Advances of N6-methyladenosine modification on circular RNA in hepatocellular carcinoma
2024-04-04CHUFengranLIULuzhengWUJincai
CHU Feng-ran, LIU Lu-zheng, WU Jin-cai,3✉
1. Clinical College of Hainan Medical University, Haikou 571199, China
2. Department of Interventional Vascular Surgery, the Second Affiliated Hospital of Hainan Medical University, Haikou 570216, China
3. Department of Hepatobiliary and Pancreatic Surgery, Hainan Affiliated Hospital of Hainan Medical University, Hainan General Hospital, Haikou 570311, China
Keywords:
ABSTRACT N6-methyladenosine(m6A)is a reversible epigenetic modification, which is one of the most abundant modifiers in eukaryotic cells and has been commonly reported in messenger RNAs and non-coding RNAs.The processing modification of m6A regulates RNA transcription,processing, splicing, degradation, and translation, and plays an important role in the biological process of tumors.Circular RNA, which lacks the 5' cap structure, has been mistakenly regarded as a "junk sequence" generated by accidental shearing during the transcription process.However, it has been found that circRNAs can be involved in tumor invasion and metastasis through microRNAs, binding proteins, translated peptides, and m6A modifications.In this paper, we reviewed the role of m6A modifications in circRNA regulation and their functions in hepatocellular carcinoma and discussed their potential clinical applications and future development in this field.
Hepatocellular carcinoma is a common malignant tumour of the digestive system in China and the world[1].Clinical findings are mostly in the middle and late stages, with poor prognosis, and the most common metastasis is intrahepatic metastasis, and extrahepatic metastasis often occurs in the late stages of the disease, which is mostly seen in the lungs, bones, abdominal cavity, brain, and so on[2].Surgical resection is still the mainstay of treatment for longterm survival of liver cancer[3-5].Postoperative metastatic recurrence is the main cause of death in liver cancer patients, and its related potential mechanisms are complicated.Therefore, it is of great clinical significance to explore the key molecular targets of liver cancer recurrence and metastasis and their mechanisms of action for the early monitoring and detection of recurrence and metastasis and the selection of targeted therapies.
CircRNAs are widely found in human peripheral blood, exosomes and even cell-free saliva, etc.CircRNAs are RNAs with a special ring structure; they do not have a special cap structure at the 5’end and a poly(A) tail at the 3’ end; they are generated by the interaction between the acceptor and donor sites of the upstream and downstream exons of the pre-mRNAs, such as by reverse splicing or by driving the loop[6, 7].During splicing, different circRNAs can be produced from the same gene through selective cyclisation,resulting in circRNA diversity.Homology studies among different species have shown that circRNAs are highly conserved and tissue specific in the evolution of species.CircRNAs have more than 20%homology between mouse and human, but the expression profiles of mouse and human are very different, and the expression is gradually up-regulated with brain development between species[8, 9].In addition to the above features, circRNAs also have many functions,for example, cir-cRNAs can play the role of microRNAs or RNAconjugated proteins, participate in the regulation of intermolecular signalling, and competitively bind to inhibit the expression of downstream genes, and so on[10].
Since its discovery in the 1970s, m6A has been regarded as one of the most common internal modifications in most eukaryotes,with an average of 3~5 m6A modifications per mRNA[11].m6A modification is a dynamic and reversible process, very similar to DNA methylation, and is controlled by the modifier enzyme complex “writers” (METTL3, METTL14, WTAP, etc.) and the demodifier enzyme “erasers” (ALKBH5, FTO, etc.), which read the m6A site through the recognising proteins “readers”, and play important biological roles[12].The deposition of m6A modifications in the transcriptome is regular, with enrichment in the CDS and 3 ‘UTR, and higher enrichment in the vicinity of the stop codon region[13].Studies have shown that the regulation of m6Amodified gene expression is closely related to normal physiological activities of organisms.The regulation of m6A-modified gene expression is closely related to the normal physiological activities of organisms, and both modified mRNAs, ribosomal RNAs (rRNAs),and circRNAs are ecologically regulated in pathophysiological processes, including tumourigenesis and metastasis[14].However,current research suggests that the deeper mechanisms of action need to be further clarified.
1.Role of m6A modifications in circRNA regulation
1.1 m6A drives circRNA translation
CircRNAs can often drive translation in a manner similar to that of IRES[15].Recent studies have shown that circRNAs can also be translated by m6A modification, a process in which m6A sites are read by recognition proteins called “readers” and interact with YTH structural domain family protein 3 (YTHDF 3) to recruit EIF4G2 to initiate translation, a process enhanced by the methyltransferase METTL3/14 and inhibited by the demethylating enzyme FTO and the sperm cell ALKBH5 (alkB homo-log 5).This process is enhanced by the methyltransferase METTL3/14 and can also be inhibited by the demethylase FTO and the sperm cell ALKBH5(alkB homo-log 5)[16].In addition, Li et al[17].found that m6A in the 3’ UTR could improve translation efficiency by binding to YTHDF1,and that m6A in the 5’ UTR could facilitate translation independent of the cap structure.In recent years, with the rapid development of genome sequencing, multisome analysis, computational prediction,and combined MeRIP-seq and RNA-seq analyses have further demonstrated that m6A-driven circRNA translation is widespread,but the mechanism by which m6A affects protein translation remains unclear.
1.2 m6A regulates circRNA degradation
Since circRNAs do not have a 5’ end cap and a 3’ end poly(A)tail, and have a special loop structure, they are not easily degraded by exonucleases.It has been reported that circRNAs with perfectly complementary miRNA targets can be degraded by Ago2 proteinmediated degradation, but circRNAs that do not have miRNA omega functions or specific microRNA targets do not work[16].However,Park et al[18].showed that m6A recognises YTHDF2 and binds to the target, recruits HRSP1, and mediates cleavage of the target molecule by the RNA endonuclease RNase P/MRP complex, a process that acts on mRNA as well as circRNA; wherein HRSP12 mediates the ligation of YTHDF2 and RNase P/MRP and facilitates the rapid degradation of the target molecule bound to YTHDF2.CircRNA degradation is regulated by multiple genes mediated by m6A modifications, and the exact mechanism remains to be explored.
1.3 m6A modification enhances circRNA stability
An s-adenosylmethionine binding motif is present in the “writers”of m6A, METTL3 and METTL14, which form a heterodimeric complex in a 1:1 ratio to enhance stability, and METTL14 plays an important role in stabilising METTL3 and recognising target RNAs[16].WTAP, as a major regulator and component of the m6A methylation complex, can help to localise METTL3 and METTL14 and enhance the stability of RNAs.Subsequent studies have shown that m6A modifications also regulate the stability of circRNAs,thereby affecting tumour progression[16].The “erasers” of m6A,ALKBH5 and YTHDF2, can also affect RNA stabilisation[19].The latest study showed that CircCPSF6, as a carcinogenic gene, could enhance the proliferation and invasion of HCC cells in vitro and in vivo.CircCPSF6 was positively correlated with ALKBH5 and negatively correlated with YTHDF2.The MeRIP assay and the construction of recombinant fluorescein enzyme (inserted with wildtype or mutant m6A sites) showed that ALKBH5 regulates the m6A modification of circCPSF6, which in turn alters the stability of the target gene circCPSF6 and regulates its expression.The degradation of circCPSF6 is also associated with m6A modification and is regulated by YTHDF2[20].Whether circCPSF6 incorporation (postsplicing process) is regulated by m6A modification or by other factors remains a mystery.
1.4 m6A modifications alter the state of downstream genes
m6A can also affect circRNA function by altering the methylation status of downstream molecules.yAP is present downstream of the Hippo pathway as one of the major response factors, and is closely related to tumour development[16].2018 Zhang et al.discovered the mechanism of circ _ 104075 up-regulated expression in HCC and stimulated the downstream mechanism of HCC tumourigenesis and development; it was found that circ _ 104075 could promote tumourigenesis through YAP binding miR-582-3p, and that m6A modification present in the 3 ‘UTR in YAP inhibited the function of circ _ 104075 by promoting The m6A modification in the 3 ‘UTR of YAP inhibits YAP function by promoting the interaction of circ _104075 with miR-382-5p[21].
1.5 Role of m6A-modified circRNAs in innate immunity
CircRNAs have a closed loop structure and are not susceptible to innate immunity.However, recent studies have shown that invasion of some exogenous circRNAs promotes innate immunity and provides protection against viral infections[22].Unmodified exogenous circRNA (circ-FOREIGN) can activate the immune system in the presence of K3-linked polyubiquitin molecules[23].Endogenous circRNA readily forms 16-26 bp RNA duplexes that resist doublestranded RNA (dsRNA) activation in innate immunity Protein kinase (PKR)[24].circSELF is generated from the ZKSCAN1 intron and its mechanism of action is related to WTAP, KIAA1429 (m6A“writers”), YTHDF2 and HNRNPC (m6A “readers”).CircSELF can be labelled by YTHDF2 as a “self” molecule, avoiding the monitoring of innate immunity and inducing unnecessary immune responses[25].These results suggest that circRNAs may play a role in the recognition of innate immunity by modifying the marker through covalent m6A.
1.6 m6A modifications mediate circRNA translocation in subcellular
Although there are relatively few studies on m6A modification of circRNA to promote subcellular transport, it has been found that m6A modification can regulate the binding of circNSUN2 to YTHDC1 from the nucleus to the cytoplasm to form the circNSUN2/IGF2BP2/HMGA2 RNA-protein ternary complex, and stabilise HMGA2, which promotes liver metastasis of colorectal cancer[26].It has also been shown that METTL14 promotes m6A modification of cir-cFUT8, which facilitates the nuclear export of circFUT8 to the cytoplasm and promotes the progression of hepatocellular carcinoma[27].
2.m6A-modified circRNA in hepatocellular carcinoma
m6A-modified circRNAs are widely involved in the growth and invasion of various malignant tumours, including HCC.
2.1 m6A-modified circRNA and liver cancer death (apoptosis,iron death)
In HCC, circMAP3K4 is an up-regulated circRNA with coding potential.IGF2BP1 recognises the m6A modification of circMAP3K4 and facilitates its translation to circMAP3K4-455aa,which inhibits cleavage and nuclear distribution of AIF and prevents apoptosis in HCC under stress.circMAP3K4-455aa mediates the degradation of the ubiquitin-proteasome E3 ligase MIB1 pathway[28].With the recent report that the m6A-modified circSAV1 triggers iron death in COPD by recruiting YTHDF1 to promote the translation of IREB2, it was mentioned that overexpression of IREB2 in hepatic stellate cells raises iron levels and causes cellular iron death[29].Iron death is a new form of programmed cell death that is distinct from apoptosis, necrosis, autophagy, and others[30].CircFGGY is up-regulated in HepG2 cells of hepatocellular carcinoma, and exogenous knockdown of circFGGY promotes iron death in HepG2 cells.The m6A modification regulates the export of circFGGY from the nucleus to the cytoplasm, and regulates the expression of FSP1 (iron death suppressor protein 1), which in turn regulates the iron death of HepG2 cells[31].cIARS (hsa _circ _0008367) is the most highly expressed circRNA in HCC cells after sorafenib treatment.A small interfering RNA against cIARS (sicI-ARS) inhibited HCC iron death and significantly suppressed the sensitivity of HCC cells to sorafenib.ALKBH5 negatively regulated HCC iron death and down-regulated ALKBH5, mediating the dissociation of the BCL-2/BECN1 complex, while si-cIARS effectively blocked this process.ALKBH5 negatively regulates HCC iron death, down-regulates ALKBH5, and mediates the dissociation of the BCL-2/BECN1 complex, and si-cIARS can effectively block this process.The inhibition of iron death in HCC cells by si-cIARS could also be mitigated by down-regulation of ALKBH5.ALKBH5 is one of the m6A-modified “erasers”[32].However, the specific role of m6A modification was not reported in this study.
2.2 m6A-modified circRNA and liver cancer invasion and metastasis
Du et al[33].found that Hsa _circ _0095868 (circMDK), an exon 5 derived from the MDK gene, overexploited miR-346 and miR-874-3p, and stimulated the PI3K/AKT/mTOR signalling pathway through the up-regulation of ATG16L1, promoting HCC cell proliferation,migration and invasion, and IGF2BP1 was significantly up-regulated in HCC in vitro in conjunction with m6A-modified circMDK and enhanced circMDK stability.IGF2BP1 binds to m6A-modified circMDK in vitro and enhances circMDK stabilisation, contributing to the significant up-regulation of circMDK in HCC.circGPR137B inhibits HCC growth and metastasis through the circGPR137B/miR-4739/FTO feedback loop.CircGPR137B mediates m6A demethylation of circ-GPR137B through upregulation of its target gene FTO, which enhances the stability and expression of circGPR137B.In addition, the expression of circGPR137B inhibited the growth and lung metastasis of hepatocellular carcinoma[34].
CircHPS5 regulates HMGA2 expression and promotes EMT and CSC phenotypes to facilitate HCC migration and proliferation.In this process, m6A modification under the influence of YTHDC1 regulates cytoplasmic export and increases HMGA2 expression[35].Recently, it has also been found that METTL3 promotes the m6A modification of hsa _circ _0058493, and that the m6A modification site of hsa _circ _0058493 binds to YTHDC1 and is exported from the nucleus to the cytoplasm of the cell in an m6A-dependent manner, which ultimately contributes to the progression of HCC[36].CircKIAA1429 accelerates the progression of HCC, and YTHDF3 promotes m6A modification and thus enhances the stability of Zeb1 (a downstream gene), thus overexpression of circKIAA1429 contributes to the development of hepatocellular carcinoma[37].m6A modification enhances circRNA-SORE stability Impacts sorafenib resistance in hepatocellular carcinoma cells[38].(see table below)
2.3 m6A-modified circRNA and liver cancer immunity
CircCCAR1 accelerates HCC growth and metastasis in vitro and in vivo.The exocrine circCCAR1 is taken up by CD8+T cells, causing functional impairment of CD8+T cells (CD8+T cell dysfunction is a major factor in HCC immune escape) through stabilisation of PD-1 protein.CircCCAR1, as a miR-127-5p omega, up-regulates its target WTAP, forming a feedback pathway consisting of the circCCAR1/miR-127-5p/WTAP axis, in which WTAP-mediated m6A modification enhances the stability of circCCAR1 via IGF2BP3[39].(see table below)
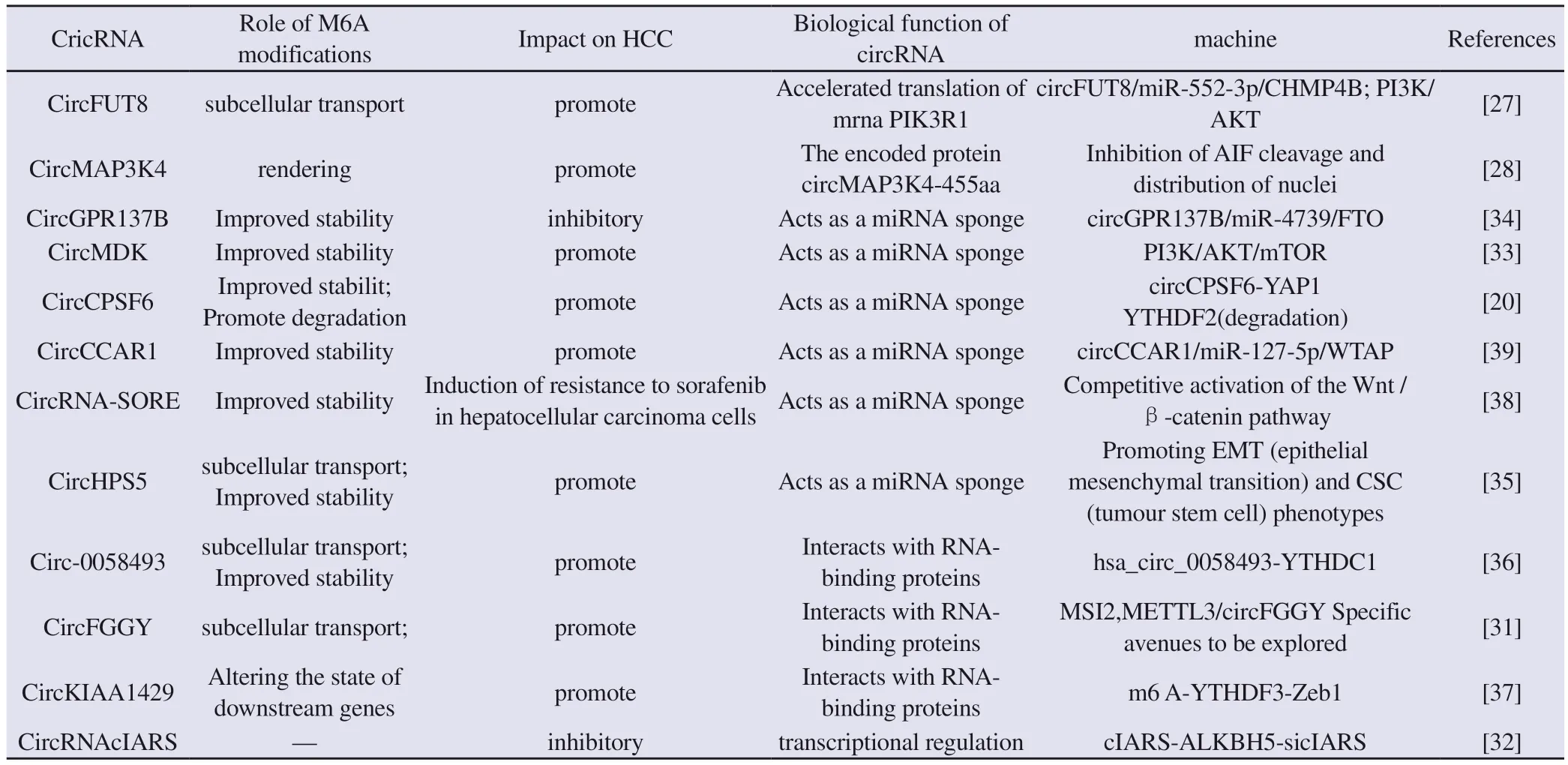
Tab 1 m6A-modified circRNA in lhepatocellular carcinoma
3.Conclusion
Nowadays, there are a lot of experimental data proving the special role of m6A-modified circRNAs in hepatocellular carcinoma and promoting in-depth research on the mechanism of hepatocellular carcinoma occurrence and development.Literature reports have shown that m6A modification affects the development of hepatocellular carcinoma by promoting circRNA translation,enhancing stability, regulating degradation, and facilitating subcellular translocation in hepatocellular carcinoma.With the wide application of high-throughput sequencing technology and bioinformatics analysis in scientific research, more and more m6Amodified circRNAs will be discovered and detected, which will bring new hope to the molecular diagnosis and treatment of hepatocellular carcinoma and promote in-depth research on the invasive and metastatic mechanisms of hepatocellular carcinoma.
Simplifying the detection of specific m6A-modified circRNA levels, efficiently extracting low-abundance circRNAs from limited samples, diagnosing HCC through the detection of different abundances of m6A-modified circRNAs in HCC, and designing accurate therapeutic agents according to the different pathways of m6A-modified circRNAs are the ultimate goals of the hepatocellular carcinoma clinic.In the future, it is likely that our understanding of how m6A modification regulates circRNAs will not be limited to the presently known aspects; other effects of m6A on circRNAs, such as processing or splicing effects, as well as the biological functions of m6A-modified circRNAs in other non-oncological diseases, remain to be explored.It is not enough to rely only on the database of m6Amodified circRNAs discovered so far to explore the mechanism of action of m6A-modified circRNAs, the combined analysis of m6A-modified circRNAs by MeRIP-seq and RNA-seq, and the analysis of m6A-modified circRNAs by qRT-PCR, RIP (RNA immunoprecipitation assays), RNA pull down, and fluorescein enzyme reports, and genetic testing.To explore new databases, to overcome technical difficulties and to discover simple and effective detection methods are the main problems we are facing now.
With the recent reports that m6A-modified circSAV1 promotes the translation of IREB2 mRNA, and the up-regulation of IREB2 induces lipid peroxidation and iron death in COPD cells, the idea that m6A-modified circRNA induces iron death in hepatocellular carcinoma cells through various modulations has been reintroduced,and it may be a hot topic of research in the next few years.
The current understanding of m6A-modified circRNAs in regulating hepatocellular carcinoma development is only the tip of the iceberg,and in-depth studies to reveal their regulatory mechanisms and biological functions are still a long way to go.However, it is of great clinical value for the future development of molecular markers and potential therapeutic targets based on the m6A-modified circRNAs in the diagnosis of hepatocellular carcinoma.
Authors’ contributions
Fengran Chu: literature search, reviewing, recording and writing;Luzheng Liu: thesis revision; Jincai Wu: research guidance, thesis revision, financial support.
All authors declare that there is no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Effect of honokiol regulating SIRT3 on chronic hypoxia-induced microglia and astrocyte polarization
- SLC6A8 promotes liver cancer cell proliferation by regulating mitochondrial autophagy through BNIP3L
- Clinical characteristics of patients with early-and late-onset optic neuromyelitis optica spectrum disease
- Distribution of gene polymorphisms associated with aspirin antiplatelet in the Han NSTEMI population
- Predictive value of systemic immunity index for sepsis in low-medium risk community-acquired pneumonia
- To analyze the differentially expressed genes in chronic rejection after renal transplantation by bioinformatics
