Promoter Hypermethylation of KiSS-1 Gene in Gastric Cancer
2010-01-08ZhiYangDongQiuDaiYunYiDu
Zhi Yang,Dong-Qiu Dai,Yun-Yi Du
Department of Surgical Oncology,the First Affiliated Hospital,China Medical University,Shenyang 110001,China
INTRODUCTION
Gastric cancer is the second most common cause of cancer death in the world[1].As its molecular basis,deep involvement of aberrant DNA methylation has been indicated by the higher incidences of aberrant DNA methylation of known tumor suppressor genes than of mutations[2].Aberrant methylation of DNA may happen in the promoter CpG island of tumor suppressor genes,where transcription of DNA into RNA begins.Transcription is the first major step in decoding DNA into a protein.An important aspect of the methylation mechanism is that it inactivates tumor suppressors genes[3].Multiple recent reviews have shown that aberrant methylation of DNA in promoter CpG island and diminished expression are present in a number of tumor related gene in gastric cancer.For example,RASSF1A,an candidate tumor suppressor gene,is hypermethylated in gastric cancer[4,5]; TIMP-3,which has also been silenced,encodes a protease inhibitor that may help inhibit tissue invasion[6].KiSS-1,a newly discovered metastasis suppressor gene,is silenced with aberrant CpG island hypermethylation in gastric cancer[7].However,the relationship between methylaiton of KiSS-1 gene and gastric cancer has not been elucidated yet.
In our study,KiSS-1 gene methylation was detected in primary tumor,the corresponding adjacent normal mucosa,metastases lymph nodes and peritoneal irrigation fluid using methylation special PCR,to find out the association between the KiSS-1 methylation and clinicopathological characteristics of gastric cancer and to demonstrate the role of testing KiSS-1 methylation of peritoneal irrigation fluid in gastric cancer diagnosis.
MATERIALS AND METHODS
Clinicopathological Data
Specimens were collected from 40 cases with its adjacent normal mucosa of gastric carcinoma undergone a surgical resection in the Surgical Oncology Department of the First Affiliated Hospital of China Medical University during July 2008-January 2009.The enrolled patients included 28 males and 12 females with a mean age of 56 years (rang,34-78 years).All of the patients did not undergo chemotherapy or radiotherapy before surgical resection.
Samples
Fifty ml of paysiological saline were injected into the Douglas cavity at the beginning of the operation and aspirated after gentle stirring,and then the peritoneal lavage fluid was collected from the cavity before a surgical resection.Half of the peritoneal lavage fluid was examined through conventional cytological methods with Papanicolaous staining,and the remaining half was centrifuged at 2000r/min.for 20min to collect the intact cells[8,9]and then kept in a liquid nitrogen tank.The tissue samples including primary tumor,corresponding paired adjacent normal mucosa and metastases lymph nodes were procured immediately after resection.Samples were stored at-70°C until DNA extraction.All the samples of the primary gastric cancer were evaluated by two experienced pathologists for diagnosis.Corresponding paired adjacent normal mucosa were obtained at least 3 cm from the distal negative surgical margin of 40 patients and the absence of malignancy was confirmed histopathologically.The lymph nodes were stained with HE method to confirm whether a true metastasis had occurred.Histological grades and tumor cell differentiation were confirmed by histopathological examination and the TNM stage was accorded to the Union International Contrele Cancer(UICC).
DNA Extraction and Bisulfite Treatment
Genome DNA was extracted by the hydroxylbenzene-chloroform extraction method,and stored at-70°C until use.Treatment of DNA samples with bisulfite converts all unmethylated cytosines to uracils,while leaving methylcytosines unaltered.This allows the subsequent differentiation of methylated and unmethylated sequences by methylation-specific polymerase chain reaction (PCR).
Methylation-specific PCR Reaction
D NA was purified using a Wizard DNA clean-up system (Promega)according to the manipulation instrument.The volume of total reaction mixture was set at 20μl,including 3μl DNA,2μl 10×PCR buffer,1.6μl dNTP,0.8μl primers (the sense and antisense chain),0.15μl Taq enzyme,and 11.65μl doubledistilled water.The reaction consisted of Predenaturing at 94°C for 5min,followed by 35 cycles,consisting of 94°C for 30 s,58°C for 30 s,72°C for 20 s,and with a final extension at 72°C for 5 min.The following primers were used for the detection of human KiSS-1 promoter methylation:M-sense,5’-AAAGTTTCGTTTCGGAGGGTTC-3’ and M-antisense,5’-CTTTTATAAAACCCGAAATAACG-3’ for the methylated sequence of the human KiSS-1 promoter; U-sense,5’-AAAGTTTTGTTTTGGAGG GTTT-3’ and U-antisense,5’-AAAGTTTTGTTTT GGAGGGTTT-3’ for the unmethylated sequence of the human KiSS-1 promoter.The predicted products for methylated and unmethylated DNA were 172 and 173 bp,respectively.The products were subjected to 2.5% agarose gel electrophoresis at 120V for 40 min and quantitated with the FluorChen 2.0 system.DNA from peripheral lymphocytes of healthy individual and water were used as negative controls.
Statiscal Analysis
Statistical analysis was performed using the SPSS13.0 package.Chi-square test was used to analyze the relationship between KiSS-1 methylation and clinicopathological characteristics in primary tumor and its matched adjacent normol mucosa,metastases lymphnodes,peritoneal lavage fluid.Fisher,s exact test was used to analyze the association between KiSS-1 methylation in peritoneal lavage fluid and peritoneal metastasis.P<0.05 was regarded to be statistically significant.
RESULTS
Methylation and Clinicopathological Characteristics
The KiSS-1 methylation of the tumro samples and its matched adjacent normal mucosa,metastases lymphnodes was detected by methylation special-PCR(Figure 1).Our data demonstrated that methylation was found in 82.5% (33/40)of primary tumor,in 80.95% (17/21)of metastases lymphnodes and in 55%(22/40)of matched adjacent normal mucosa.Methylation of in gastric carcinoma and lymphnode was more frequent than in non-neoplastic gastiric mucosa,and there were significant differences between the former two and the latter.KiSS-1 methylation had no significant relation with the clinicopathological characteristics(Table 1).
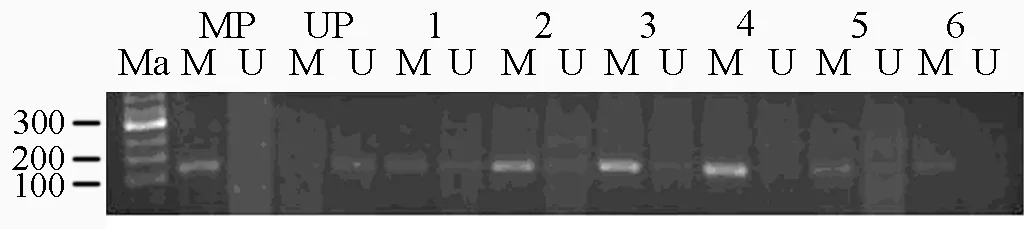
Figure 1.MSP results: M: methylation result; U: unmethylation result; Ma: 50bp DNA Ladder Marker; MP: methylation positive control; UP: unmethylation positive control.
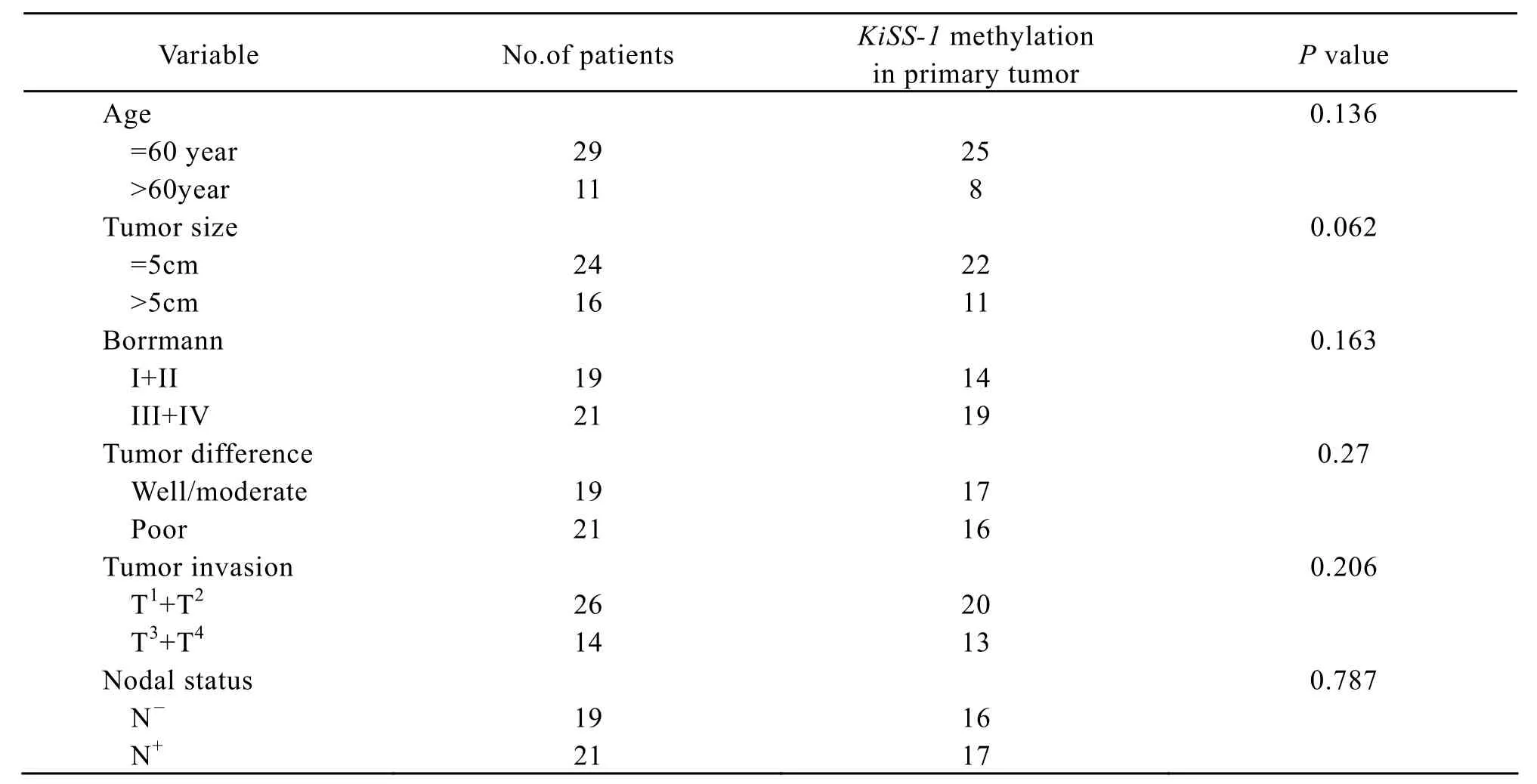
Table l.The association between KiSS-1 methylation and clinicopathological characteristics in gastric cancer
Relationship between Peritoneal Metastasis and Methylation of Peritoneal Lavage Fluid
In the study,we observed that the promoter of KiSS-1 gene was hypermethylated,at a ratio of 42.5%(17/40).Among the 19 cases,9 cases had peritoneal metastasis.The accuracy of KiSS-1 methylation in peritoneal lavage fluid for diagnosing peritoneal metastasis was 70%,with a sensitivity of 77.8%,a specificity of 67.7%,PPV 41.2%,and NPV 91.3% (Table 2).

Table 2.sociation between KiSS-1 methylation in peritobeal lavage and peritoneal metastasis
As shown in Table 3,the presence of KiSS-1 methylation in peritoneal lavage fluid was significantly correlated with tumor invasion(P=0.043).The results of the experiment also showed that KiSS-1 methylation in peritoneal lavage fluid was found more frequently in tumor with lymph node metastasis than in tumor without lymph node metastasis.
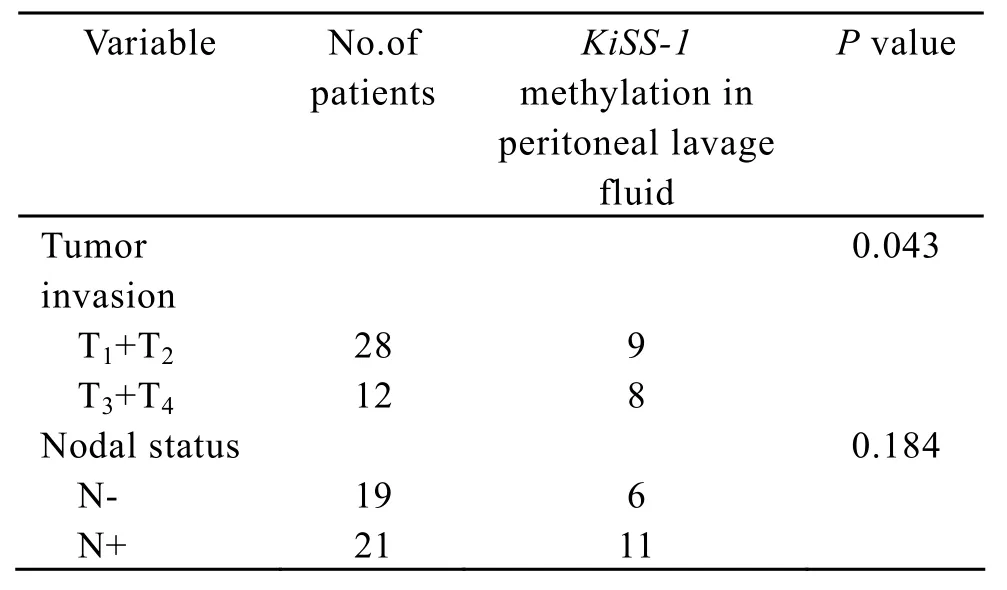
Table 3.Association between KiSS-1 methylation in peritoneal lavage fluid and clinicopathologica factors
DISCUSSION
Epigenetic alteration,conducted by the DNA methyltransferases (DNMTs) catalyzing the methylation of the 5 position of the cytosine ring using S-asenosylmethionine as the donor molecule for the methyl group,has been shown to play an important role in tumorigenesis and progression[3].Methylation of promoter CpG islands leads to transcriptional silencing of their downstream genes.In various human cancers,silencing of tumor suppressor genes,such as CDKN2A(p16),CDH1(Ecadherin)and MLH1,is known to be one of the major mechanisms for their inactivation,along with mutations and LOH.KiSS-1 is a metastasissuppressing gene found in human melanoma cell lines in 1996 and peptides derived from its product are natural ligands of orphan protein-coupled receptor GPR54.Lots of researches have suggested that KiSS-1 gene is associated closely with metastases of melanoma[10],breast cancer[11,12],thyroid cancer[13],esophageal cancer[14],gastric cancer[15]and pancreatic cancer[16].The mechanisms of the decreased expression of KiSS-1 appear to be multifactorial,and more studies need to be elucidated.To expore the novel methylation-silenced genes in gastric cancer,Yamashita et al[7]carried out a chemical genomic screening,a genome-wide search for genes upregulated by treatment with a demethylating agent,5-aza-2′-deoxycytidine (5-aza-dC).After 5-aza-dC treatment of a gastric cancer cell line (AGS)579 genes were upregulated 16-fold or more,using an oligonucleotide microarray with 39000 genes.The KiSS-1 was upregulated 54.8-fold,and its methylation was found in 80%(8/10)of primary gastric cancers.This result is approximates to our results.
In this study,we studied the association between KiSS-1 methylation and primary tumor,corresponding paired adjacent normal mucosa and metastases lymph nodes in gastric cancer.The results of this experiment indicate that the methlytion in primary tumor (82.5%)and metastases lymph nodes (80.95%)was much higher than corresponding paired adjacent normal mucosa (55%),and there were significant differences between the former two and the latter,respectively.KiSS-1 methylation was not correlated with the clinico- pathological characteristics.
Methylation alteration can be found not only in solid cancer tissues but also in various remote samples derived from patients with cancer,such as sputum for lung cancer[17,18],urine for urologic tumors[19],saliva for head and neck squamous cell carcinoma[20],breast fluid[21]and serum or plasma for almost all types of cancer[22-26].More recent studies have been carried out to detect differences in various remote samples and demonstrated that DNA methylation could be act as a promising biomarker in early detection and prognosis[27].In our study,the association between KiSS-1 methylation in peritoneal lavage fluid and peritoneal metastasis was examined.We observed that the promoter of KiSS-1 gene was hypermethylated at a ratio of 42.5% (17/40).The presence of KiSS-1 methylation in peritoneal lavage fluid was significantly correlated with tumor invasion(P= 0.043).The reason may be that the incidence of free cancer cells fall off into peritoneal cavity increase according to the depth of tumor invasion.Among the 17 cases with promoter hypermethylation,9 cases had peritoneal metastasis including cytologically positive peritoneal lavage.KiSS-1 promoter hypermethylation in peritoneal lavage fluid showed a higher sensitivity (77.8%)for the diagnosis of peritoneal dissemination than cytological examination of peritoneal lavage fluid only.As known,the methylation alteration attractively acts as a biomarker for some reasons.Firstly,the methylation signal can be detected at low concentrations.Secondly,the methylation pattern and the underlying DNA are more stable than RNA[28].However,methylation alteration in peritoneal lavage fluid showed a lower specificity for diagnosis of peritoneal dissemination.Possible explanations are as follows.Firstly,most of the cells in peritoneal lavage are peritoneal mesothelial cells which may cause false positive of the KiSS-1 methylation.Secondly,the discrepancy of the methlytion profile exists sometimes between the peritoneal lavage samples and the cancer tissue.In order to solve these problems,a serial tests of KiSS-1 methylation and other examinations,for example,carcino-embryonic antigen in peritoneal lavage,could be used for diagnosing peritoneal dissemination to enhance specificity.The results of the experiment also show that KiSS-1 methylation in peritoneal lavage fluid is more frequently found in tumor with lymph node metastasis than in tumor without lymph node metastasis,although the difference between them did not reach a statistical significance.These findings suggest that the KiSS-1 methylation in peritoneal lavage fluid can be considered to be a biomarker for predicting peritoneal metastasis.
In summary,hypermethylation of KiSS-1 promoter is a common event in the development and progress of human gastric cancer,it may provide useful information for the diagnosis and assessment of occult metastasis in lymphonode.KiSS-1 methylation in peritoneal lavage fluid could act as a biomarker to evaluate peritoneal metastasis.However,more research should be carried out to further explore the mechanism of KiSS-1 hypermethylation in gastric cancer,and the promoter hypermethylation of KiSS-1 gene provides a new idea for prevention and treatment of gastric cancer.
[1]Parkin DM,Bray F,Ferlay J,et al.Global cancer statistics,2002[J].CA Cancer J Clin 2005; 55: 74-108.
[2]Ushijima T,Sasako M.Focus on gastric cancer[J].Cancer Cell 2004; 5: 121-5.
[3]Herman JG,Bsylin SB.Gene silencing in cancer in association with promoter hypermethylation[J].N Engl J Med 2003; 349: 2042-54.
[4]Oliveira C,Velho S,Domingo E,et al.Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer[J].Oncogene 2005; 24: 7630-4.
[5]Shen WJ,Dai DQ,Teng Y,et al.Regulation of demethylation and re-expression of RASSF1A gene in gastric cancer cell lines by combined treatment of 5-Aza-CdR and NaB[J].World J Gastroenterol 2008; 14: 595-600.
[6]Kang GH,Shim YH,Jung HY,et al.CpG island methylation in premalignant stages of gastric carcinoma[J].Cancer Res 2001; 61: 2847-51.
[7]Yamashita S,Tsujino Y,Moriguchi K,et al.Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2’ -deoxycytidine treatment and oligonucleotide microarry[J].Cancer Sci 2006; 97:64-71.
[8]Katsuragi K,Yashiro M,Sawada T,et al.Prognostic impact of PCR-based identification of isolated tumour cells in the peritoneal lavage fluid of gastric cancer patients who underwent a curative R0 resection[J].Br J Cancer 2007; 97:550-6.
[9]Kamiyama H,Noda H,Takata O,et al.Promoter hypermethylation of tumor-related genes in peritoneal lavage and the prognosis of patients with colorectal cancer[J].J Surg Oncol 2009; 100: 69-74.
[10]Shirasaki F,Takata M,Hatta N,et al.Loss of expression of the metastasis suppressor gene KiSS1 during melanoma progression and its association with LOH of chromosome 6q16.3-q23[J].Cancer Res 2001; 61: 7422-5.
[11]Stark AM,Tongers K,Maass N,et al.Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases[J].J Cancer Res Clin Oncol 2005;131: 191-8.
[12]Yi X,Li CY,Zhang SH,et al.Relationship and clinical significance of KiSS-1,nuclear factor kappa B (NF-kappaB),p50,and matrix metalloproteinase 9 expression in breast cancer[J].Zhonghua Bing Li Xue Za Zhi(in Chinese)2008;37: 238-42.
[13]Ringel MD,Hardy E,Bernet VJ,et al.Metastin receptor is ov erexpressed in papillary thyroid cancer and activates MAP Kinase in thyroid cancer cells[J].J Clin Endocrinol Metab 2002,87: 2399-402.
[14]Li N,Li SS,Zhang HY,et al.Effect of KISS-1 on invasive potential and proliferation of esophageal squamous carcinoma cell line EC-1[J].Zhonghua Bing Li Xue Za Zhi(in Chinese)2009; 38: 263-7.
[15]Guan-Zhen Y,Ying C,Can-Rong N,et al.Reduced protein expression of metastasis-related genes (nm23,KISS1,KAI1 and p53)in lymph node and liver metastases of gastric cancer[J].Int J Exp Pathol 2007; 88: 175-83.
[16]Nagai K,Doi R,Katagiri F,et al.Prognostic value of metastin expression in human pancreatic cancer[J].J Exp Clin Cancer Res 2009; 28: 9.
[17]Belinsky SA.Gene-promoter hypermethylation as a biomarker in lung cancer[J].Nat Rev Cancer 2004;4:707-17.
[18]Topaloglu O,Hoque MO,Tokumaru Y,et al.Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer[J].Clin Cancer Res 2004; 10: 2284-8.
[19]Hoque MO,Begum S,Topaloglu O,et al.Quantitative detection of promoter hypermethylation of multiple genes in the tumor,urine,and serum DNA of patients with renal cancer[J].Cancer Res 2004; 64: 5511-7.
[20]Carvalho AL,Jeronimo C,Kim MM,et al.Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma[J].Clin Cancer Res 2008; 14: 97-107.
[21]Fackler MJ,Malone K,Zhang Z,et al.Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid[J].Clin Cancer Res 2006; 12: 3306-10.
[22]Usadel H,Brabender J,Danenberg KD,et al.Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue,serum,and plasma DNA of patients with lung cancer[J].Cancer Res 2002; 62: 371-5.
[23]Koike H,Ichikawa D,Ikoma H,et al.Comparison of methylation-specific polymerase chain reaction (MSP)with reverse transcriptase-polymerase chain reaction (RT-PCR)inperipheral blood of gastric cancer patients[J].J Surg Oncol 2004; 87: 182-6.
[24]Laird PW.The power and the promise of DNA methylation markers[J].Nat Rev Cancer 2003; 3: 253-66.
[25]Sidransky D.Emerging molecular markers of cancer[J].Nat Rev Cancer 2002; 2: 210-9.
[26]Miyamoto K,Ushijima T.Diagnostic and therapeutic applications of epigenetics[J].Jpn J Clin Oncol 2005; 35:293-301.
[27]Tost J.DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker[J].Methods Mol Biol 2009; 507: 3-20.
[28]Cottrell SE.Molecular diagnostic applications of DNA methylation technology[J].Clin Biochem 2004; 37:595-604.
杂志排行
Chinese Journal of Cancer Research的其它文章
- Methylation and Demethylation of Ink4 Locus in Cancer Development
- Apoptotic B16-F1 Cells Coated with Recombinant Calreticulin Mediated Anti-tumor Immune Response in Mice
- Polymorphisms of UGT1A7 and XRCC1 are Associated with an Increased Risk of Hepatocellular Carcinoma in Northeast China
- Risk Factors of Precancerous Gastric Lesions in A Population at High Risk of Gastric Cancer
- Stanniocalcin-1 Detection of Peripheral Blood in Patients with Colorectal Cancer
- Recurrent Patterns and Factors Involved in Node-negative Advanced Gastric Cancer
