o2突变引起糯玉米籽粒淀粉积累差异研究
2024-04-28韩洁楠刘晓丽上官小川周婷芳郝转芳翁建峰雍洪军周志强徐晶宇李新海李明顺
韩洁楠 张 泽, 刘晓丽 李 冉 上官小川, 周婷芳, 潘 越 郝转芳 翁建峰 雍洪军 周志强 徐晶宇 李新海, 李明顺,*
突变引起糯玉米籽粒淀粉积累差异研究
韩洁楠1张 泽1,2刘晓丽1李 冉1上官小川1,2周婷芳1,2潘 越1郝转芳1翁建峰1雍洪军1周志强1徐晶宇2李新海1,2李明顺1,*
1中国农业科学院作物科学研究所, 北京 100081;2黑龙江八一农垦大学农学院, 黑龙江大庆 163319
糯玉米是主要鲜食玉米类型,()基因导入可增加籽粒赖氨酸含量, 但同时引起籽粒皱缩、淀粉含量下降等, 限制了其育种应用。为发掘优良糯玉米受体, 以籽粒饱满圆型近等基因系(-NIL)糯2/和皱缩型黄糯2/为研究材料, 通过对鲜食期、成熟期的百粒重和籽粒成分测定, 发现淀粉和可溶性糖含量不同可能是导致2份糯玉米-NILs表型差异的主要原因。利用实时荧光定量PCR技术分析, 发现授粉后10~24 d两糯玉米-NILs中6个淀粉合成基因动态表达模式不同, 其中、、和差异较大。分析胚乳转录组数据, 发现两糯玉米-NILs中24个海藻糖和糖基水解酶编码基因和48个胚乳修饰基因变化不同, 以上结果表明淀粉合成关键基因前期表达量高, 后期与对照无差异, 且糖代谢基因表达变化有利于淀粉合成可能是糯2/淀粉含量和百粒重不受突变影响, 籽粒性状明显优于黄糯2/的重要原因, 同时多个胚乳修饰基因的差异表达可能与该结果直接相关。本研究结果可为突变体在玉米育种中的应用提供重要参考。
糯玉米; 糯2/; 黄糯2/; 籽粒饱满度; 淀粉; 糖代谢; 差异基因
玉米是全球种植面积最大的农作物, 具有产量高、增产潜力大、用途广等特点, 是人类食物、动物饲料及工业原料的重要来源[1]。玉米籽粒赖氨酸匮乏, 是制约营养价值的第一限制性氨基酸, 因此以玉米为主粮或饲用需额外添加赖氨酸。()为调控籽粒发育的核心转录因子, 其隐性突变体胚乳中赖氨酸含量比普通玉米提高约70%[2]。但突变导致胚乳变粉质、容重及产量低、易感病。研究发现聚合胚乳修饰基因可使突变体籽粒恢复为硬质、产量提高, 从而选育出优质蛋白玉米(quality protein maize, QPM)[3-4]。QPM有效改善了南美、非洲和亚洲等以玉米为主食的发展中国家人群营养不良症, QPM饲料也提高了畜禽蛋白利用率[5]。将突变基因导入糯玉米可培育优质蛋白糯玉米, 但与连锁的不良性状导致糯玉米商品性差, 与普通QPM一样优质蛋白糯玉米选育也需要积累胚乳修饰基因。
糯玉米起源于我国, 为9号染色体()基因突变引起。我国糯玉米品种资源丰富, 广泛分布在云南、广西一带[6-7]。糯玉米适口性好、易消化、营养成分高于普通玉米, 是鲜食玉米的主力军[8]。糯玉米若作青穗鲜食, 对果穗食用品质和营养品质有严格的要求。食用品质主要是风味、柔嫩性、含糖量、糯性及皮渣率等; 营养品质主要包括蛋白质含量、氨基酸组成、淀粉含量等[9-10]。分析我国40份温带糯玉米自交系赖氨酸含量为0.14%~0.39%, 平均含量仅为0.23%[11], 与人体及畜禽需求的0.5%差异较大[12]。为获得兼具糯玉米和优质蛋白玉米优良特性的糯玉米品种, 研究人员将基因导入糯玉米自交系。Misra等[13]发现双突变体赖氨酸及游离氨基酸含量显著提高。张述宽等[14]选育出18个高赖氨酸糯玉米自交系, 氨基酸含量高于QPM, 具有较好的抗病性和农艺性状。Sinkangam等[15]通过回交获得11个高赖氨酸糯玉米, 赖氨酸、可溶性糖和支链淀粉含量显著高于普通糯玉米。Zhou等[16]发现双突变体醇溶蛋白、氨基酸合成、胁迫和信号转导等相关蛋白质积累发生变化, 支链淀粉含量增加。Dang等[17]运用双单倍体技术发现聚合和基因可提高赖氨酸含量, 同时对糯玉米品质影响较小。
对多个突变体进行转录组分析发现, 一些基因仅在特定突变体/背景中表达水平有差异; 不同突变体中一致存在的差异表达基因, 表达水平因背景差异而明显不同[18]。玉米种内变异丰富, 因此突变引起的胚乳基因转录水平表达差异较大[19-23]。Jia等[20]发现W64A/中多个淀粉合成基因转录及蛋白水平上调表达, 认为、上调表达引起淀粉结晶度改变; Zhang等[22]发现W64A/中淀粉合成基因下调表达, 蛋白表达减少; 本课题组发现CAL58/中下调表达,、上调表达, SuSy酶活性降低, AGPase酶活性增加(数据未发表)。通过分子辅助育种可聚合和基因, 提高籽粒赖氨酸含量, 但双突自交系籽粒容重低、易感病、产量下降, 不适宜直接选育品种[24-25]。突变体胚乳修饰基因数量多, 多数分子机制不清楚, 缺乏相应的分子标记, 因此将及其修饰基因同时导入糯玉米自交系困难较大。通过比较多份糯玉米近等基因系(-NILs), 我们发现籽粒表型明显不同[16,26], 表明存在优良糯玉米受体, 在导入基因后籽粒产量与相应普通糯玉米产量接近。本研究以籽粒表型显著不同的-NILs黄糯2/和糯2/为材料, 通过比较鲜食期和成熟期籽粒组成、淀粉合成基因表达和转录组差异基因等, 为发掘优良糯玉米受体亲本提供依据。
1 材料与方法
1.1 田间种植和取样
2020年5月初将试验材料种植于中国农业科学院作物科学研究所北京昌平试验基地, 3 m行长, 25 cm株距, 每材料设置3个小区, 每小区种植5行, 自交授粉并记录授粉日期。用于转录组测序及定量分析的胚乳样品通过人工剥取, 每个材料取3个重复, 每重复取3个果穗置于冰上保鲜, 取果穗中部的籽粒, 在冰上用镊子剥取胚乳并迅速放进液氮中冷冻暂存, 后置于–80℃冰箱中备用。授粉后25 d收获鲜食期果穗, 每小区随机选取3个穗, 混作一个生物学重复, 部分新鲜籽粒保存备用, 另一部分籽粒在自然条件下晾干、脱粒, 去除杂损粒后装袋保存备用。其余果穗授粉后自然成熟并晾干, 分小区收获并脱粒后, 装袋保存。
1.2 10K液相芯片技术检测背景回复率
检测样品为回交6代自交3代以上的糯玉米回交转育植株, 取田间种植下植株新叶样品送石家庄博奥迪有限公司进行分析。在黄糯2一组材料中共检测到11,444个核心SNP位点, 其中相同位点有11,359个, 差异位点有85个, 背景恢复率为99.26%; 糯2一组材料中共检测到11,420个核心SNP, 差异位点有313个, 相同位点有11,107个, 背景恢复率为97.26%, 表明两者与理论值98.4%接近。
1.3 百粒重、种皮厚度、可溶性糖、氨基酸及淀粉含量测定
百粒重用万分之一天平称量, 每小区随机称量3次100粒种子重量, 取平均值记作一个重复。鲜食期籽粒种皮厚度测定: 取授粉后25 d饱满一致籽粒用FAA固定, 剥取籽粒胚背面果皮, 在水和甘油混合液中软化, 后置于27℃干燥, 用螺旋测微仪对每块果皮测量3次, 取10粒果皮平均值记作一个重复。成熟期籽粒种皮厚度测定: 取饱满一致籽粒10粒混合, 于沸水中煮, 冷却后将背胚面种皮剥下, 吸水纸擦干后, 用螺旋测微尺对每块种皮测量3次, 取平均值记作一个重复, 具体测定方法参见刘晓丽等[27]。
赖氨酸测定依据《植物中游离氨基酸的测定》(GB/T 30987-2020)中全自动氨基酸分析仪法, 每份样品测2次, 取平均值记作一个重复, 委托青岛科创质量检测有限公司进行。可溶性糖和总淀粉测定分别根据植物可溶性糖试剂盒(蒽酮比色法)以及Megazyme淀粉总量检测试剂盒(K-TSTA)说明书操作, 每份样品测2次, 取平均值记作一个重复。
1.4 醇溶蛋白的提取和SDS-PAGE电泳
将供试材料成熟籽粒分别研磨成粉, 称取0.05 g置于离心管中, 加入70%乙醇400 μL (含2%巯基乙醇), 室温摇晃2~3 h, 12,000转min–1离心10 min, 吸取上清100 μL, 加入10% SDS溶液10 μL, 80℃烘干, 加入100 μL ddH2O重悬并加入Loading buffer煮沸, 吸取5 μL样品进行10% SDS-PAGE凝胶电泳, 最后用固定液、染色液和脱色液处理直至条带清晰可见, 详细步骤参见刘晓丽等[27]。
1.5 转录组测序
对糯2/、黄糯2/及其轮回亲本每个材料设置3次生物学重复, 共计12个样品进行转录组测序, 获得90.73 GB数据量。各样品Clean Data均达到6.42 GB, Q30碱基百分比在92.93%以上。将各样品Clean Reads与B73参考基因进行序列比对, 比对率为86.05%~90.17%, 表明转录组测序的质量较高。对12个样品做系统进化树分析, 糯2/与糯2、黄糯2/与黄糯2聚类在2个分支上, 表明所用材料和转录组数据具有较高可靠性。
1.6 RNA提取及cDNA合成
使用TransZolUP Plus RNA提取试剂盒(北京全式金生物技术股份有限公司)提取玉米胚乳RNA, 详细操作步骤见说明书。使用FastKing gDNA Dispelling RT SuperMix试剂盒(天根生化科技(北京)有限公司)将RNA反转为cDNA, 详细操作步骤见说明书。得到的cDNA保存于–20℃冰箱。
1.7 荧光定量引物设计及定量验证
以Ubiquitin为内参基因, 对淀粉合成及糖代谢差异表达基因进行定量分析, 根据基因ID, 在NCBI数据库中下载基因序列。将基因序列导入Primer 5.0设计定量引物, 委托北京华大基因科技有限公司合成。将cDNA稀释作为模板, 通过普通PCR扩增目的条带, 得到的PCR产物进行琼脂糖凝胶电泳, 在紫外线照胶仪中观察条带是否特异, 本文所用定量引物信息见附表1。
采用cDNA模板2 μL, 10 μL 2×SuperReal PreMix Plus, 0.6 μL引物, 6.8 μL RNase-free ddH2O配制qRT-PCR反应体系。每份样品均设置3个生物学重复和3个技术重复。使用BIO-IQ5荧光定量PCR仪进行三步法PCR反应, 程序如下: 预变性95℃ 15 min,变性95℃ 10 s, 退火60℃ 10 s, 延伸72℃ 10 s, 40个循环。数据处理采用相对定量的分析方法采用2–ΔΔCT法, 计算公式为: 表达量比值=2–{[待测组目的基因平均CT值–待测组内参基因平均CT值]–[对照组目的基因平均CT值–对照组内参基因平均CT值]}
1.8 数据分析
数据通过Microsoft Excel 2007和SPSS 21.0进行统计分析。
2 结果与分析
2.1 糯玉米o2-NILs籽粒表型分析
前期我们以优质蛋白玉米CA339为基因供体, 通过回交转育创制出多份糯玉米-NILs[16,25]。通过比较发现黄糯2/与糯2/籽粒表型显著不同, 黄糯2/为典型的突变体表型, 籽粒明显皱缩, 而糯2/籽粒呈饱满圆润状, 皱缩不明显, 但两者胚乳均呈粉质态(图1-A)。与各自轮回亲本相比(对照), 黄糯2/百粒重和淀粉含量显著降低, 糯2/籽粒则无差异, 明显高于黄糯2/并且该差异不受环境影响(图1-B)。进一步测定籽粒成分, 鲜食期与对照相比黄糯2/总淀粉含量和百粒重降低6.12%和1.11%, 可溶性糖含量不变, 赖氨酸含量增加54.28%; 而糯2/籽粒总淀粉含量增加6.12%, 赖氨酸增加38.10%, 百粒重和可溶性糖含量不受影响。这与我们前期发现的突变对多份糯玉米鲜食期品质影响较大, 黄糯2/和糯2/为代表性差异材料的结果一致[27]。成熟期黄糯2/籽粒中可溶性糖增加12.50%、总淀粉含量降低6.24%, 百粒重降低8.80%, 赖氨酸含量增加79.17%; 但糯2/籽粒总淀粉含量、可溶性糖及百粒重均不受影响, 赖氨酸含量增加58.33% (表1)。糯2/籽粒表型与对照相似, 但粉质胚乳占比明显增加, 因此胚乳质地不同于QPM。黄糯2/胚乳中醇溶蛋白亚基27 kD γ-zein显著降低, 糯2/胚乳中27 kD γ-zein含量显著高于对照, 与CA339接近(图2)。此外, 15 kD β-zein、19 kD和22 kD α-zein含量在糯玉米-NILs均明显降低, 16 kD γ-zein在黄糯2/和糯2/变化相反。前期我们发现赵OP-6/SY1-2/与糯2/相似, 27 kD γ-zein明显高于对照, 赵OP-6/淀粉含量增加, 籽粒较饱满、圆润而SY1-2/淀粉含量降低, 籽粒呈明显皱缩状[27], 表明27 kD γ-zein蛋白增加是糯玉米-NILs籽粒恢复饱满状的必要非充分条件, 淀粉含量也是影响籽粒饱满度的重要因素。
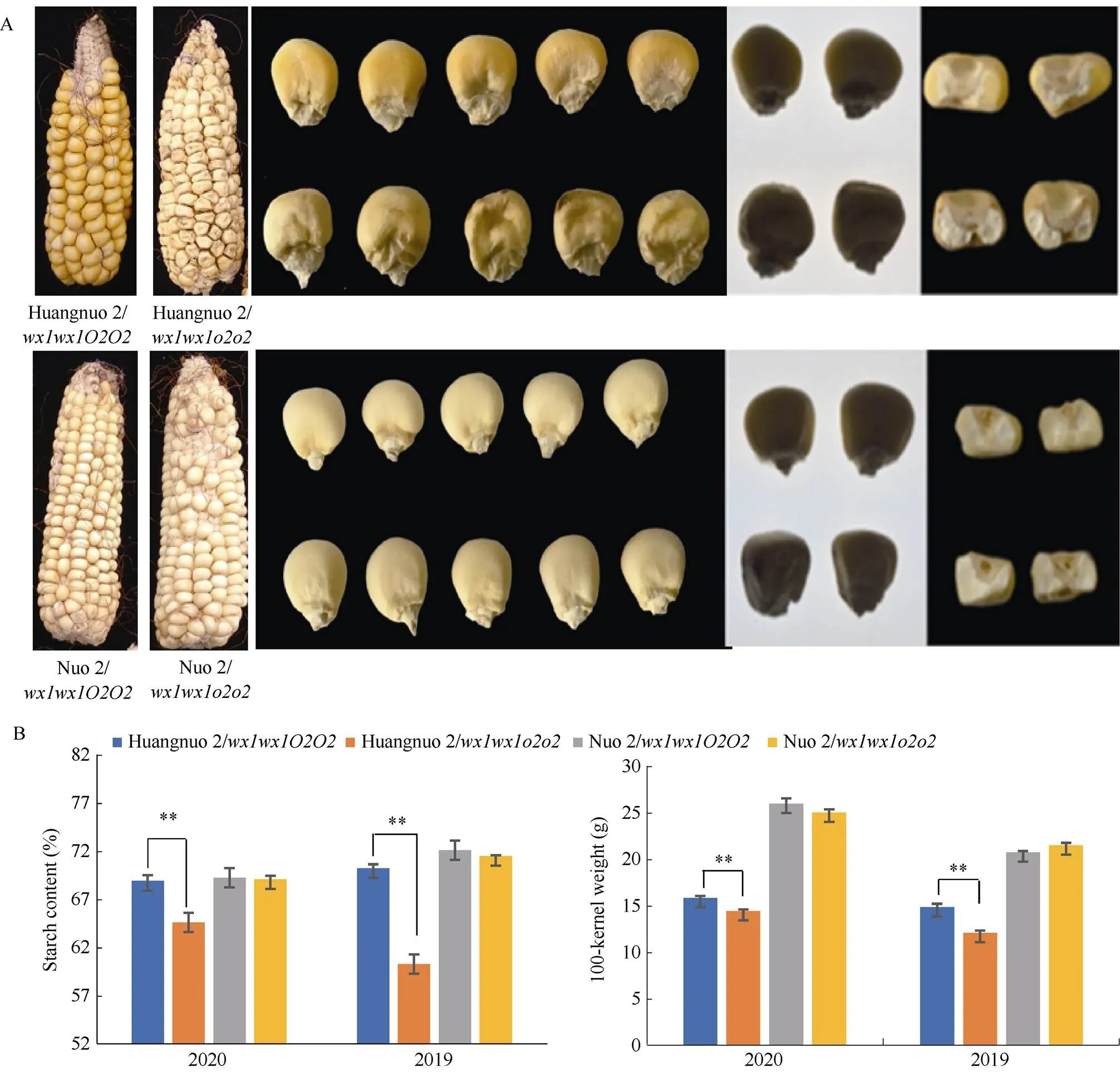
图1 黄糯2/wx1wx1o2o2和糯2/wx1wx1o2o2籽粒表型
(A): 籽粒表型比较; (B): 不同年代下籽粒淀粉含量和百粒重比较。**表示在0.01概率水平差异显著。
(A): kernel phenotype; (B): starch content and 100-kernel weight in different years. ** indicates significant difference at the 0.01 probability level.
2.2 糯玉米o2-NILs淀粉合成关键基因的动态表达分析
淀粉合成积累是一个持续过程, 显著影响籽粒饱满度。取授粉后10 d (10 DAP)、15 DAP、20 DAP、24 DAP胚乳对淀粉合成基因表达水平进行比较(图3)。与各自对照相比, 10 DAP时两糯玉米NILs中、、、和表达量均无显著变化;和在黄糯2/中上调1.70倍和1.55倍, 在糯2/中上调1.99倍和3.22倍;在黄糯2/中上调1.52倍, 在糯2/中无显著变化。15 DAP时,和在两糯玉米NILs中无显著变化;和在黄糯2/中下调1.69倍和2.22倍, 但在糯2/中无显著变化;、、和在黄糯2/中无显著变化, 在糯2/中则分别是对照的5.79、8.88、3.63和4.27倍。20 DAP时, 与对照相比黄糯2/中、、和无变化,和下调1.72倍和1.30倍,上调1.95倍和1.85倍; 糯2/中仅表达量下调3.13倍, 剩余基因均无变化。24 DAP时, 相较于对照黄糯2/中表达量下调1.37倍,、和上调1.48、1.45和1.54倍,、、和表达量无显著变化; 而糯2/中所有供试基因无变化。综上表明两糯玉米NILs在授粉后15~24 d淀粉合成基因表达差异显著, 其中、、和在3个时期发生显著变化, 差异最明显, 推测是导致两糯玉米NILs籽粒淀粉含量不同的主要原因。

表1 鲜食期和成熟期糯玉米o2-NILs籽粒成分测定结果
*和**分别表示在0.05和0.01概率水平差异显著。
*and**indicate significant difference at the 0.05 and 0.01 probability levels, respectively.
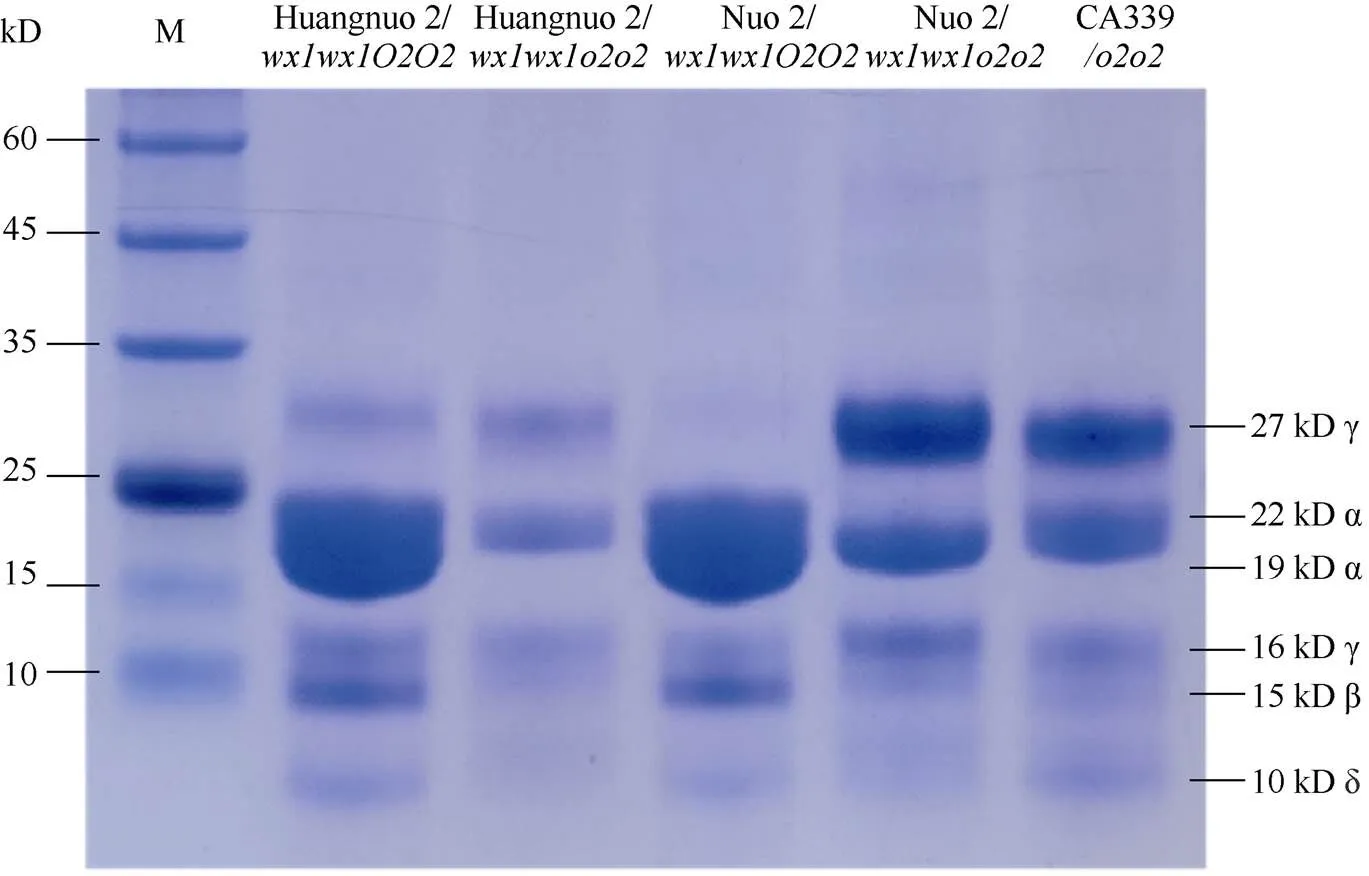
图2 糯玉米胚乳醇溶蛋白亚基分析
2.3 糯玉米o2-NILs胚乳糖代谢路径差异表达基因分析
为进一步探究突变导致黄糯2/和糯2/籽粒淀粉积累不同原因, 对两材料20 DAP胚乳进行转录组测序。以各自轮回亲本为对照分析黄糯2/和糯2/差异表达基因(differently expressed genes, DEGs), 黄糯2/中共有2466个DEGs, 其中1290个基因上调表达, 1176个基因下调表达; 糯2/中共有1685个DEGs, 其中1124个基因上调表达, 561个基因下调表达(图4-A)。糯2/中DEGs远低于黄糯2/, 表明突变对胚乳发育的影响依赖于基因型, 糯2/受影响程度可能低于黄糯2/。对DEGs进行KEGG路径富集, 黄糯2/DEGs主要富集到内质网蛋白质加工、碳代谢、DNA复制、淀粉和蔗糖代谢路径(图4-B); 糯2/主要富集到碳代谢、淀粉和蔗糖代谢、甘氨酸、丝氨酸和苏氨酸代谢、苯丙烷生物合成和糖酵解5个路径(图4-C)。
授粉后20 d两糯玉米-NILs共筛选出34个淀粉和糖代谢相关DEGs (图5-A), 淀粉合成相关DEGs有AGPase小亚基、淀粉合成酶SSIII、葡萄糖变位酶、异淀粉酶和α淀粉酶编码基因共计7个。剩余27个DEGs为糖代谢相关基因(图5-B), 其中4个DEGs为两糯玉米-NILs共有基因, 分别编码丙酮酸磷酸双激酶(PPDK)、海藻糖磷酸合酶(TPP)和果胶酯酶, 在两糯玉米-NILs中一致下调。15个DEGs仅在黄糯2/中差异表达, 其中12个基因下调, 3个基因上调, 包括5个海藻糖六磷酸合酶(TPS)基因、3个TPP基因、4个糖基水解酶基因、2个果糖激酶、1个果胶酯酶; 7个DEGs在糯2/中特异表达, 其中5个基因下调表达, 2个基因上调, 包括2个TPP编码基因和5个糖基水解酶基因。对8个DEGs进行RT-PCR验证, 发现与RNA-seq结果一致(图6)。
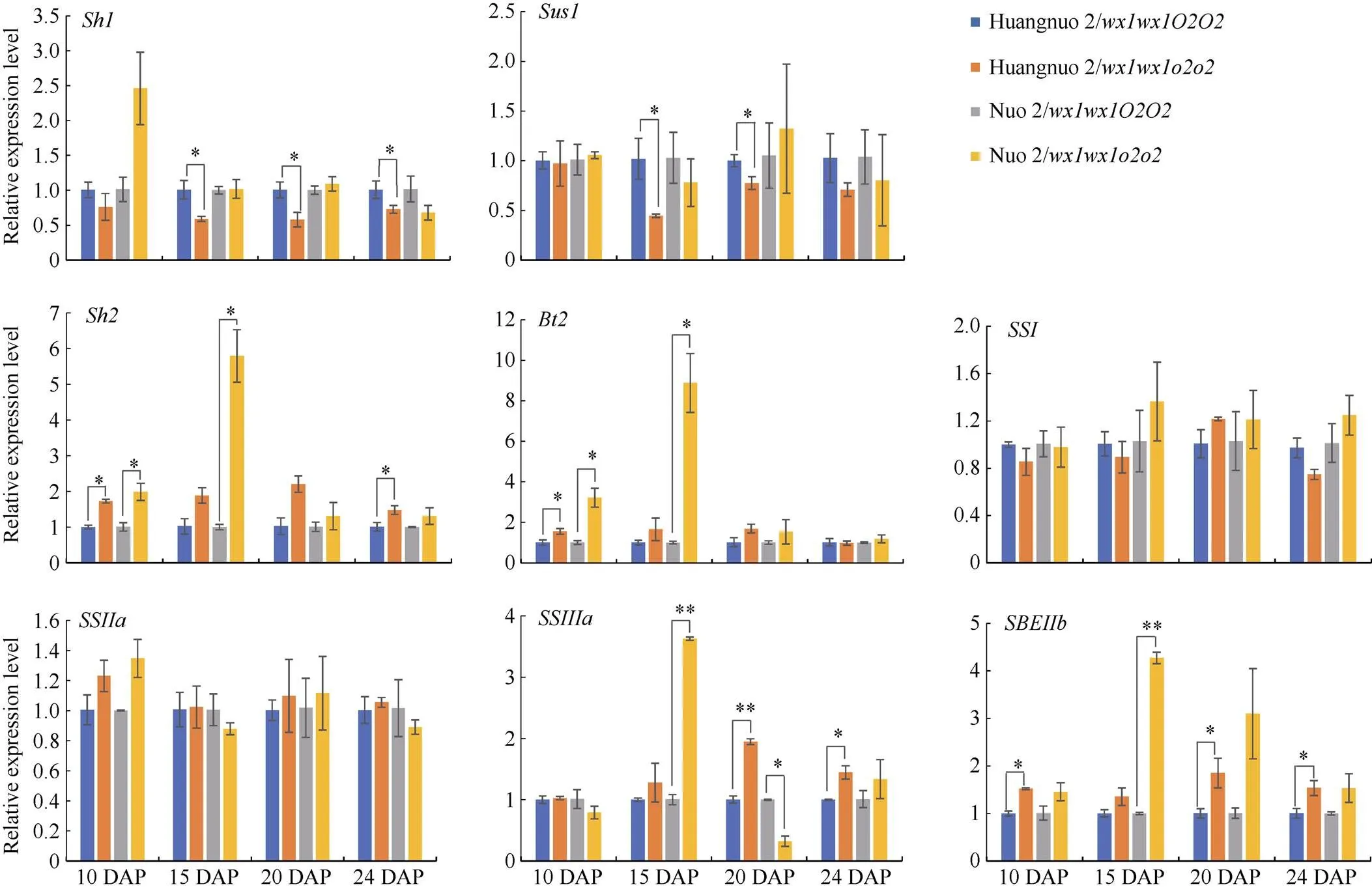
图3 糯玉米o2-NILs淀粉合成路径关键基因差异表达量分析
*和**分别表示在0.05和0.01概率水平差异显著。DAP: 授粉后天数。
* and ** indicate significant difference at the 0.05 and 0.01 probability levels, respectively.DAP: days after pollination.
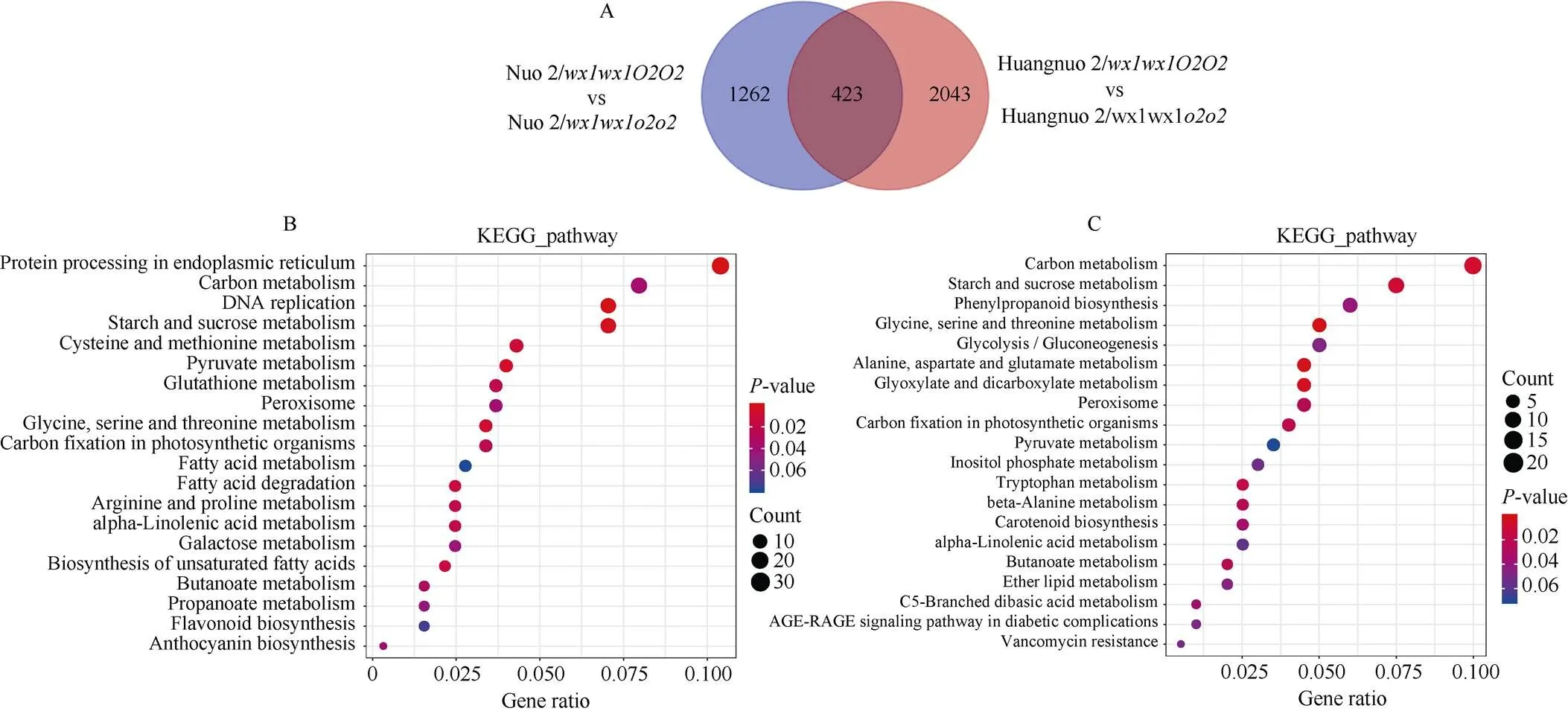
图4 糯玉米o2-NILs差异表达基因KEGG富集分析
(A): 两糯玉米-NILs差异表达基因数目; (B): 黄糯2/差异表达基因KEGG富集分析; (C): 糯2/差异表达基因KEGG富集分析。
(A): the number of DEGs of the two waxy maize-NILs; (B): KEGG enrichment of differentially expressed genes in Huangnuo 2/; (C): KEGG enrichment of differentially expressed genes in Nuo 2/
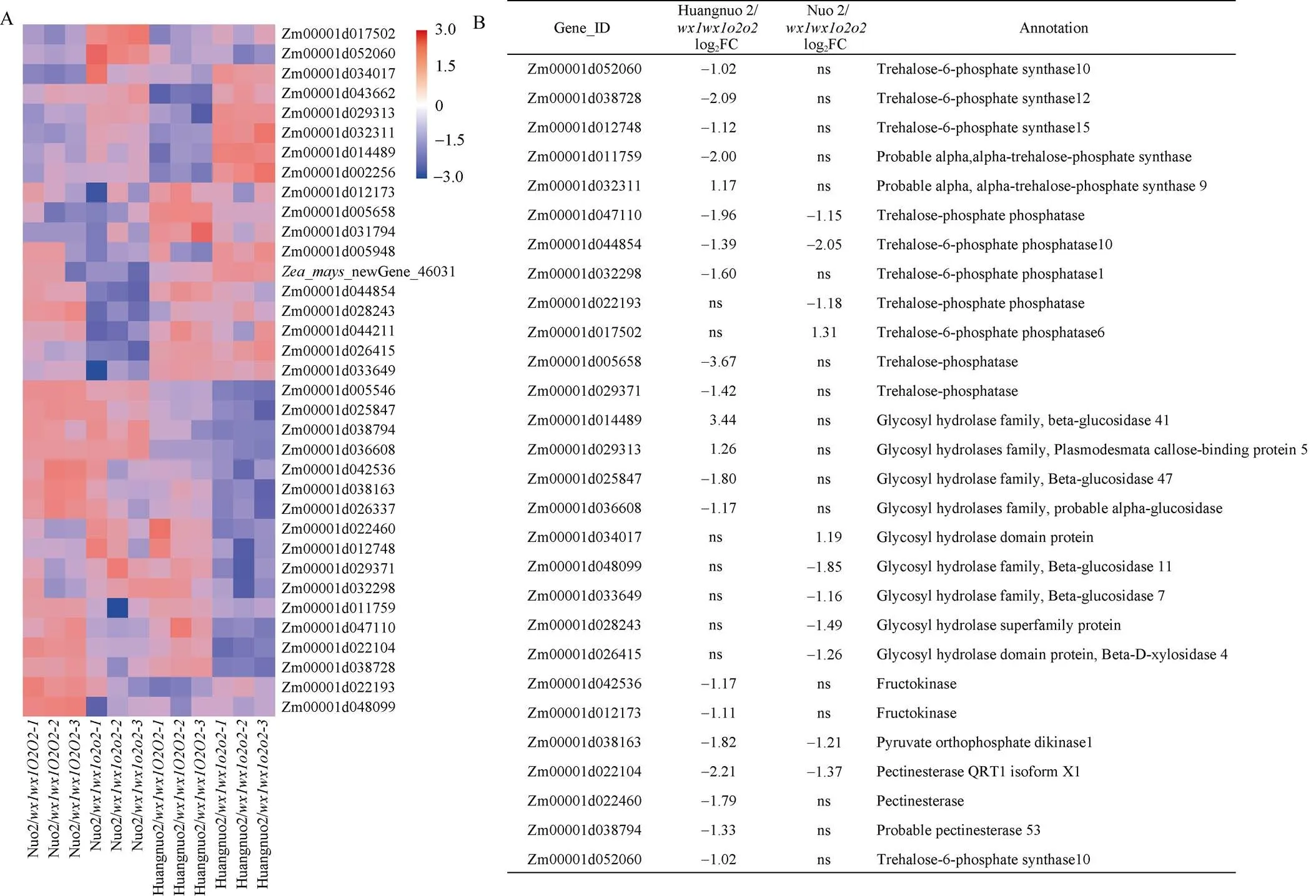
图5 授粉后20 d糯玉米o2-NILs淀粉和糖代谢路径差异基因分析
(A): 糯玉米-NILs淀粉和糖代谢相关差异基因分析; (B): 糯玉米-NILs糖代谢差异基因分析。ns代表无显著性差异。
(A): the analysis of differentially expressed genes in starch and sugar metabolism pathways of waxy maize-NILs; (B): the analysis of differentially expressed genes in sugar metabolic pathways of waxy maize-NILs. ns: no significant difference.
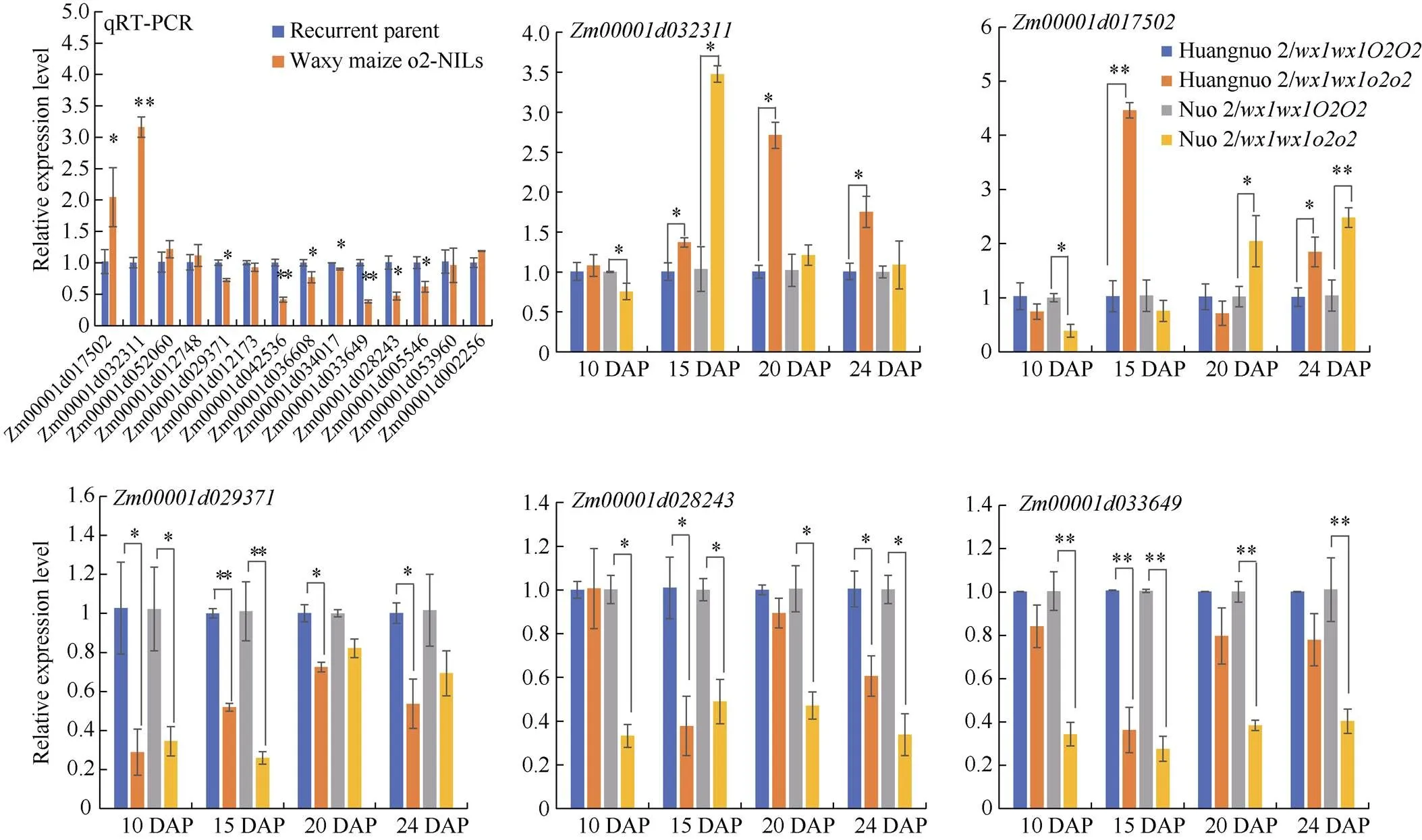
图6 糯玉米o2-NILs糖代谢差异基因定量分析
*和**分别表示在0.05和0.01概率水平差异显著。DAP: 授粉后天数。
* and ** indicate significant difference at the 0.05 and 0.01 probability levels, respectively.DAP: days after pollination.
糖代谢相关DEGs以海藻糖合成和糖基水解酶基因为主, 暗示糖代谢在两糯玉米-NILs中发生显著变化。动态表达分析黄糯2/中TPS编码基因在10 DAP时表达量无变化, 但此后上调表达, 分别上调1.37、2.71和1.75倍, 糯2/中10 DAP时其表达量下调1.32倍, 15 DAP上调3.38倍, 20 DAP和24 DAP时无差异。TPP编码基因在黄糯2/中10 DAP时表达量无变化, 15 DAP时上调4.46倍, 24 DAP时上调1.85倍, 在糯2/中10 DAP时表达量下调2.56倍, 20 DAP和24 DAP上调2.04倍和2.48倍; 另一TPP编码基因在黄糯2/中显著下调(1.40~3.33倍), 在糯2/中10 DAP和15 DAP下调2.86倍和3.85倍, 后2个时期不受影响。编码葡聚糖-内-1,3-β葡糖苷酶, 黄糯2/中15 DAP和24 DAP时表达量下调2.63和1.64倍, 但在10 DAP和20 DAP不受影响, 在糯2/中4个时期均下调表达(2.04~3.03倍);编码另一个糖基水解酶, 黄糯2/中15 DAP时表达量下调2.78倍, 其余3个时期表达量与对照无差异, 在糯2/中4个时期表达量均显著下调(2.50~3.45倍)。
2.4 糯玉米o2-NILs胚乳醇溶蛋白和氨基酸相关差异表达基因分析
黄糯2/中30个醇溶蛋白相关基因下调2.31~99.30倍(log2FC = –1.21~ –6.63), 糯2/中有24个醇溶蛋白基因下调2.30~ 45.21倍(log2FC = –1.20~ –5.50) (附表2)。其中22个为两糯玉米-NILs共有DEGs, 19个基因编码19 kD α-zein和22 kD α-zein,编码15 kD β-zein, 20个基因在两糯玉米-NILs中均下调表达, 与-NILs籽粒19 kD、22 kD和15 kD α-zein显著降低结果一致;编码16 kD γ-zein, 仅在黄糯2/中表达量下降,与其16 kD α-zein降低一致(图2)。、、和为赖氨酸降解基因(附表3), 其下调1.2~17.1倍(log2FC = –1.01 ~ –4.10)使赖氨酸降解受阻, 可能是糯玉米-NILs赖氨酸含量增加的主要原因[28](表1)。
3 讨论
玉米突变后籽粒表现不同, 例如导入自交系B46籽粒变化显著, 而导入M14籽粒基本无变化[18]。谭华等[29]将导入多个普通玉米自交系后发现-NILs赖氨酸含量大幅提高, 容重增加, 热带和亚热带种质百粒重多数下降, 但在温带种质中增加。籽粒皱缩、胚乳粉质且不透光是突变体的典型表现。淀粉是籽粒主要贮藏物质, 占胚乳干重的70%, 淀粉积累不足被认为是籽粒皱缩、百粒重低的重要原因。淀粉的合成是一个复杂连续过程, 授粉后10 d籽粒中淀粉开始大量合成, 20 d左右达到最大速率[30-32], 这一过程受蔗糖合成酶(SuSy)、腺苷二磷酸葡萄糖焦磷酸化酶(AGPase)、淀粉合成酶(SSs)、淀粉分支酶(SBE)和去分支酶(DBE)等关键酶调控, 编码基因表达量的改变对籽粒淀粉含量影响显著[33-35]。本研究发现黄糯2/和糯2/籽粒皱缩程度与胚乳淀粉合成基因表达不同密切相关(图1、图4和表1)。SuSy是双向催化酶, 主要功能是分解蔗糖, 为淀粉和蛋白质合成提供底物[36]。玉米K0326Y/中突变, SuSy活性降低, 淀粉合成受抑制, 籽粒皱缩[37]。黄糯2/中表达水平显著下降, 籽粒明显皱缩, 而糯2/中表达不受影响, 籽粒饱满程度与对照接近。是SuSy的另一个编码基因, 表达量变化与相似, 黄糯2/中下调而糯2/中无变化, 表明黄糯2/胚乳中淀粉合成底物供应可能受到抑制, 但糯2/中不受影响。AGPase是淀粉合成限速酶, 影响碳向淀粉途径的分配[38], 胚乳中主要编码基因是和。正常籽粒授粉后蔗糖比重逐渐降低, 淀粉比重增加, 收获时以淀粉为主, 而突变导致籽粒中多聚糖链合成受抑制, 蔗糖含量增加, 淀粉积累速率受抑制, 籽粒变为皱缩状[39]。10 DAP时糯2/和黄糯2/中和显著上调, 但15 DAP时仅糯2/中上调表达(图3), 表明淀粉积累初期(10 DAP)糯玉米NILs中AGPase表达增加, 但只有糯2/保持较高水平至15 DAP。SSs通过催化ADPG葡萄糖基转移到葡聚糖的非还原末端来延长支链淀粉的长度, 糯玉米由于()基因突变, 直链淀粉合成受抑制, 只能合成支链淀粉; SBE将α-1,4糖苷键切开, 将截短的葡聚糖链与C6羟基链接, 形成支链淀粉的分支[40]。与对照比, 两糯玉米-NIL中、表达水平无差异, 推测胚乳中短葡聚糖链的合成不受影响; 糯2/中15 DAP时上调表达, 与和变化一致, 表明前期糯2/可能积累更多支链淀粉, 与鲜食期籽粒淀粉含量高于对照相符; 黄糯2/中也上调表达, 推测胚乳中淀粉合成底物不足为主要限制因素, 因此籽粒淀粉含量低于对照。根据淀粉合成基因表达模式推测, 前期(0~15 DAP)糯2/籽粒淀粉合成速率较高, 20 DAP时恢复至对照水平, 但黄糯2/自15 DAP淀粉合成持续低于对照水平。
淀粉的合成还与糖代谢密切相关, 例如海藻糖合成、糖酵解等均可与淀粉合成底物或中间产物相互转化。海藻糖-6-磷酸酶(TPS)催化UDPG和葡萄糖-6-磷酸(G6P)合成海藻糖-6-磷酸(T6P), 进而在海藻糖-6-磷酸磷酸酶(TPP)作用下合成海藻糖。G6P和UDPG是淀粉合成和碳代谢底物, T6P为G6P和UDPG库容标志物, 在植物生长及碳利用中有重要作用[41]。豌豆缺乏T6P, 种子大小和淀粉含量受影响,表型与孟德尔研究的皱缩豌豆种子相似[42]; 过表达大肠杆菌基因可增加拟南芥叶片淀粉合成[43]; Hu等[44]发现增强TPS或降低TPP活性可增加玉米籽粒淀粉含量。T6P还与蔗糖非发酵蛋白激酶(SnRK1)互作双向调控蔗糖和淀粉合成[45]。植物TPS和TPP酶由多个基因编码[46], 20 DAP时糯2/中基因表达不受影响, 多个基因下调, 推测糯2/TPS活性不受影响,但TPP活性降低, 有利于淀粉合成; 而黄糯2/中多数和编码基因下调, 推测黄糯2/TPS和TPP活性均降低, 不利于淀粉生成(图5-B)。进一步分析和动态表达模式说明海藻糖途径少数基因的变化与淀粉合成速率直接相关(图6)。果糖激酶在糖酵解途径发挥重要作用, 可与蔗糖合酶协同调控蔗糖合成与降解[47], 番茄幼果中通过调节蔗糖输入, 对淀粉积累产生影响[48-49]。本研究发现突变仅引起2个果糖激酶基因在黄糯2中下调表达, 是两糯玉米-NILs糖酵解受影响不同的表现。结构多样的糖苷水解酶在糖和糖缀合物水解与合成中扮演重要角色, 编码基因众多, 例如拟南芥有393个编码基因[50]。α-葡萄糖苷酶参与淀粉及糖原代谢, β-木糖酶与β-葡萄糖苷酶为纤维素酶, 降解纤维素生成纤维二糖和葡萄糖[51], 以上编码基因在两糯玉米-NILs多数下调, 说明突变可能抑制了胚乳的糖苷水解, 使胚乳发育和细胞壁代谢受影响。果胶是大分子多糖, 与植物生长发育、逆境应答等生物学过程密切联系[52], 番茄中催化果胶多聚体形成的半乳糖醛酸转移酶编码基因突变, 果实果胶结构改变, 淀粉和果实含量降低[53]。果胶酯酶是三大果胶酶之一, 随机切除水溶性果胶分子的酯键, 产生游离羧基[54], 苹果果胶甲酯酶在粉质化不同的果实中呈现表达差异[55]。突变导致果胶酯酶基因下调表达, 可能与糯玉米-NILs胚乳质地改变相关。磷酸丙酮酸双激酶(PPDK)是CO2固定关键酶, 胚乳中高表达可与淀粉合成酶稳定结合, 协同调控后期籽粒灌浆[56], 两糯玉米NILs中一致下调, 可能导致淀粉酶活性降低。
醇溶蛋白形成后储存于蛋白体中, 若醇溶蛋白与淀粉同步合成, 则紧密结合形成硬质胚乳占比高的硬粒玉米。糯玉米-NILs醇溶蛋白合成受阻, 淀粉合成基因也受影响(附表2和图3), 导致成熟籽粒中储藏物质降低, 胚乳几乎全部变为粉质状(图1)。27-kD γ-zein调控蛋白体形成数量, QPM胚乳中γ27蛋白表达量增加, 是公认的修饰基因, 并且修饰基因数量与γ27蛋白表达正相关[57-58], 糯2/中27 kD γ-zein表达量高于黄糯2/(图2), 暗示糯2/中胚乳修饰基因数量多。通过定位群体Holding发掘到多个胚乳修饰基因位点, 主效基因位于7号染色体; 包括27-kD γ-zein、葡萄糖转运蛋白、焦磷酸依赖型果糖-6-磷酸1-磷酸转移酶α亚基()、蛋白质磷酸酶2C ()等16个基因位于连锁区间内, 这些基因在QPM中多数上调表达[59-60]。Li等[61]发现热激蛋白()、HSP伴侣蛋白()、丝氨酸/精氨酸富含蛋白质编码基因()在QPM中也呈上调表达。本研究发现黄糯2/中39个胚乳修饰基因差异表达, 包括32个、5个、1个和1个其中仅4个基因上调; 糯2/中13个胚乳修饰基因差异表达, 包括6个、6个和1个, 其中8个基因上调(附表4)。表达增加使PFP催化活性提高, 加速糖酵解, 改善突变引起的能量匮乏, 与硬质透明胚乳恢复相关[59-62];参与RNA剪切, 拟南芥中受胁迫时其上调表达[63], 水稻中在内质网应激下被激活, 可促进未折叠蛋白在液泡和内质网之间的传递[64]。上述基因在糯玉米-NILs均下调表达, 是籽粒表型不同于普通糯玉米的重要原因。功能与脱落酸信号途径有关, 在优质蛋白玉米K0326Y/中上调, 可能与其他修饰基因共同起作用[57], 但在糯玉米-NILs变化趋势不一致。在QPM中上调, 对缓解突变引起未折叠单重组或蛋白质聚集引起的应激效应有重要作用[61-62], 糯2/中仅1个下调, 而黄糯2/中有30个, 这可能与糯2/籽粒淀粉和百粒重不受影响直接相关,需进一步研究。
4 结论
将基因导入糯玉米可显著提高籽粒赖氨酸含量, 改善营养价值, 但与基因连锁的不良性状限制了优质蛋白糯玉米选育。与皱缩型近等基因系黄糯2/相比, 糯2/玉米成熟期淀粉含量及百粒重明显提高, 籽粒圆润饱满。研究发现糯2/与黄糯2/中以、、和为主的淀粉合成基因、以海藻糖和糖基水解酶为主的糖代谢基因表达模式明显不同, 推测糯2/10~15 DAP淀粉合成速率高, 后与对照无差异, 但黄糯2/自15 DAP起淀粉合成速率持续低于对照, 且糯2/中糖代谢基因的变化更有利于淀粉合成。根据转录组差异表达基因分析结果, 认为胚乳修饰基因, 尤其是热激蛋白编码基因的差异表达可能与两糯玉米-NILs籽粒淀粉和百粒重不同直接相关。
[1] Ellis R P, Cochrane M P, Dale M F B, Duffus C M, Lynn A, Morrison I M, Prentice R D M, Swanston J S, Tiller S A. Starch production and industrial use., 1998, 77: 289–311.
[2] Mertz E T, Bates L S, Nelson O E. Mutant gene that changes protein composition and increases lysine content of maize endosperm., 1964, 145: 279–280.
[3] Paez A V, Helm J L, Zuber M S. Lysine content of opaque2 maize kernels having different phenotypes., 1969, 9: 251–253.
[4] Gibbon B C, Larkins B A. Molecular genetic approaches to developing quality protein maize., 2005, 21: 227–233.
[5] 石德权. 优质蛋白玉米. 北京: 中国农业出版社, 1995. Shi D Q. High Quality Protein Maize. Beijing: China Agriculture Press, 1995 (in Chinese).
[6] 曾孟潜. 我国糯质玉米的亲缘关系. 作物品种资源, 1987, (3): 4. Zeng M Q. The affinities of glutinous maize in China., 1987, (3): 4 (in Chinese).
[7] Zheng H J, Wang H, Yang H, Wu J H, Shi B, Cai R, Xu Y B, Wu A Z, Luo L J. Genetic diversity and molecular evolution of Chinese waxy maize germplasm., 2013, 8: e66606.
[8] 赵久然, 卢柏山, 史亚兴, 徐丽. 我国糯玉米育种及产业发展动态. 玉米科学, 2016, 24(4): 67–71. Zhao J R, Lu B S, Shi Y X, Xu L. Dynamics of breeding and industrial development of glutinous maize in China., 2016, 24(4): 67–71 (in Chinese with English abstract).
[9] Azanza F, Klein B P, Juvik J A. Sensory characterization of sweet maize lines differing in physical and chemical composition., 1996, 61: 253–257.
[10] Simla S, Lertrat K, Suriharn B. Carbohydrate characters of six vegetable waxy maize varieties as affected by harvest time and storage duration., 2010, 9: 463–470.
[11] 杨引福, 郭强, 陈婧, 郑小亚, 蔺崇明. 中国温带糯玉米自交系遗传及品质性状分析. 西北农业学报, 2009, 29: 2213–2220. Yang Y F, Guo Q, Chen J, Zheng X Y, Lin C M. Analysis of genetic quality traits in temperate glutinous maize inbred lines in China., 2009, 29: 2213–2220 (in Chinese with English abstract).
[12] Young V R, Scrimshaw N S. Significance of Dietary Protein Source in Human Nutrition: Animal and/or Plant Proteins? online edn. New York: Oxford Academic, 1998. pp 205–221.
[13] Misra P S, Jambunathan R, Mertz E T, Glover D V, Barbosa H M, McWhirter K S. Endosperm protein synthesis in maize mutants with increased lysine content., 1972, 176: 1425–1427.
[14] 张述宽, 滕辉升, 苏琪, 杨耀迥. 应用SSR辅助选择技术选育优质蛋白糯玉米自交系. 广西农业科学, 2009, 40: 1279–1283. Zhang S K, Teng H S, Su Q, Yang Y J. Application of SSR-assisted selection technology to select high-quality protein glutinous maize inbred lines., 2009, 40: 1279–1283 (in Chinese with English abstract).
[15] Sinkangam B, Stamp P, Srinives P, Jompuk P, Mongkol W, Porniyom A, Dang N C, Jompuk C. Integration of quality protein in waxy maize by means of simple sequence repeat markers.,2011, 51: 2499–2504.
[16] Zhou Z Q, Song L Y, Zhang X X, Li X H, Yan N, Xia R P, Zhu H, Weng J F, Hao Z F, Zhang D G, Yong H J, Li M S, Zhang S H. Introgression of opaque2 into waxy maize causes extensive biochemical and proteomic changes in endosperm., 2016, 11: e0161924.
[17] Dang N C, Munsch M, Aulinger I, Renlai W, Stamp P. Inducer line generated double haploid seeds for combined waxy and opaque 2 grain quality in subtropical maize (L.)., 2012, 183: 153–160.
[18] Jia H W, Nettleton D, Peterson J M, Vazquez-Carrillo G, Jannink J L, Scott M P. Comparison of transcript profiles in wild-type and o2 maize endosperm in different genetic backgrounds., 2007, 47(S1): 45–59.
[19] Frizzi A, Caldo R A, Morrell J A, Wang M, Lutfiyya L L, Brown W E, Malvar T M, Huang S S. Compositional and transcriptional analyses of reduced zein kernels derived from themutation and RNAi suppression., 2010, 73: 569–585.
[20] Jia M, Wu H, Clay K L, Jung R, Larkins B A, Gibbon B C. Identification and characterization of lysine-rich proteins and starch biosynthesis genes in themutant by transcriptional and proteomic analysis., 2013, 13: 60.
[21] Li C B, Qiao Z Y, Qi W W, Wang Q, Yuan Y, Yang X, Tang Y P, Mei B, Lyu Y D, Zhao H, Xiao H, Song R. Genome-wide characterization of-acting DNA targets reveals the transcriptional regulatory framework ofin maize., 2015, 27: 532–545.
[22] Zhang Z Y, Zheng X X, Yang J, Messing J, Wu Y R. Maize endosperm-specific transcription factorsand PBF network the regulation of protein and starch synthesis., 2016, 113: 10842–10847.
[23] Zhan J P, Li G S, Ryu C-H, Ma C, Zhang S S, Lloyd A, Hunter B G, Larkins B A, Drews G N, Wang X F, Yadegari R.regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm., 2018, 30: 2425–2446.
[24] 陈亮, 张德贵, 史振声, 赵刚, 白丽, 张世煌, 李明顺.突变基因()对玉米产量和产量配合力的影响. 玉米科学, 2011, 19(1): 8–13. Chen L, Zhang D G, Shi Z S, Zhao G, Bai L, Zhang S H, Li M S. Effect ofmutant gene () on yield and yield fitness of maize., 2011, 19(1): 8–13 (in Chinese with English abstract).
[25] 宋丽雅, 陈亮, 何聪芬, 赵刚, 白鹏飞, 陈岩, 常驰.突变基因对玉米组合品质的影响. 安徽农业科学, 2012, 40: 9607–9609. Song L Y, Chen L, He C F, Zhao G, Bai P F, Chen Y, Chang C. Effect ofmutant gene on the quality of maize combinations., 2012, 40: 9607–9609 (in Chinese with English abstract).
[26] 周昱婕, 韩洁楠, 王美娟, 刘晓丽, 李明顺.基因对糯玉米子粒品质的影响分析. 玉米科学, 2021, 29(2): 29–34. Zhou Y J, Han J N, Wang M J, Liu X L, Li M S. Analysis of the effect ofgene on kernel quality of glutinous maize., 2021, 29(2): 29–34 (in Chinese with English abstract).
[27] 刘晓丽, 韩洁楠, 李冉, 郭增辉, 张德贵, 李明顺.对糯玉米籽粒食味和营养品质的影响分析. 玉米科学, 2023, 31(4): 52–58. Liu X L, Han J N, Li R, Guo Z H, Zhang D G, Li M S. Analysis of the effect ofon flavour and nutritional quality of glutinous maize kernels., 2023, 31(4): 52–58 (in Chinese with English abstract).
[28] Wang W, Dai Y, Wang M C, Yang W P, Zhao D G. Transcriptome dynamics of double recessive mutant,, reveals the transcriptional mechanisms in the increase of its lysine and tryptophan content in maize., 2019, 10: 316.
[29] 谭华, 邹成林, 吴永升, 郑德波, 莫润秀, 黄爱花, 韦新兴, 蒋维萍, 韦慧, 黄开健. 不同遗传背景普通玉米种质导入基因效应探讨. 广东农业科学, 2015, 42(23): 127–132. Tan H, Zou C L, Wu Y S, Zheng D B, Mo R X, Huang A H, Wei X X, Jiang W P, Wei H, Huang K J. Exploration of the effect of introducinggene in common maize germplasm with different genetic backgrounds., 2015, 42(23): 127–132 (in Chinese with English abstract).
[30] Prioul J L, Mechin V, Lessard P, Thévenot C, Grimmer M, Chateau-Joubert S, Coates S, Hartings H, Kloiber-Maitz M, Murigneux A, Sarda X, Damerval C, Edwards K J. A joint transcriptomic, proteomic and metabolic analysis of maize endosperm development and starch filling., 2008, 6: 855–869.
[31] Chen J, Zeng B, Zhang M, Xie S J, Wang G K, Hauck A, Lai J S. Dynamic transcriptome landscape of maize embryo and endosperm development., 2014, 166: 252–264.
[32] Ji C, Xu L N, Li Y J, Fu Y X, Li S, Wang Q, Zeng X, Zhang Z Q, Zhang Z Y, Wang W Q, Wang J C, Wu Y R. The-transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize., 2022, 15: 468–487.
[33] Li N, Zhang S J, Zhao Y J, Li B, Zhang J R. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize., 2011, 233: 241–250.
[34] Jiang L L, Yu X M, Qi X, Yu Q, Deng S, Bai B, Li N, Zhang A, Zhu C F, Liu B, Pang J S. Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose., 2013, 22: 1133–1142.
[35] Hu S T, Wang M, Zhang X, Chen W K, Song X R, Fu X Y, Fang H, Xu J, Xiao Y N, Li Y R, Bai G H, Li J S, Yang X H. Genetic basis of kernel starch content decoded in a maize multi-parent population., 2021, 19: 2192–2205.
[36] Cobb B G, Hannah L C. Shrunken-1 encoded sucrose synthase is not required for sucrose synthesis in the maize endosperm., 1988, 88: 1219–1221.
[37] Deng Y T, Wang J C, Zhang Z Y, Wu Y R. Transactivation of Sus1 and Sus2 byis an essential supplement to sucrose synthase-mediated endosperm filling in maize., 2020, 18: 1897–1907.
[38] Denyer K, Dunlap F, Thorbjørnsen T, Keeling P, Smith A M. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial., 1996, 112: 779–785.
[39] Jennings P H, McCombs C L. Effects of sugary-1 and shrunken-2 loci on kernel carbohydrate contents, phosphorylase and branching enzyme activities during maize kernel ontogeny., 1969, 8: 1357–1363.
[40] Tetlow I J, Beisel K G, Cameron S, Makhmoudova A, Liu F, Bresolin N S, Wait R, Morell M K, Emes M J. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes., 2008, 146: 1878–1891.
[41] Paul M J, Watson A, Griffiths C A. Trehalose 6-phosphate signalling and impact on crop yield., 2020, 48: 2127–2137.
[42] Meitzel T, Radchuk R, McAdam E L, Thormählen I, Feil R, Munz E, Hilo A, Geigenberger P, Ross J J, Lunn J E, Borisjuk L. Trehalose 6-phosphate promotes seed filling by activating auxin biosynthesis., 2021, 229: 1553–1565.
[43] Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P. Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase., 2005, 102: 11118–11123.
[44] Hu S T, Wang M, Zhang X, Chen W K, Song X R, Fu X Y, Fang H, Xu J, Xiao Y N, Li Y R, Bai G H, Li J S, Yang X H. Genetic basis of kernel starch content decoded in a maize multi-parent population., 2021, 19: 2192–2205.
[45] Fernandez O, Vandesteene L, Feil R, Baillieul F, Lunn J E, Clément C. Trehalose metabolism is activated upon chilling in grapevine and might participate ininduced chilling tolerance., 2012, 236: 355–369.
[46] Leyman B, Dijck P V, Thevelein J M. An unexpected plethora of trehalose biosynthesis genes in., 2001, 6: 510–513.
[47] Davies H V, Shepherd L V, Burrell M M, Carrari F, Urbanczyk-Wochniak E, Leisse A, Hancock R D, Taylor M, Viola R, Ross H, McRae D, Willmitzer L, Fernie A R. Modulation of fructokinase activity of potato () results in substantial shifts in tuber metabolism., 2005, 46: 1103–1115.
[48] Schaffer A A, Petreikov M. Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation., 1997, 113: 739–746.
[49] German M A, Dai N, Matsevitz T, Hanael R, Petreikov M, Bernstein N, Ioffe M, Shahak Y, Schaffer A A, Granot D. Suppression of fructokinase encoded by LeFRK2 in tomato stem inhibits growth and causes wilting of young leaves., 2003, 34: 837–846.
[50] Urbanowicz B R, Bennett A B, Del Campillo E, Catalá C, Hayashi T, Henrissat B, Höfte H, McQueen-Mason S J, Patterson S E, Shoseyov O, Teeri T T, Rose J K. Structural organization and a standardized nomenclature for plant endo-1,4-beta-glucanases (cellulases) of glycosyl hydrolase family 9., 2007, 144: 1693–1696.
[51] 潘利华, 罗建平. β-葡萄糖苷酶的研究及应用进展. 食品科学, 2006, 27: 803–807. Pan L H, Luo J P. Progress of research and application of β-glucosidase., 2006, 27: 803–807 (in Chinese with English abstract).
[52] 陈凯莉, 许轲, 张贤聪, 王亚楠, 汪志辉, 王迅. 果实中果胶代谢相关酶基因的研究进展. 园艺学报, 2017, 44: 2008–2014. Chen K L, Xu K, Zhang X C, Wang Y N, Wang Z H, Wang X. Progress of pectin metabolism-related enzyme genes in fruits., 2017, 44: 2008–2014 (in Chinese with English abstract).
[53] Godoy F D, Bermúdez L, Lira B S, Souza A P D, Elbl P, Dcmarco D, Alseekh S, Insani M, Buckeridge M, Almeida J, Grigioni G, FernieA R, Carrari F, Rossi M. Galacturonosyl transferase 4 silencing alters pectin composition and carbon partitioning in tomato., 2013, 64: 2449–2466.
[54] 傅海, 赵佳, 李伟, 孙科, 王希信. 果胶酶研究进展及应用. 生物化工, 2020, 6(5): 148–153. Fu H, Zhao J, Li W, Sun K, Wang X X. Research progress and application of pectinase., 2020, 6(5): 148–153 (in Chinese with English abstract).
[55] Segonne S M, Bruneau M, Celton J M, Gall S L, Francin-Allami M, Juchaux M, Laurens F, Orsel M, Penou J P. Multiscale investigation of mealiness in apple: an atypical role for a pectin methylesterase during fruit maturation., 2014, 14: 375.
[56] Hennen-Bierwagen T A, Lin Q, Grimaud F, Planchot V, Keeling PL, James M G, Myers A M. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts., 2009, 149: 1541–1559.
[57] Wang W, Niu S Z, Dai Y, Wang M C, Li Y, Yang W P, Zhao D G. Themutantsand opaque16 disclose lysine change in waxy maize as revealed by RNA-seq., 2019, 9: 12265.
[58] Lopes M A, Takasaki K, Bostwick D E, Helentjaris T, Larkins B A. Identification of two opaque2 modifier loci in quality protein maize., 1995, 247: 603–613.
[59] Holding D R, Hunter B G, Chung T, Gibbo B C, Ford C F, Bharti A K, Messing J, Hamaker B R, Larkins B A. Genetic analysis ofmodifier loci in quality protein maize., 2008, 117: 157–170.
[60] Holding D R, Hunter B G, Klingler J P, Wu S, Guo X M, Gibbon B C, Wu R L, Schulze J M, Jung R, Larkins B A. Characterization ofmodifier QTLs and candidate genes in recombinant inbred lines derived from the K0326Y quality protein maize inbred., 2011, 122: 783–794.
[61] Li C S, Xiang X L, Huang Y C, Zhou Y, An D, Dong J Q, Zhao C X, Liu H J, Li Y B, Wang Q, Du C G, Messing J, Larkins B A, Wu Y R, Wang W Q. Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize., 2020, 11: 17.
[62] Guo X M, Ronhovde K, Yuan L L, Yao B, Soundararajan M P, Elthon T, Zhang C, Holding D R. Pyrophosphate-dependent fructose-6-phosphate 1-phosphotransferase induction and attenuation ofgene expression during endosperm modification in quality protein maize., 2012, 158: 917–929.
[63] Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S. Differential expression of alternatively spliced mRNAs ofSR protein homologs, atSR30 and atSR45a, in response to environmental stress., 2007, 48: 1036–1049.
[64] Ohta M, Takaiwa F. Emerging features of ER resident J-proteins in plants., 2014, 9: e28194.
附表1 定量引物信息列表
Table S1 Information of RT-PCR primers
基因名称Gene ID引物序列Primer sequence (5'–3')片段长度Production size (bp)退火温度Annealing temperature (℃) Zm00001d045042(Sh1)TGTTTCACCGCAATTCGCA19060 AGACAGGTGAACGAGCAGGC Zm00001d029091(Sus1)GAAGCGTTCGGTCTCACC16260 GAAGAAGTCGGCCATCAG Zm00001d050032(Bt2)CTATGACCGTTTTGCTCCAAT19560 GCACCCATTAGTAAACTGTCCTC Zm00001d044129(Sh2)CGTGTCAGCTCTGGATGTGAA21060 AGCCTCTTGGATGCCCTTA Zm00001d045261(SSI)GACATTAATGATTGGAACCCTGCC18760 GAATGAGATCAATGCCTTTCTG Zm00001d037234(SSIIa)CTGCACTCCTGCCTGTTTAT17560 GGATCGTACAGCTCGAAATGTT Zm00001d000002(SSIIIa)CAAAAGGGGATCCACCTGATCA20860 CAGAGCCAGCGTATATCAGAT Zm00001d016684(SBEIIb)TAGACTTATCACAATGGGTTTAGGA22760 GCATTGCCTGATCAAACTCTTG Zm00001d017502GTCGCAGGACTCGTGAAACA20660 TTGAAGGCGTCTTCGTCTGT Zm00001d032311GTATGGCACTATTTGGACGCG18660 GACCAGTGTGTGAATCAGCTTG
(续附表1)
基因名称Gene ID引物序列Primer sequence (5'–3')片段长度Production size (bp)退火温度Annealing temperature (℃) Zm00001d052060TGAATCGTCTGTGCGAGGACC16460 CCACCTTGTGAAGTAACCGTGCT Zm00001d012748GGTTACTTCTTGAGGTGGTCT20860 GCATCTCCTTTGCCTGTGAG Zm00001d029371CCCGCCGAGGTCAAGGAGTT16060 TGCCGGTGCAGCAGGTAGGT Zm00001d012173ATCACCGCCACGAAGAAGGG21060 TAGGAACTAGGGCAAGCAAA Zm00001d042536GCCTTCCTTCAAAGTACAACA22060 CTCTATTAGCTGCAAGACCTCC Zm00001d036608ACCTCCGCTACTCCATCAACACCA23460 CCCTTGCGAACGGGTAGAACG Zm00001d034017TCTCCACCGTCATGATCTCCTA20060 AATGCCAGCAAGAATCGAAGCC Zm00001d033649GGAGAAGTATGGGAATCCAACG21360 TACCCTGCCAGCCACTCGAAG Zm00001d028243GCCTTCAACGCCTACTACCACG14460 GGTGCCGTTCATCAACGACGTC Zm00001d005546CAGCACGAGTGTTCTTGGGATC19960 GCGGTTGAGCGAAGCAGAGT Zm00001d053960GCTGGAGCTTTTGGTCAGTTTGC19060 GGTCGCACCAGATACTGAAATCT Zm00001d002256CGACAGAGCCATAACCACAT17260 AAACGAGCCTGATTTCCCTA Zm00001d015327(Ubiquitin)TAAGCTGCCGATGTGCCTGCGTCG20660 CTGAAAGACAGAACATAATGAGCACAG
附表2 糯玉米-NILs醇溶蛋白相关DEGs
Table S2 DEGs related to zein protein in waxy maize-NILs
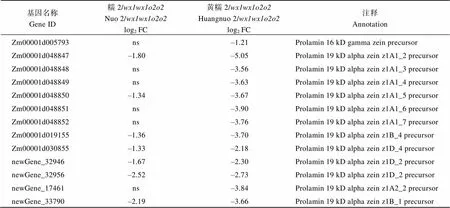
基因名称Gene ID糯2/wx1wx1o2o2Nuo 2/wx1wx1o2o2log2 FC黄糯2/wx1wx1o2o2Huangnuo 2/wx1wx1o2o2log2 FC注释Annotation Zm00001d005793ns–1.21Prolamin 16 kD gamma zein precursor Zm00001d048847–1.80–5.05Prolamin 19 kD alpha zein z1A1_2 precursor Zm00001d048848ns–3.56Prolamin 19 kD alpha zein z1A1_3 precursor Zm00001d048849ns–3.63Prolamin 19 kD alpha zein z1A1_4 precursor Zm00001d048850–1.34–3.67Prolamin 19 kD alpha zein z1A1_5 precursor Zm00001d048851ns–3.90Prolamin 19 kD alpha zein z1A1_6 precursor Zm00001d048852ns–3.76Prolamin 19 kD alpha zein z1A1_7 precursor Zm00001d019155–1.36–3.70Prolamin 19 kD alpha zein z1B_4 precursor Zm00001d030855–1.33–2.18Prolamin 19 kD alpha zein z1D_4 precursor newGene_32946–1.67–2.30Prolamin 19 kD alpha zein z1D_2 precursor newGene_32956–2.52–2.73Prolamin 19 kD alpha zein z1D_2 precursor newGene_17461ns–3.84Prolamin 19 kD alpha zein z1A2_2 precursor newGene_33790–2.19–3.66Prolamin 19 kD alpha zein z1B_1 precursor
(续附表2)
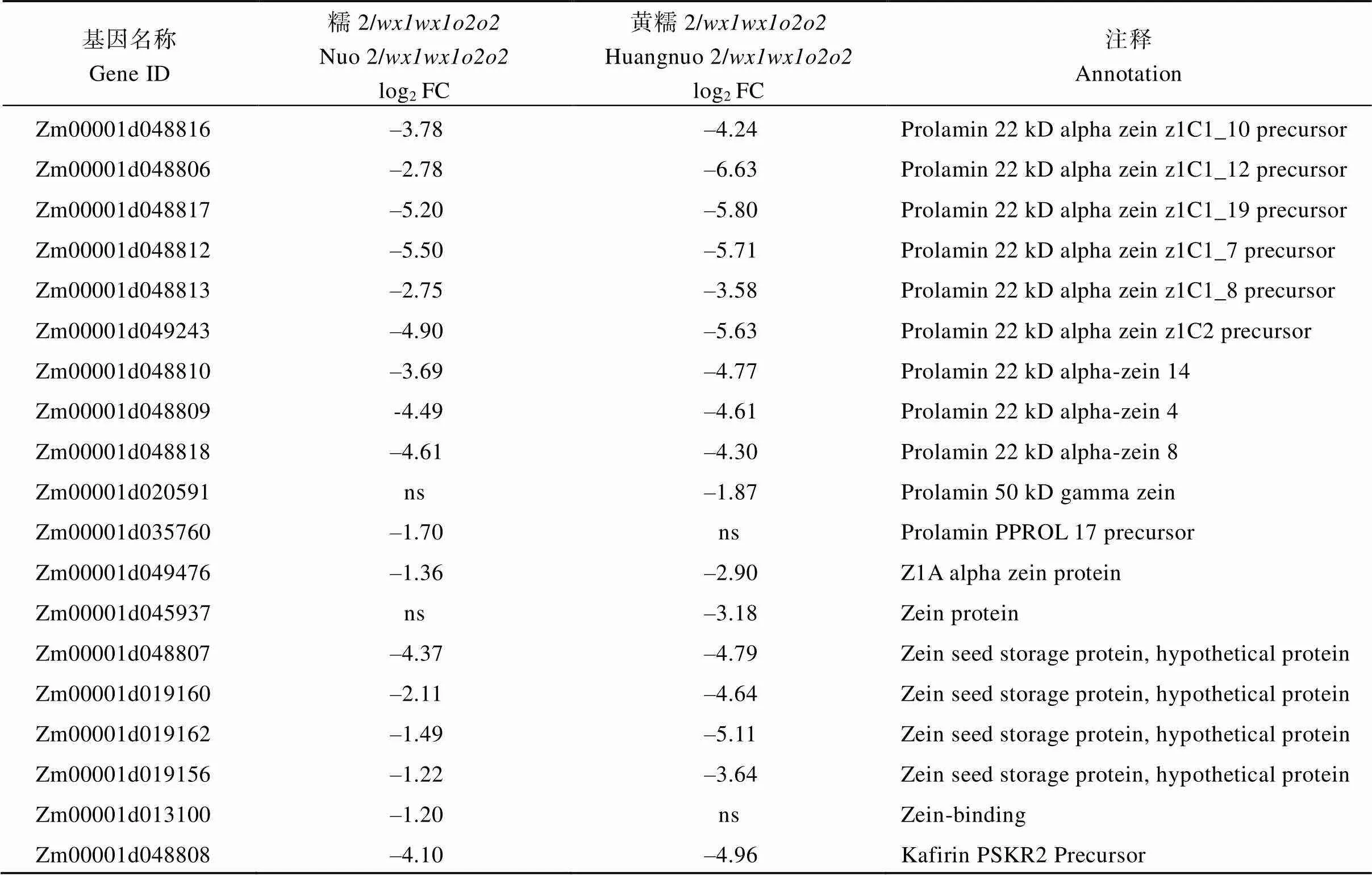
基因名称Gene ID糯2/wx1wx1o2o2Nuo 2/wx1wx1o2o2log2 FC黄糯2/wx1wx1o2o2Huangnuo 2/wx1wx1o2o2log2 FC注释Annotation Zm00001d048816–3.78–4.24Prolamin 22 kD alpha zein z1C1_10 precursor Zm00001d048806–2.78–6.63Prolamin 22 kD alpha zein z1C1_12 precursor Zm00001d048817–5.20–5.80Prolamin 22 kD alpha zein z1C1_19 precursor Zm00001d048812–5.50–5.71Prolamin 22 kD alpha zein z1C1_7 precursor Zm00001d048813–2.75–3.58Prolamin 22 kD alpha zein z1C1_8 precursor Zm00001d049243–4.90–5.63Prolamin 22 kD alpha zein z1C2 precursor Zm00001d048810–3.69–4.77Prolamin 22 kD alpha-zein 14 Zm00001d048809-4.49–4.61Prolamin 22 kD alpha-zein 4 Zm00001d048818–4.61–4.30Prolamin 22 kD alpha-zein 8 Zm00001d020591ns–1.87Prolamin 50 kD gamma zein Zm00001d035760–1.70nsProlamin PPROL 17 precursor Zm00001d049476–1.36–2.90Z1A alpha zein protein Zm00001d045937ns–3.18Zein protein Zm00001d048807–4.37–4.79Zein seed storage protein, hypothetical protein Zm00001d019160–2.11–4.64Zein seed storage protein, hypothetical protein Zm00001d019162–1.49–5.11Zein seed storage protein, hypothetical protein Zm00001d019156–1.22–3.64Zein seed storage protein, hypothetical protein Zm00001d013100–1.20nsZein-binding Zm00001d048808–4.10–4.96Kafirin PSKR2 Precursor
附表3 糯玉米-NILs赖氨酸降解相关DEGs
Table S3 DEGs related to lysine degradation in waxy maize-NILs

基因名称Gene_ID糯2/wx1wx1o2o2Nuo 2/wx1wx1o2o2log2 FC黄糯2/wx1wx1o2o2Huangnuo 2/wx1wx1o2o2log2 FC注释Annotation Zm00001d020984–2.43–4.10Probable sarcosine oxidase Zm00001d003983–2.48nsAldehyde dehydrogenase family 7 member A1 Zm00001d008432ns–1.01Putative acetyl-CoA acetyltransferase cytosolic 2 Zm00001d052079ns–2.65Lysine-ketoglutarate reductase/saccharopine dehydrogenase1
附表4 糯玉米-NILs胚乳修饰相关DEGs
Table S4 DEGs related to endosperm modification in waxy maize-NILs
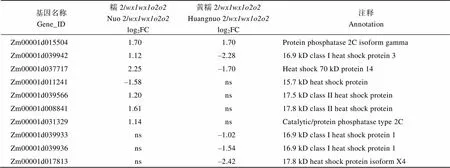
基因名称Gene_ID糯2/wx1wx1o2o2Nuo 2/wx1wx1o2o2log2FC黄糯2/wx1wx1o2o2Huangnuo 2/wx1wx1o2o2log2FC注释Annotation Zm00001d0155041.701.70Protein phosphatase 2C isoform gamma Zm00001d0399421.12–2.2816.9 kD class I heat shock protein 3 Zm00001d0377172.25–1.70Heat shock 70 kD protein 14 Zm00001d011241–1.58ns15.7 kD heat shock protein Zm00001d0395661.20ns17.5 kD class II heat shock protein Zm00001d0088411.61ns17.8 kD class II heat shock protein Zm00001d0313291.14nsCatalytic/protein phosphatase type 2C Zm00001d039933ns–1.0216.9 kD class I heat shock protein 1 Zm00001d039936ns–1.5416.9 kD class I heat shock protein 1 Zm00001d017813ns–2.4217.8 kD heat shock protein isoform X4
(续附表4)
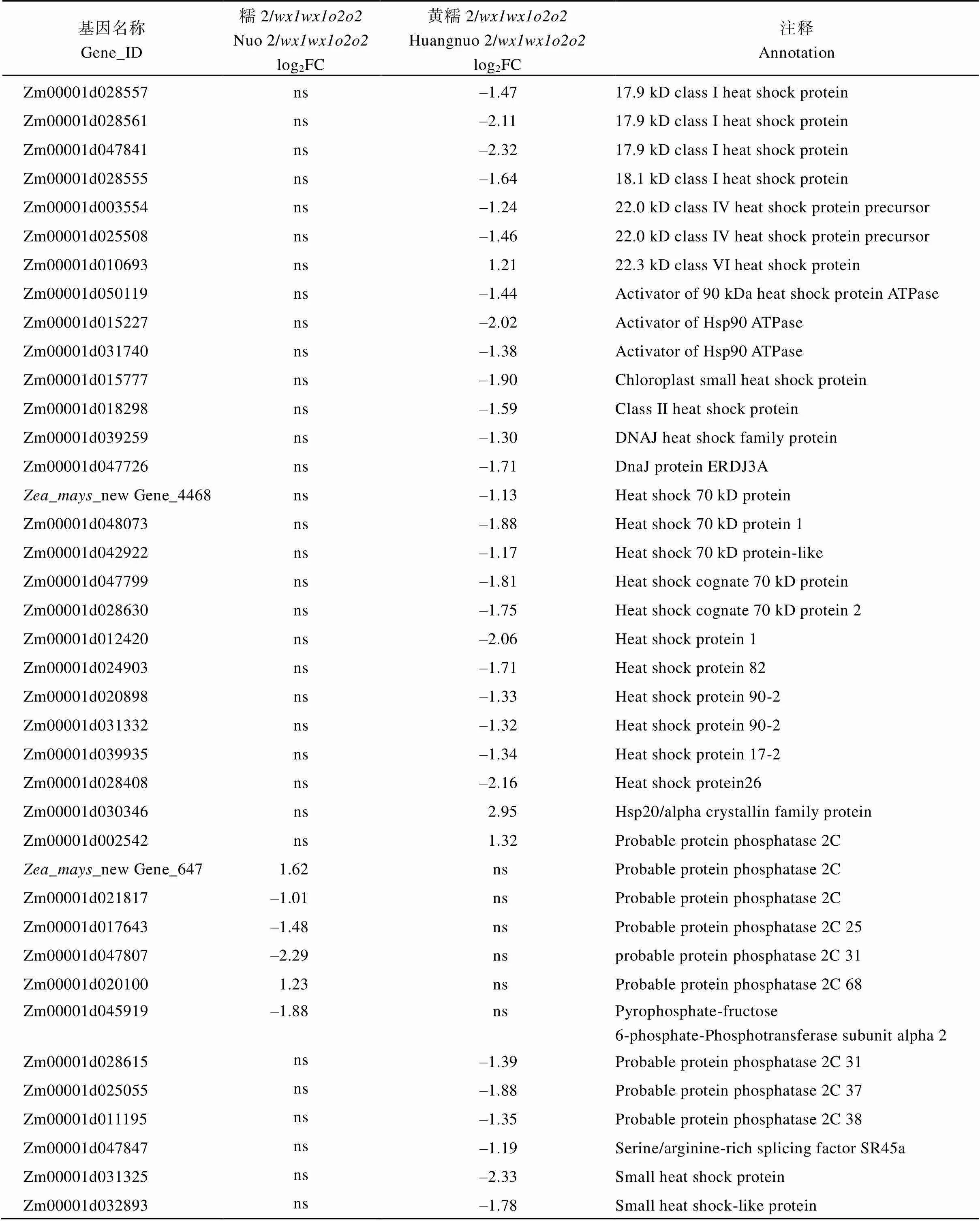
基因名称Gene_ID糯2/wx1wx1o2o2Nuo 2/wx1wx1o2o2log2FC黄糯2/wx1wx1o2o2Huangnuo 2/wx1wx1o2o2log2FC注释Annotation Zm00001d028557ns–1.4717.9 kD class I heat shock protein Zm00001d028561ns–2.1117.9 kD class I heat shock protein Zm00001d047841ns–2.3217.9 kD class I heat shock protein Zm00001d028555ns–1.6418.1 kD class I heat shock protein Zm00001d003554ns–1.2422.0 kD class IV heat shock protein precursor Zm00001d025508ns–1.4622.0 kD class IV heat shock protein precursor Zm00001d010693ns1.2122.3 kD class VI heat shock protein Zm00001d050119ns–1.44Activator of 90 kDa heat shock protein ATPase Zm00001d015227ns–2.02Activator of Hsp90 ATPase Zm00001d031740ns–1.38Activator of Hsp90 ATPase Zm00001d015777ns–1.90Chloroplast small heat shock protein Zm00001d018298ns–1.59Class II heat shock protein Zm00001d039259ns–1.30DNAJ heat shock family protein Zm00001d047726ns–1.71DnaJ protein ERDJ3A Zea_mays_new Gene_4468ns–1.13Heat shock 70 kD protein Zm00001d048073ns–1.88Heat shock 70 kD protein 1 Zm00001d042922ns–1.17Heat shock 70 kD protein-like Zm00001d047799ns–1.81Heat shock cognate 70 kD protein Zm00001d028630ns–1.75Heat shock cognate 70 kD protein 2 Zm00001d012420ns–2.06Heat shock protein 1 Zm00001d024903ns–1.71Heat shock protein 82 Zm00001d020898ns–1.33Heat shock protein 90-2 Zm00001d031332ns–1.32Heat shock protein 90-2 Zm00001d039935ns–1.34Heat shock protein 17-2 Zm00001d028408ns–2.16Heat shock protein26 Zm00001d030346ns2.95Hsp20/alpha crystallin family protein Zm00001d002542ns1.32Probable protein phosphatase 2C Zea_mays_new Gene_6471.62nsProbable protein phosphatase 2C Zm00001d021817–1.01nsProbable protein phosphatase 2C Zm00001d017643–1.48nsProbable protein phosphatase 2C 25 Zm00001d047807–2.29nsprobable protein phosphatase 2C 31 Zm00001d0201001.23nsProbable protein phosphatase 2C 68 Zm00001d045919–1.88nsPyrophosphate-fructose 6-phosphate-Phosphotransferase subunit alpha 2 Zm00001d028615ns–1.39Probable protein phosphatase 2C 31 Zm00001d025055ns–1.88Probable protein phosphatase 2C 37 Zm00001d011195ns–1.35Probable protein phosphatase 2C 38 Zm00001d047847ns–1.19Serine/arginine-rich splicing factor SR45a Zm00001d031325ns–2.33Small heat shock protein Zm00001d032893ns–1.78Small heat shock-like protein
Analysis of differential accumulation of starch in waxy maize grain caused by themutation gene
HAN Jie-Nan1, ZHANG Ze1,2, LIU Xiao-Li1, LI Ran1, SHANG-GUAN Xiao-Chuan1,2, ZHOU Ting-Fang1,2, PAN Yue1, HAO Zhuan-Fang1, WENG Jian-Feng1, YONG Hong-Jun1, ZHOU Zhi-Qiang1, XU Jing-Yu2, LI Xin-Hai1,2, and LI Ming-Shun1,*
1Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China;2College of Agriculture, Heilongjiang Bayi Agricultural University, Daqing 163319, Heilongjiang, China
The primary variety of fresh maize known as waxy maize undergoes a transformation with the introduction of the() mutant gene, resulting in an increased lysine content, thus improving the grain's nutritional composition. Yet, themutation brings about desirable agronomic traits such as wrinkle formation and a decrease in starch content, which restrict its use in breeding applications. To explore the high performing waxy maizereceptors, we capitalized on the use ofnear-isogenic line (-NIL), specifically the plump and round grain type Nuo 2/and its wrinkled counterpart, Huangnuo2/. Measurements of 100-grain weight and grain composition at the fresh ear and mature stages showed that there was difference in starch and soluble sugar content, which might be the primary cause of kernel phenotype variation between the two waxy maize-NILs. Genetic analysis of starch synthesis in the two-NILs was performed using qRT-PCR technique revealed that six gene-regulated trends fluctuated between 10 and 24 days after pollination, among which,,, andgenes were significant differences. Endosperm transcriptomes indicated that 24 genes encoding trehalose and glycosyl hydrolases and 48 genes involved in endosperm modification exhibited distinct changes between the two-NILs. There was no detectable alteration in the 100-grain weight or the starch content of Nuo 2/, which may well be tied to the early high-level expression of the primary starch synthesis gene, leaving later stages unchanged compared with the control. Furthermore, the shifts in the expression of sugar metabolism genes was beneficial to starch synthesis, which may be an important reason why starch content and 100-kernel weight of Nuo 2/were unaffected by themutation, and grain traits were significantly better than the superior grain traits compared with Huangnuo 2/. These results may be directly related to the differential expression of multiple endosperm modifying genes. The results of this study can provide important reference for the future utilization ofmutants in maize breeding.
waxy maize; Nuo 2/; Huangnuo 2/; kernel fullness degree; starch; sucrose metabolism; different expression genes
10.3724/SP.J.1006.2024.33046
本研究由国家重点研发计划项目(2021YFD1201004)和财政部和农业农村部国家现代农业产业技术体系建设专项(玉米, CARS-02)资助。
This study was supported by the National Key Research and Development Program of China (2021YFD1201004) and the China Agriculture Research System of MOF and MARA (Maize, CARS-02).
李明顺, E-mail: limingshun@caas.cn
E-mail: hanjienan@caas.cn
2023-08-01;
2023-10-23;
2023-11-13.
URL: https://link.cnki.net/urlid/11.1809.S.20231110.0845.002
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
