin-based chemotherapy and having measureable lesions and available paraffin-embedded tissue samples.The study was approved by Beijing cancer hospital Instientional Review Board.
Assessment of Objective Response and Clinical Outcomes
The tumor response were assessed and confirmed by computer tomography(CT),brain magnetic resonance imaging(MRI) and bone scintigraphy scan at baseline and after every two treatment cycles.Designations of complete response (CR),partial response (PR),stable disease(SD),and progressive disease(PD),were based on the standardized response definitions established by response evaluation criteria in solid tumors(RECIST).The response rate (RR)was defined as total number of CR and PR patients divided by the number of evaluable patients.Time to progression(TTP)was calculated from the date of initiation of chemotherapy to the date of disease progression.Overall survival(OS)was calculated as the time between the beginning of chemotherapy and death or censored at last follow-up.
Realtime PCR Detection
Sample pretreatment
Preliminarily,archive slides of the collected tumor samples were reviewed by pathologists.From each paraffin block of representative tumor areas,serial sections with a thickness of 4um were prepared.Malignant cells were selected under microscope magnification (from ×5 to ×10)and dissected from the slide simply by using a scalpel[12].
RNA isolation and complementary DNA synthesis
RNA was isolated from formalin-fixed,paraffinembedded specimens using a novel,proprietary procedure (MiRNeasy FFPE).After RNA isolation,cDNA was derived from each sample according to a kit described procedure(using the company kit)and stored in -20°C.
Reverse transcription-polymerase chain reaction
Relative complementary DNA quantitation for ERCC1,RRM1 and an internal reference gene(β-actin)was done using SYBR green real-time fluorescent quantitative PCR detection method[Stepone Sequence Detection System Applied Biosystems].
The sequences of the primers used were as follows:
ERCC1 forward 5’-GGGAATTTGGCGACGTA ATTC-3’,
reverse 5’-GCGGAGGCTGAGGAACAG-3’,
RRM1 forward 5’-AAGAGCAGCGTGCCAGA GAT -3’,
reverse 5’-ACACATCAAAGACCAGTCCT GATTAG -3’,
β-actin (internal reference gene)forward 5’-TGA GCGCGGCTACAGCTT -3’,
reverse 5’-TCCTTAATGTCACGCACGATT -3’,[9,15,22].
All primers used in this study were intron spanning to avoid genomic DNA contamination.The PCR mixture consisted 2×POWER SYBR GREEEN PCR MASTER MIX 7.5 µl,cDNA 3ul,water 4.5 µl to a final volume of 15 µl (all reagents were from PE Applied Biosystems,Foster City,CA).
Cycling conditions were 50°C for 2 min and 95°C for 10 min and followed by 45 cycles at 95°C for 15 s and 60°C for 1 min (Fluorescence signal was collected)and the melt curve was done to verify the products of PCR.Relative gene expression levels are expressed as ratios (differences between the Ct values)between two absolute measurements (genes of interest/internal reference gene).HT-29 total RNA(from)were used as control calibrators on each plate.Data were analysis by the method of 2-ΔΔCT.By adopting cut-off values according to median expression levels,we divided ERCC1 and RRM1 into high expression and low expression[10,12,1]
Statistical Analysis
To test significant associations between the continuous variable ‘gene expression’ and dichotomous variables (patient’s age,sex,tumor stage,etc.),the Mann-Whitney test was used.The Kruskal-Wallis test was used to test significant associations with multiple variables as histology.Spearman correlation coefficients were calculated to estimate the correlation between ERCC1 and RRM1 mRNA levels.Kaplan-Meier survival curves and the logrank test were used to analyze univariate distributions for survival.Cox’s proportional hazards multivariate analysis was used to evaluate which of the significant factors at the univariate analysis had a significant influence on survival.Statistical significance was set at P=0.05.
RESULTS
Patient Characteristics
There were 142(88.75%)specimens were successfully amplified,and the remaining 18 samples were not quantifiable because of the minimal amounts of available tissue or because of large amounts of necrosis in the tumor sample.The characteristics of the 142 patients are shown in Table 1.All the patients received cisplatin-based chemotherapy.One hundred and eight were stage IV and 34 stage IIIB disease.At the time of the survival analysis,89 patients had died and 53 were still alive.Median follow-up time was 10.97 months.Overall median survival time (MST)is 13.27 months.
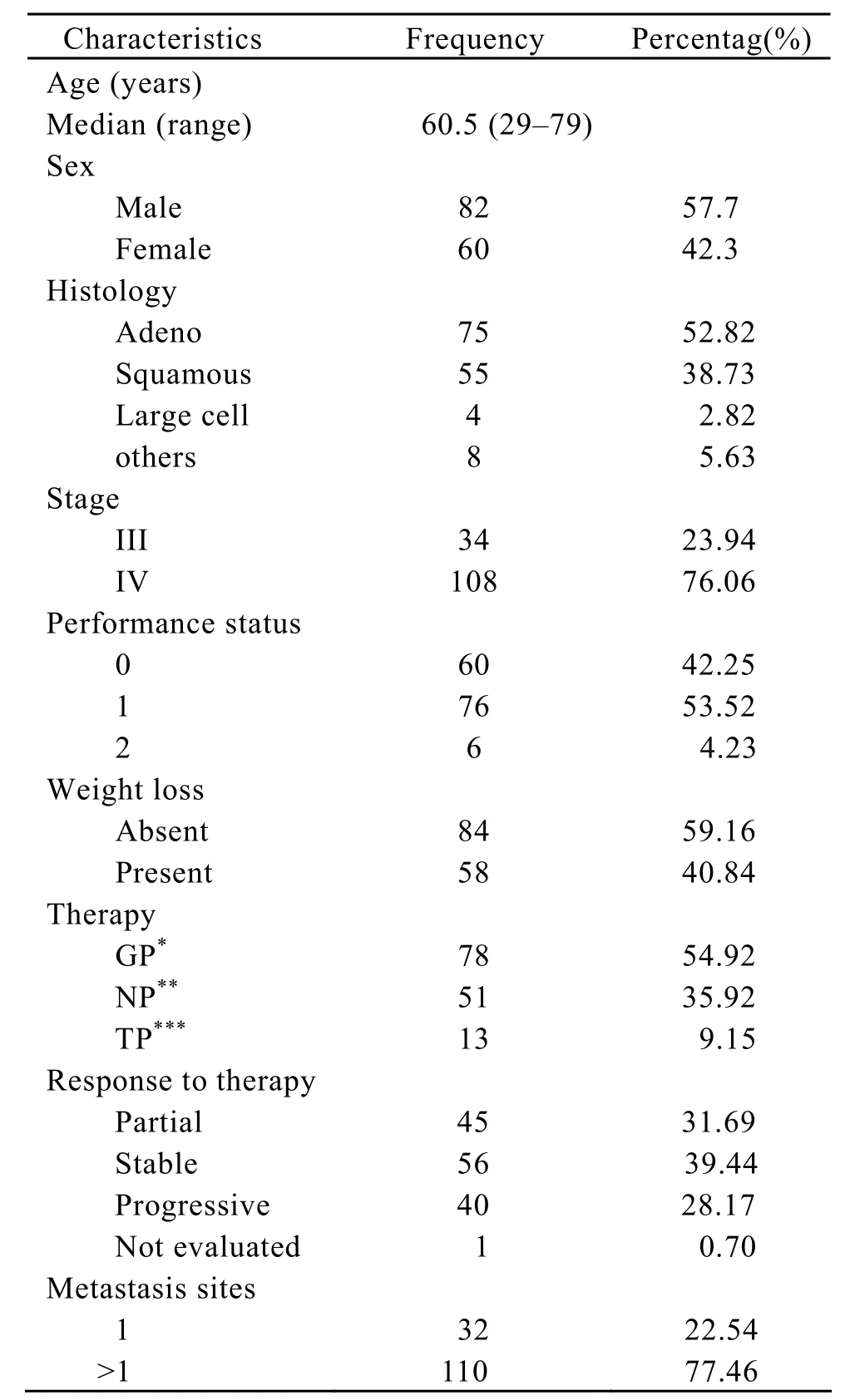
Table 1.Patients’ characteristics
ERCC1,RRM1 Expression Levels in Advanced NSCLC
Real-time PCR was used to detect mRNA quantification of ERCC1 /RRM1 and β-actin was used as the internal reference gene.ERCC1 expression levels ranged from 0.01 to 78.79 (median 0.63,mean 4.25 and standard deviation (SD)9.23)and RRM1 from 0.00 to 30.91 (median 0.63,mean 0.87 and SD 3.36).By adopting cut-off values according to median expression levels,no correlation was found between age (P=0.440),sex (P=0.07),histology (P=0.283),stage (P=0.668),presence of weight loss (P=0.07,all by Mann-Whitney test,except that Kruskal-Wallis test was used for histology)and ERCC1 expression.Similar results were observed between RRM1 gene expression and any of the clinical- pathological factors considered.In addition,no correlation with gene expression profiles and response to chemotherapy was found (P=0.261,Kruskal-Wallis test).We found a correlation between ERCC1 and RRM1 (rs=0.380,P<0.0001),by means of the Spearman’s rank correlation method.(The results of Matching ERCC1 and RRM1 expression levels were obtained by the real-time PCR for every sample).
There was a better disease control rate in patients with low expression of ERCC1 than that of high expression.But we did not find relations between disease control rate and the expression of RRM1(Table 2).

Table 2.Response and DCR
Correlation of ERCC1 and RRM1 mRNA Levels with TTP and Overall Survival
We used Median ERCC1 gene expression value as a cut-off at univariate model and found that MST in patients with low ERCC1 mRNA levels was significantly longer in overall population than in patients with higher levels (18.63 versus 13.69 months,log-rank 5.73,P=0.017,Figure 1).

Figure 1.Kaplan-Meier survival analysis ERCC1
Similar analysis was carried out on RRM1 mRNA expression levels,but we did not found the survival benefit of the patients with low expression level of RRM1.(19.07 versus 14.88,log-rank 1.66,P=0.197,Figure 2).
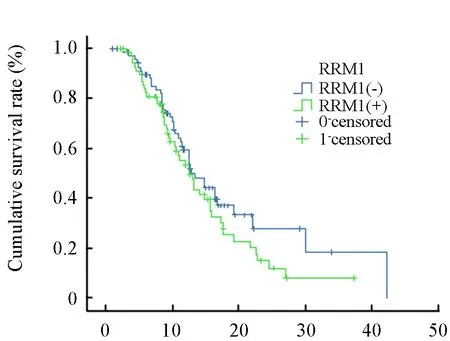
Figure 2.Kaplan-Meier survival analysis RRM1
Cut-off values for each of the clinical and pathological factors (age,sex,stage,etc.,see Table 2)were selected,according to the median value for continuous variables,and univariate analysis was carried out to identify those factors significantly associated with overall survival.The results show that the level of ERCC1 expression,the presence of weight loss and target therapy,were significant prognostic factors for survival (Covariate means 0.496,0.394 and 0.27 respectively,P=0.001,0.012,and 0.037 respectively).All the other selected factors were not statistically correlated with survival.A Cox regression analysis was carried out to assess whether ERCC1 and RRM1 are prognostic factor of survival independently from other variables found to be significant at the univariate analysis.The results show that low ERCC1 level (P=0.001),but not RRM1 level,together with the presence of weight loss (P=0.012),and target therapy (P=0.037)are independent prognostic factors for survival (Table 3).
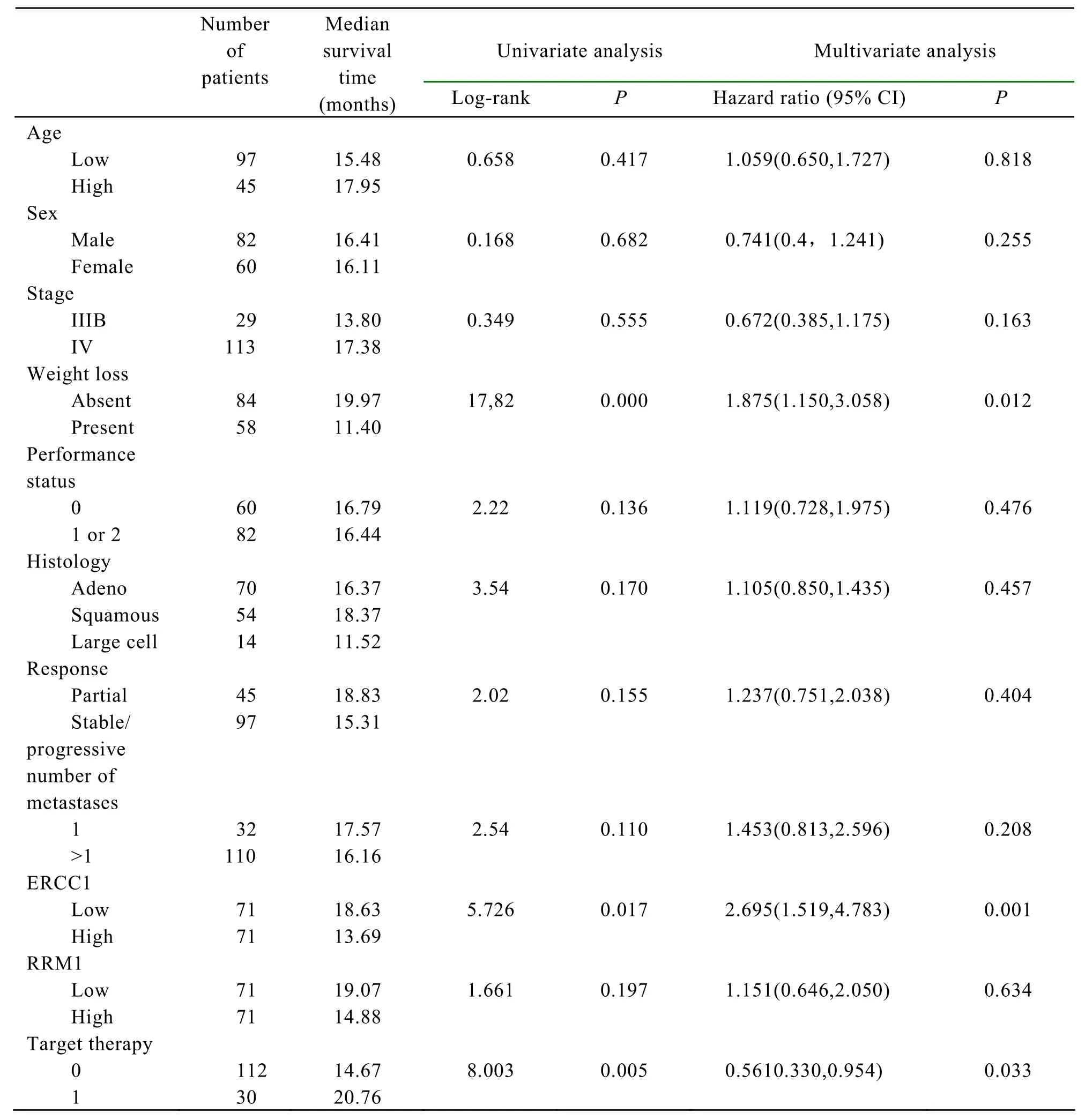
Table 3.Factors associated with overall survival.
The similar analysis was done for TTP and the similar result was obtained.TTP in patients with low ERCC1 mRNA levels was significantly longer in overall population than in patients with higher levels(5.84 versus 4.64 months,log-rank 6.98,P=0.008,Figure 3)and we did not found the TTP benefit of the patients with low expression level of RRM1.(5.40 versus 5.08,log-rank 0.716,P=0.398,Figure 4).

Figure 3.Kaplan-Meier TTP analysis of ERCC1

Figure 4.Kaplan-Meier TTP analysis of RRM1
DISCUSSION
It has been more popular to personalize chemotherapy based on the molecular determinants of the tumor and augment response rates and improve survival of the advanced NSCLC patients.Published data showed that ERCC1 and RRM1 are the important components of the NER pathway and their mRNA expressions play an important role in predicting cisplatin and gemcitabine resistance and treatment outcome[10,17,18].In adjuvant regimen patients with low RRM1 level benefit significantly from cisplatin/gemcitabine administered[11].Experimental studies also have indicated that the increasing removal of CDDP-induced DNA adducts and relative CDDP resistance are associated with high ERCC1expression levels[19],and ERCC1-defective cells or knockout mice are highly sensitive to DNA cross-linking agents[20,21].Lee et al.showed that transferring ERCC1 into a UV repair-deficient (ERCC1[-])Chinese hamster ovary cell line got DNA adduct repair capability and CDDP resistance[22].
Our data suggest that the intra-tumor ERCC1 levels are associated with the effectiveness of cisplatin-based therapy because ERCC1 expression influences ERCC1- mediated DNA adduct repair activity[19-23].We found significant correlation between ERCC1 expression levels and survival.Our data revealed that DNA repair genes may play a crucial role in the prognosis of advanced NSCLC patients.And there were statistically significant associations between the ERCC1 and RRM1 expression in advanced NSCLC patients,and the ERCC1 expression was associated with the response of the platinum-based chemotherapy and the survival.ERCC1 may be used as a marker of personalized chemotherapy to decide which patient will be mostly benefit from the platinum-based chemotherapy.
In the multivariable Cox regression model analysis we found that low level of ERCC1 expression,the presence of weight loss and target therapy were significant prognostic factors for survival.From published data performance status was independent prognostic factors for survival,but we did not get the same result.The reasons probably were that most of the enrolled patients were PS 0/1,and no patients were PS 3/4.
As well known,it is very difficult to transfer research findings to clinical practice in lung cancer because of several technical and practical obstacles.For example,the tissue diagnosis is usually made from a limited amount of bronchial tissue obtained at the fiber-bronchoscope and paraffin-embedded,formalin-fixed tissues and there exists the absence of standardized inter-laboratory quality-control procedures.Our results further confirmed the feasibility to assess gene expression with the formalin-fixed,paraffin- embedded specimens: we extracted and amplicate the gene expressions of the original 88.75% of the specimens successfully.This was a fair good yield of paraffin-embedded,formalinfixed tissues.
In conclusion,this retrospective study further validates that the high gene expression of ERCC1 is a risk factor for survival in patients treated with cisplatin-based therapy for non-small cell lung cancer.Additionally,the present study further validates the suitability of gene expression detection using limited amounts of neo-plastic tissue obtained through fiber-bronchoscope or transthoracic FNA biopsies,FFPE.On the bases of the available evidence,the prospective,large sample,multi-institutional,and randomized trials for selecting chemotherapy according to the ERCC1 and RRM1 expression will be put into practice.
[1]Non-small Cell Lung Cancer Collaborative Group.Chemotherapy in non-small cell lung cancer: a meta-analysis using updated data on individual patients from 52 randomized clinical trials[J].Br Med J 1995; 311: 899-909,
[2]Parkin DM,Yang L,Li LD,et al.Mortality time trends and the incidence and mortality estimation and projection for lung cancer in China[J].Chin J Lung Cancer(in Chinese)2005; 8: 274-8.
[3]Scagliotti GV,De Marinis F,Rinaldi M,et al.Phase III randomized trial comparing three-platinum-based doublets in advanced non-small cell lung cancer[J].J Clin Oncol 2002; 20: 4285-91.
[4]Lawley PD,Phillips DH.DNA adducts from chemotherapeutic agents[J].Mutat Res 1996; 355:13-40.
[5]Reardon JT,Vaisman A,Chaney SG,et al.Efficient nucleotide excision repair of cisplatin,oxaliplatin,and bis-aceto-ammine-dichlorocyclohexylamineplatinum(IV)(JM216)platinum intrastrand DNA diadducts[J].Cancer Res 1999; 59: 3968-71.
[6]Li Q,Yu JJ,Mu C,et al.Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells[J].Anticancer Res 2000; 20: 645-52.
[7]Metzger R,Leichman CG,Danenberg KD,et al.ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy[J].J Clin Oncol 1998; 16: 309-16.
[8]Shirota Y,Stoehlmacher J,Brabender J,et al.ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy[J].J Clin Oncol 2001; 19: 4298-304.
[9]Joshi MB,Shirota Y,Danenberg KD,et al.High gene expression of TS1,GSTP1,and ERCC1 are risk factors for survival in patients treated with trimodality therapy for esophageal cancer[J].Clin Cancer Res 2005; 11: 2215-21.
[10]Lord RV,Brabender J,Gandara D,et al.Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer[J].Clin Cancer Res 2002; 8:2286-91.
[11]Simon GR,Sharma S,Cantor A,et al.ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer[J].Chest 2005; 127: 978-83.
[12]Ceppi P,Volante M,Novello S,et al.ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine[J].Ann Oncol 2006,17: 1818-25
[13]Goan YG,Zhou B,Hu E,et al.Overexpression of ribonucleotide reductase as a mechanism of resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell line[J].Cancer Res 1999; 59: 4204-7.
[14]Dumontet C,Fabianowska-Majewska K,Mantincic D,et al.Common resistance mechanisms to deoxynucleoside analogues in variants of the human erythroleukaemic line K562[J].Br J Haematol 1999;106: 78-85.
[15]Cao MY,Lee Y,Feng NP,et al.Adenovirus-mediated ribonucleotide reductase R1 gene therapy of human colon adenocarcinoma[J].Clin Cancer Res 2003; 9:4553-61.
[16]Davidson JD,Ma L,Flagella M,et al.An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines[J].Cancer Res 2004; 64: 3761-6.
[17]Rosell R,Danenberg KD,Alberola V,et al.Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients[J].Clin Cancer Res 2004; 10: 1318-25.
[18]Simon G,Sharma A,Li X,et al.Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small cell lung cancer[J].J Clin Oncol.2007; 25: 2741-6.
[19]Li Q,Yu JJ,Mu C,et al.Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells.Anticancer Res.,2000,20: 645-52.
[20]Westerveld A,Hoeijmakers JH,van Duin M,et al.Molecular cloning of a human DNA repair gene[J].Nature (Lond.)1984; 310: 425-9.
[21]Melton DW,Ketchen AM,Nunez F,et al.Cells from ERCC1-deficient mice show increased genome instability and a reduced frequency of S-phasedependent illegitimate chromosome exchange but a normal frequency of homologous recombination[J].J Cell Sci 1998; 111: 395-404.
[22]Lee KB,Parker RJ,Bohr V,et al.Cisplatin sensitivity/resistance in UV repair-deficient Chinese hamster ovary cells of complementation groups 1 and 3[J].Carcinogenesis (Lond.)1993; 14: 2177-80.
[23]Chaney SG,Sancar A.DNA repair: enzymatic mechanisms and relevance to drug response[J].J Natl Cancer Inst 1996; 88: 1346-60.
杂志排行
Chinese Journal of Cancer Research的其它文章
- Decrease of Peripheral Blood CD8+/CD28- Suppressor T Cell Followed by Dentritic Cells Immunomodulation among Metastatic Breast Cancer Patients
- Chinese Journal of Cancer Research Guidelines for Authors
- Inhibition of Proliferation Induced by Cyclin D1 Gene Silence in Human Renal Carcinoma ACHN Cells
- Interleukin-18 Suppresses Angiogenesis and Lymphangiogenesis in Implanted Lewis Lung Cancer
- Association of Lysosome Associated Protein Transmembrane 4 Beta Gene Polymorphism with the Risk of Pancreatic Cancer
- Recurrent Patterns and Factors Involved in Node-negative Advanced Gastric Cancer
