Polymorphisms of UGT1A7 and XRCC1 are Associated with an Increased Risk of Hepatocellular Carcinoma in Northeast China
2010-01-08ZhifangJiaHongyingSuXuelianLiXinXuZhihuaYinPengGuanBaosenZhou
Zhi-fang Jia,Hong-ying Su,Xue-lian Li,Xin Xu,Zhi-hua Yin,Peng Guan Bao-sen Zhou*
1Department of Epidemiology and Key Laboratory of Cancer Etiology and Intervention of Liaoning Province,School of Public Health,China Medical University,Shenyang 110001,China;
2Department of Intervention,the First Affiliated Hospital,China Medical University,Shenyang 110001,China
INTRODUCTION
Hepatocellular carcinoma (HCC)is one of the most common malignancies and the third cause of death from cancer in the world.The situation in China is even worse,for about 55% of the world new HCC cases come from China[1]and the incidence rate of HCC in China is increasing[2,3].Chronic infection with either hepatitis B virus (HBV)or hepatitis C virus(HCV),dietary aflatoxin exposure and alcohol consumption are important risk factors for HCC[4].However,only a minority of people at risk eventually develops HCC and it is likely that there may be some modified mechanisms involved during the development of HCC.
The human uridine 5’-diphosphate (UDP)-glucuronosyltransferases (UGTs)are an enzyme superfamily that catalyzes the glucuronidation of both endogenous compounds such as bilirubin and steroid hormones,as well as exogenous compounds such as environmental carcinogens (including polycyclic aromatic hydrocarbons and heterocyclic amines)and dietary constituents[5,6].UGT1A7 is one of the products by alternative splicing of exon one to the four common exons(exon2-exon5)of UGT1A gene and it metabolizes a variety of clinically and toxicologically important compounds such as benzo(a)pyrene[7].So far,several missense variants of UGT1A7 have been discovered at codon 115 (Gly→Ser),129(Asn→Lys),131(Arg→Lys),139(Glu→Asp),and 208(Trp→Arg)resulting in 11 polymorphic alleles (UGT1A7*1--*11)[8].Previous study has demonstrated that polymorphisms of codon 129 and 131 are in complete linkage dysequilibrium.Compared to the wild type UGT1A7*1 allele,other variants such as the UGT1A7*2 allele(Lys129Lys131Trp208)and UGT1A7*3 allele (Lys129Lys131Arg208)encode lower catalytic-activity proteins against benzopyrene metabolites[7].Several studies have suggested a link between polymorphism of UGT1A7 and HCC susceptivity[9-11].However,the role of the UGT1A7 still needs further study as other studies gained controversial conclusions[12,13].
In addition,DNA repair genes are increasingly studied because of their pivotal role in maintaining genome integrity[14].The X-ray repair crosscomplementing group 1 (XRCC1)functions as a scaffold protein and plays a central role in two interconnected DNA repair pathways: base excision repair (BER)and single-strand break repair(SSBR)[15,16].Many single nucleotide polymorphisms(SNPs)have been found in XRCC1,and the most extensively studied SNP is Arg399Gln on exon 10.The Arg399Gln allele is situated within the BRCT-1 region harboring the ADPRT binding domain and it has been suggested to be associated with altered DNA repair capacity with an substitution of Arg with Gln[17].
In this study,we detected the genetic polymorphisms of UGT1A7 and XRCC1 genes in a group of hospital-derived HCC patients and their age and gender frequency-matched controls,which all came from Northeast China.This study was a hospital-based, case-control study under an established protocol.
MATERIALS AND METHODS
Patients
Blood samples were collected from consecutive HCC patients hospitalizing for the first time in the Department of Intervention,the First Affiliated Hospital of China Medical University from October,2007 to April,2008.A total of 136 HCC patients were included who were diagnosed by liver biopsy,or by the findings of radiological features suggestive of HCC in at least two image examinations including abdominal ultrasound,contrast enhanced dynamic computed tomography(CT),magnetic resonance imaging(MRI)and hepatic angiography,or by a single positive imaging technique associated with serum α-fetoprotein level(AFP)>400 μg/L and with no evidence of other tumors.Once recruiting HCC patients,we selected a group of age(±5)and sex frequency-matched controls without clinically apparent liver diseases in the same hospital who attended the physical examination center.Serum hepatitis B surface antigen (HBsAg)and anti-HCV antibody were tested to determine the infection of hepatitis B or hepatitis C by microparticle enzyme immunoassay using commercial assay kits (AxSYM,Abbott,USA).All subjects were investigated with a questionnaire including demographic characteristics,history of disease,consumption of tobacco and alcohol,family history of HCC and other tumors by trained investigators.This study was approved by the Medical Ethical Committee of China Medical University,and informed consent was obtained from all participants.
Determination of UGT1A7 and XRCC1 Genotype
Blood samples were collected in EDTA tubes and stored at -20°C until DNA extraction.Genome DNA was isolated by potassium iodide technique.
For UGT1A7,as other mutations are rare in humans[18],we screened the mutations in codons 129,131 and 208 using polymerase chain reaction(PCR)-based genotyping techniques including allele-specific PCR (AS-PCR)and PCR-restriction fragment length polymorphism (RFLP) as described[19,20].Firstly,sequences of UGT1A7 containing codons 129/131 and 208 were amplified using primers UF11 and UR2 (Table 1)to increase specificity as the UGT1A7 gene shared a high level of homology with other UGT related genes.The 25 μl reaction mixture contained 1μl genome DNA,2μl 10×PCR buffer, 1.6μl each deoxynucleoside triphosphate (dNTPs),4 pmol each primer and 0.5 units of Taq DNA polymerase (Primers were synthesized by Invitrogen,Beijing,China.Others were obtained from TaKaRa,Dalian,China).The PCR condition was 94°C 7 min followed by 25 cycle of 94°C for 30 s,56°C for 30 s and 72°C for 40 s and an ultimate extension at 72°C for 7 min.Secondly,as mutations at codons 129 and 131 were in complete linkage dysequilibrium and closed to each other,allele-specific PCR was applied to determine the sequence variants at codons 129 and 131.Two independent parallel amplifications were performed with different forward primers UF12 and UF13 but the same reverse primer UR2 using the 1000-fold-diluted product of the fist PCR as template.The PCR reaction mixture and condition were similar to the first amplification expect for annealing temperature of 57°C.The products of two pairs of primers were both 454 bp which UF12-UR2 represented existence of the wild haplotype of Asn129Arg131while UF13-UR2 the mutant haplotype of Lys129Lys131.Finally,the second PCR products were digested with the restriction enzyme RsaⅠ(Invitrogen,Beijing,China)for 2 h at 37°C to detect polymorphism of codon 208.The existence of mutant Arg208 gave two fragments:198 bp and 256 bp,which was not observed in the wild Trp208.
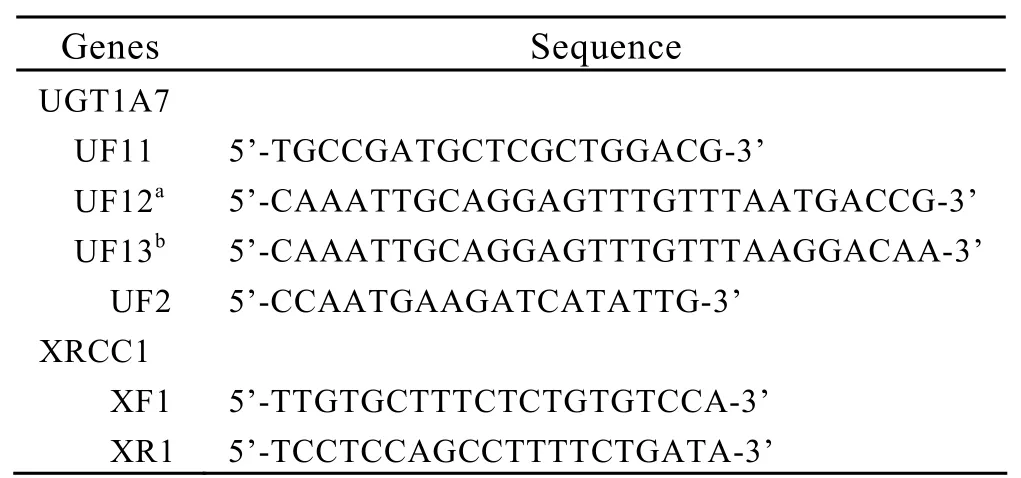
Table 1.Primer sequences for UGT1A7 and XRCC1
For XRCC1,PCR-RFLP was carried out to detect the status of codon 399 by primers XF1 and XR1.We undertook PCR using 5 μl genome DNA,2.5μl 10×PCR buffer, 2μl each deoxynucleoside triphosphate (dNTPs),5 pmol each primer and 0.6 units of Taq DNA polymerase in a final volume of 25μl.The reaction mixture was denatured at 94°C for 5 min followed by 30 cycles of denatured at 94°C for 30 s,annealing at 57°C for 30 s,elongation at 72°C for 45 s,and a final extension at 72°C for 7 min.The PCR products were then digested with the restriction enzyme Msp I.The wild type of Arg399Arg resulted in two fragments: 377bp and 238bp,whereas the homozygote of Gln399Gln only one 615 bp fragment.
All products were separated by electrophoresis on 2% agarose gels and observed on an electrophoresis gel imaging analysis system (Alpha Innotech,USA)with GeneFinder (Bio-Vision,China)staining.
Statistical Analysis
Categorical variables such as sex,alcohol intake were estimated with Pearson χ2-test or Fish’s exact test.Odds ratios (ORs)with 95% confidence interval(CI)were calculated by logistic regression analysis for allele frequencies and genotype distribution of UGT1A7 and XRCC1.The significant risk factors identified from univariate analysis were evaluated subsequently by the multivariate logistic regression model.A two-tailed P value <0.05 was considered to be statistically significant.
RESULTS
Characteristics of Subjects
A total of 136 HCC patients and 136 frequency-matched controls were enrolled in this study.Table 2 listed the basic characteristics of the subjects.There were no significant differences between the two groups in distribution of age,gender,cigarette smoking and alcohol intake.However,the HCC patients had significantly higher rates of HBV or HCV infection and family history of HCC than controls (Table 2).
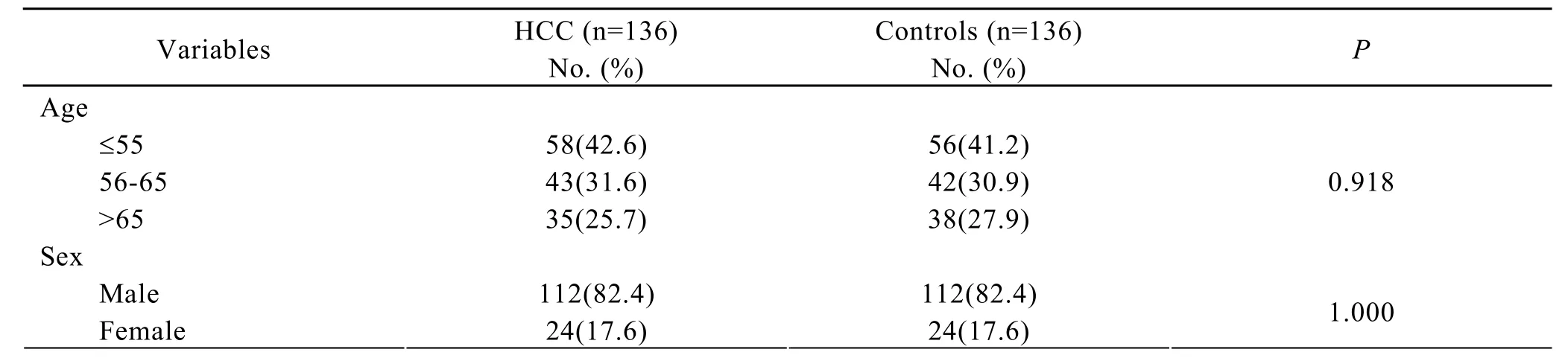
Table 2.Characteristics of subjects
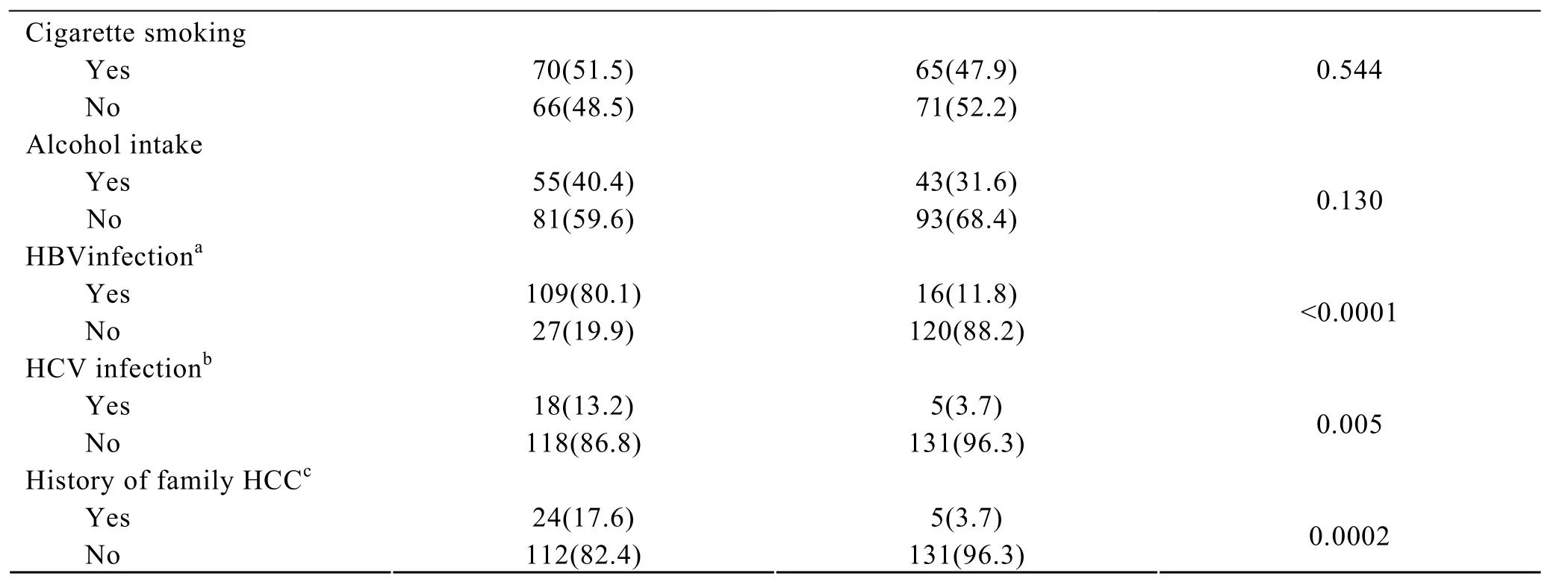
OR(95% CI): a,30.28(15.49-59.20); b,4.00(1.44-11.10); c,5.61(2.07-15.20)
UGT1A7 Polymorphisms and HCC Risk
UGT1A7 alleles were designated as UGT1A7*1(wild type, Asn129Arg131Trp208), UGT1A7*2(Lys129Lys131Trp208),UGT1A7*3 (Lys129Lys131Arg208),UGT1A7*4 (Asn129Arg131Arg208)according to the previous study[7].In this study,we failed to detect any individual carrying the UGT1A7*4 allele.The distributions of UGT1A7 genotypes and alleles were shown in Table 3.The frequencies of UGT1A7*1/*2 and*3/*3 genotypes in HCC patients (36% and 15%)were statistically higher than those in controls (26%and 4%),with ORs of 2.09 (95%CI:1.10-3.97)and 5.67 (95%CI:1.76-18.30)compared with the*1/*1 genotype,respectively.As for the UGT1A7 alleles,we found that both UGT1A7*2 and UGTA7*3 allele could slightly but significantly increase the risk for HCC,with ORs of 1.60 (95%CI:1.03-2.49)and 1.77(95%CI:1.15-2.74),respectively (Table 3).Furthermore,as previous studies have shown that UGT1A7 isoenzymes encoded by variant UGT1A7 alleles(UGT1A7*2,*3)exhibit lower catalytic activity than that by UGT1A7*1 allele[7],we designated the UGT1A7*1 allele as a higher-activity allele (H allele)and UGT1A7*2,*3 lower-activity alleles (L allele)and categorized UGT1A7 genotype into high (H/H),intermediate (H/L)and low-activity variant (L/L)according to the enzyme activity.When HCC risk was compared with the high catalytic genotype (H/H),significant increase was shown for the intermediate(OR=1.73,95%CI:1.01-2.96)and the low-activity genotype (OR=2.55,95%CI:1.28-5.08).An inverse dose-response relationship between the decreased enzymatic activity of UGT1A7 genotype and increased HCC risk was observed (P=0.005).

Table 3.Genotype and allele frequencies of UGT1A7 polymorphisms in subjects
XRCC1 Polymorphisms and HCC Risk
The distributions of XRCC1 polymorphisms were shown in Table 4.The heterozygous genotype Arg/Gln could slightly increase the risk for HCC(OR=2.16,95%CI:1.29-3.61).However,this effect was not observed for homozygous Gln/Gln genotype.When considering the allele frequency,the mutant Gln allele statistically increased the HCC risk(OR=1.65,95%CI: 1.14-2.73).
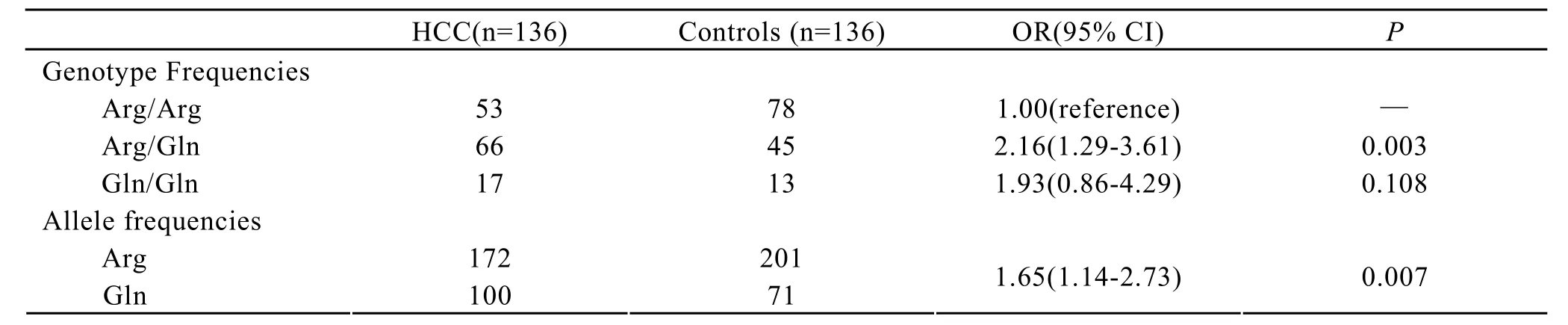
Table 4.Genotype and allele frequencies of XRCC1 polymorphisms in subjects
Multivariate Analysis
We further evaluated factors involved in HCC risk using multivariate logistic regression model(Table 5).In addition to polymorphisms of UGT1A7 and XRCC1,subjects with HBV or HCV infection and family history of HCC incurred significantly higher risks for HCC (OR=68.07,95%CI:28.03-165.26;OR=30.97,95%CI:8.06-118.94 and OR=10.62,95%CI: 2.22-50.77; respectively).

Table 5.Multivariate regression analysis
DISCUSSION
In this study,we examined the polymorphisms of UGT1A7 and XRCC1 in the Northeastern Chinese population with or without HCC.Our study revealed that both UGT1A7 and XRCC1 polymorphisms were independent risk factors of HCC.We also evaluated other factors related to HCC in multivariate model.
UGTs constitute a major cellular defense through its conjugation of hydrophobic compounds both endogenous and exogenous to form water soluble glucuronides which facilitate renal or biliary excretion.Polymorphisms of UGT1A7 could alter the enzymatic activity of the encoded protein[7]and have been reported to be involved in colorectal cancer[20-23],lung cancer[24,25], breast cancer[26], pancreatic cancer[27]and HCC[9,11,28].Vogel et al[9]first reported that UGT1A7*3 allele could significantly increase HCC risk but Stucker et al[12]found that the association existed only when the case group was restricted to the viral origin.However,Borentain et al[13]failed to find any association between them.In our study,we observed that carriage of UGT1A7*3/*3 or*1*2 genotype was significantly associated with higher HCC risk (OR=5.67,95%CI:1.76-18.30;OR=2.09,95%CI:1.10-3.97,respectively)and both UGT1A7*2 and*3 alleles were linked to increased HCC risk (OR=1.60,95%CI:1.03-2.49; OR=1.77,95%CI:1.15-2.74,respectively).When we designated the UGT1A7*1 allele as a higher-activity allele (H allele)and UGT1A7*2,*3 as lower-activity allele (L allele)and categorized UGT1A7 genotypes into high(H/H),intermediate (H/L)and low-activity variants(L/L)according to the enzyme activity,an reverse dose-response relationship was found between low-activity genotype and increased risk of HCC(OR=1.73,95%CI:1.01-2.96; OR=2.55 95%CI:1.28-5.08,respectively.P for trend 0.005).Our results were compatible with previous studies which indicated that polymorphisms of UGT1A7 might be a marker of increased risk for HCC[9-11,28].
Recent studies suggested that the polymorphism of DNA repair gene XRCC1 at codon 399 with a substitution of glutamine (Gln)for arginine (Arg)could alter the activity of DNA repair and was associated with risk of various cancers[29-32].Yu et al[33]did not observe an independent increase in HCC risk of Gln allele although a trend was present.However,Kirk et al[34]observed a 3-fold increased risk of HCC in patients with heterozygote genotype(Arg/Gln).Our study found a similar result that Arg/Gln genotype was significantly associated with HCC (OR=2.16,95%CI:1.29-3.61).The variant allele Gln could slightly increase the HCC risk (OR=1.65,95CI:1.14-2.73).However,we could not observe a statistical association of the homogeneous of Gln/Gln which might due to the low sample size.
As HCC was considered to be a multifactorial disease,we further performed multivariate logistic regression analysis to evaluate HCC risk.Besides the polymorphisms of UGT1A7 and XRCC1,HBV infection,HCV infection and family history of HCC also entered into the multivariate model.This result confirms the multifactorial characteristics of hepatocarcinogenesis.In addition to the environmental factors such as virus infection,genetic predisposition to HCC deserves to be paid enough attention.
As far as we know,this was the first study to examine the UGT1A7 polymorphisms in HCC patients coming from Chinese mainland.However,we are aware that there may be potential limitation in our study.We could not evaluate the roles of gender and age which have been suggested to be associated with HCC risk because of the frequency-matched cases and controls by gender and age.However,the gender ratio(male:female)in HCC group was 4.7:1,which may indicate that HCC was more prevalent in male gender as we selected patients consecutively.
In summary,polymorphisms of UGT1A7 and XRCC1 are independently associated with HCC risk in addition to HBV or HCV infection and family history of HCC.The UGT1A7*2 or*3 allele as well as XRCC1 Gln allele at codon 399 might be used as markers related to higher risk of HCC.
[1]Parkin DM,Bray F,Ferlay J,and Pisani P.Global Cancer Statistics,2002[J].CA Cancer J Clin 2005; 55:74-108.
[2]Cai L,Binh G,Parkin D,et al.Cancer trends in Asian Pacific rim region[J].Tumor (in Chinese)2004; 24:422-6.
[3]Jing LB,Yu GH,Guo CS,et al.An analysis of cancer incidence and mortality in some cities,Liaoning province[J].Bulletin of Chinese Cancer (in Chinese)2003; 12: 393-4.
[4]Farazi PA,DePinho RA.Hepatocellular carcinoma pathogenesis: from genes to environment[J].Nat Rev Cancer 2006; 6: 674-87.
[5]Radominska-Pandya A,Czernik PJ,Little JM,et al.Structural and functional studies of UDP-glucuronosyltransferases[J].Drug Metab Rev 1999;31: 817-99.
[6]Tukey RH, Strassburg CP.Human UDPGlucuronosyltransferases: Metabolism,Expression,and Disease[J].Annu Rev Pharmacol Toxicol 2000;40: 581-616.
[7]Guillemette C,Ritter JK,Auyeung DJ,et al.Structural heterogeneity at the UDP-glucuronosyltransferase 1 locus: functional consequences of three novel missense mutations in the human UGT1A7 gene[J].Pharmacogenetics 2000; 10: 629-44.
[8]Verlaan M,Drenth JP,Truninger K,et al.Polymorphisms of UDP-glucuronosyltransferase 1A7 are not involved in pancreatic diseases[J].J Med Genet 2005; 42: e62.
[9]Vogel A,Kneip S,Barut A,et al.Genetic link of hepatocellular carcinoma with polymorphisms of the UDP-glucuronosyltransferase UGT1A7 gene[J].Gastroenterology 2001; 121: 1136-44.
[10]Wang Y,Kato N,Hoshida Y,et al.UDP-glucuronosyltransferase 1A7 genetic polymorphisms are associated with hepatocellular carcinoma in japanese patients with hepatitis C virus infection[J].Clin Cancer Res 2004; 10: 2441-6.
[11]Tseng CS,Tang KS,Lo HW,et al.UDP-glucuronosyltransferase 1A7 genetic polymorphisms are associated with hepatocellular carcinoma risk and onset age[J].Am J Gastroenterol 2005; 100: 1758-63.
[12]Stücker I,Loriot MA,N'Koutchou G,et al.UDP-glucuronosyltransferase UGT1A7 genetic polymorphisms in hepatocellular carcinoma: a differential impact according to seropositivity of HBV or HCV markers?[J].BMC Cancer 2007; 7: 214.
[13]Borentain P,Gérolami V,Ananian P,et al.DNA-repair and carcinogen-metabolising enzymes genetic polymorphisms as an independent risk factor for hepatocellular carcinoma in Caucasian livertransplanted patients[J].Eur J Cancer 2007; 43:2479-86.
[14]Horton JK,Watson M,Stefanick DF,et al.XRCC1 and DNA polymerase [beta] in cellular protection against cytotoxic DNA single-strand breaks[J].Cell Res 2008; 18: 48-63.
[15]Yu Z,Chen J,Ford BN,et al.Human DNA repair systems: an overview[J].Environ Mol Mutagen 1999;33: 3-20.
[16]Petermann E,Keil C,Shiao LO.Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER[J].DNA Repair 2006; 5:544-55.
[17]Matullo G,Palli D,Peluso M,et al.XRCC1,XRCC3,XPD gene polymorphisms,smoking and 32P-DNA adducts in a sample of healthy subjects[J].Carcinogenesis 2001; 22: 1437-45.
[18]Villeneuve L,Girard H,Fortier LC,et al.Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs[J].J Pharmacol Exp Ther 2003; 307: 117-28.
[19]Ando M,Ando Y,Sekido Y,et al.Genetic polymorphisms of the UDP-glucuronosyltransferase 1A7 gene and irinotecan toxicity in Japanese cancer patients[J].Jpn J Cancer Res 2002; 93: 591-7.
[20]Chen K,Jin M,Zhu Y,et al.Genetic polymorphisms of the uridine diphosphate glucuronosyltransferase 1A7 and colorectal cancer risk in relation to cigarette smoking and alcohol drinking in a Chinese population[J].J Gastroenterol Hepatol 2006; 21:1036-41.
[21]Strassburg CP,Vogel A,Kneip S,et al.Polymorphisms of the human UDP-glucuronosyltransferase (UGT)1A7 gene in colorectal cancer[J].Gut 2002; 50: 851-6.
[22]Butler LM,Duguay Y,Millikan RC,et al.Joint effects between UDP-glucuronosyltransferase 1A7 genotype and dietary carcinogen exposure on risk of colon cancer[J].Cancer Epidemiol Biomarkers Prev 2005; 14: 1626-32.
[23]van der Logt EM,Bergevoet SM,Roelofs HM,et al.Genetic polymorphisms in UDP-glucuronosyltransferases and glutathione S-transferases and colorectal cancer risk[J].Carcinogenesis 2004; 25:2407-15.
[24]Araki J,Kobayashi Y,Iwasa M,et al.Polymorphism of UDP-glucuronosyltransferase 1A7 gene: A possible new risk factor for lung cancer[J].Eur J Cancer.2005;41: 2360-5.
[25]Feng FY,Liang G,Lu WF,et al.Correlation of polymorphisms of UDP-glucuronosyltransferase 1A7 gene to genetic susceptibility of lung cancer[J].Chin J Cancer(in Chinese)2005; 24: 1085-90.
[26]Ralph DA,Zhao LP,Aston CE,et al.Age-specific association of steroid hormone pathway gene polymorphisms with breast cancer risk[J].Cancer 2007; 109: 1940-8.
[27]Ockenga J,Vogel A,Teich N,et al.UDP glucuronosyltransferase (UGT1A7) gene polymorphisms increase the risk of chronic pancreatitis and pancreatic cancer[J].Gastroenterology 2003; 124:1802-8.
[28]Kong SY,Ki CS,Yoo BC,et al.UGT1A7 haplotype is associated with an increased risk of hepatocellular carcinoma in hepatitis B carriers[J].Cancer Sci 2008;99: 340-4.
[29]Lao T,Gu W,and Huang Q.A meta-analysis on XRCC1 R399Q and R194W polymorphisms,smoking and bladder cancer risk[J].Mutagenesis 2008; 23:523-32.
[30]Li M,Yin Z,Guan P,et al.XRCC1 polymorphisms,cooking oil fume and lung cancer in Chinese women nonsmokers[J].Lung Cancer 2008; 62: 145-51.
[31]Stern MC,Conti DV,Siegmund KD,et al.DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese health study[J].Cancer Epidemiol Biomarkers Prev 2007; 16:2363-72.
[32]Ratnasinghe LD,Abnet C,Qiao Y-L,et al.Polymorphisms of XRCC1 and risk of esophageal and gastric cardia cancer[J].Cancer Lett 2004; 216:157-64.
[33]Yu MW,Yang SY,Pan IJ,et al.Polymorphisms in XRCC1 and glutathione S-transferase genes and hepatitis B-related hepatocellular carcinoma[J].J Natl Cancer Inst 2003; 95: 1485-8.
[34]Kirk GD,Turner PC,Gong Y,et al.Hepatocellular carcinoma and polymorphisms in carcinogenmetabolizing and DNA repair enzymes in a population with aflatoxin exposure and hepatitis B virus endemicity[J].Cancer Epidemiol Biomarkers Prev 2005; 14: 373-9.
杂志排行
Chinese Journal of Cancer Research的其它文章
- Methylation and Demethylation of Ink4 Locus in Cancer Development
- Apoptotic B16-F1 Cells Coated with Recombinant Calreticulin Mediated Anti-tumor Immune Response in Mice
- Risk Factors of Precancerous Gastric Lesions in A Population at High Risk of Gastric Cancer
- Stanniocalcin-1 Detection of Peripheral Blood in Patients with Colorectal Cancer
- Promoter Hypermethylation of KiSS-1 Gene in Gastric Cancer
- Recurrent Patterns and Factors Involved in Node-negative Advanced Gastric Cancer
