SLC6A8 promotes liver cancer cell proliferation by regulating mitochondrial autophagy through BNIP3L
2024-04-04LUYuanTANGJunLIWenchuanXUZuomingLUOZongjiangMAJiashengPUJian
LU Yuan, TANG Jun, LI Wen-chuan, XU Zuo-ming, LUO Zong-jiang, MA Jia-sheng, PU Jian
Affiliated Hospital of Youjiang Medical University for Nationalities, Baise 533000, Guangxi, China
Keywords:
ABSTRACT Objective: To investigate the effect of SLC6A8 on the proliferation of liver cancer cells by regulating mitophagy through BNIP3L.Methods: The expression of the SLC6A8 gene in liver cancer tissues was analyzed using the TCGA database, and its correlation with BNIP3L expression was assessed.RT-PCR and Western blot techniques were employed to detect the expression of SLC6A8 and BNIP3L in human liver cancer Huh-7 and Hep3B cells.Immunofluorescence labeling and CCK-8 assay were used to observe mitochondrial autophagy and cell proliferation rate in SLC6A8-overexpressing Huh-7 and Hep3B cells.Evaluate the proliferation rate of SLC6A8-overexpressing Huh-7 and Hep3B cells after silencing BNIP3L using the CCK-8 detection method.Results: SLC6A8 was significantly overexpressed in liver cancer tissues and positively correlated with BNIP3L expression.Overexpression of SLC6A8 significantly promoted mitochondrial autophagy and proliferation of Huh-7 and Hep3B cells.Additionally, SLC6A8 overexpression significantly enhanced the expression of BNIP3L mRNA and protein.Upon BNIP3L silencing, the proliferative effect of SLC6A8 overexpression on liver cancer cells was reversed.Conclusion: High expression of SLC6A8 in liver cancer tissues is positively correlated with BNIP3L, and overexpression of SLC6A8 promotes mitochondrial autophagy and liver cancer cell proliferation.Silencing BNIP3L can reverses the effect of overexpression of SLC6A8 on liver cancer cell proliferation.This provides new targets and strategies for the treatment of liver cancer.
1.Introduction
Hepatocellular carcinoma (HCC) is a prevalent form of primary liver cancer, often diagnosed at an advanced, unresectable stage.It has high incidence and mortality rates, constituting the third leading cause of cancer-associated deaths[1].In 2020, global statistics estimated 905,700 people were diagnosed with HCC, with 830,200 succumbing to liver cancer[2].Presently, patients with HCC have a spectrum of treatment options: curative therapy (liver transplantation, surgical resection, or ablation/radioembolic therapy)and palliative care (transarterial chemoembolization (TACE),systemic therapy).However, specific effective treatments for BCLC stage D patients are yet to be discovered [3,4].
Mitophagy, a conserved cellular degradative process, is responsible for the clearance of dysfunctional or surplus mitochondria to maintain mitochondrial quality and cellular requirements [5].Studies have revealed that tumor cells rely on functional mitochondria to promote growth[6] and dictate cancer functionality and fate through mitophagic renewal[7].For instance, STOML2 may enhance metastatic capabilities of HCC and modulate sensitivity towards Lenvatinib by promoting PINK1-mediated mitophagy[8].Likewise,Ketoconazole may induce HCC apoptosis by exacerbating mitophagy[9].Thus, the state of mitochondria is vital for the progression of HCC.Recent studies imply that chemotherapy and targeted-drug induced mitochondrial damage is mitigated by heightened mitophagy, underpinning a close relationship between mitophagy and chemotherapy resistance and recurrence[10-12].These findings emphasize the critical role of mitophagy in HCC progression and treatment, revealing its potential as a therapeutic target.
SLC6A8 (Solute carrier family 6 member 8), a gene encoding for creatine-specific transporter, transports creatine into the cell in a Na+and Cl-dependent manner[13] The role of the SLC6A8 gene has been reported multiple times in various cancers[14-16].Notably, in HCC,downregulation of SLC6A8 has been found to inhibit invasiveness and migration of human hepatoma cells Huh-7 and Hep3B[17]BNIP3L(BCL2/adenovirus E1B interacting protein 3-like) , a mitophagy receptor, mediates the selective removal of mitochondria[18].However, whether SLC6A8 modulates mitophagy through BNIP3L remains unreported and deserves further investigation.
This study aims to investigate the role of overexpressed SLC6A8,transduced via lentivirus, in modulating mitophagy through BNIP3L and its influence on promoting hepatoma cell proliferation.This would advance the understanding of the regulatory mechanisms and implications of mitophagy in HCC.It thereby provides a theoretical basis for understanding the roles and interaction mechanisms of SLC6A8 and BNIP3L in HCC and enhances the study on mitophagy.These insights could contribute to uncovering HCC pathogenesis and offer novel targets and strategies for HCC therapy.
2.Materials and Methods
2.1 Experimental cells
Human hepatocellular carcinoma cell lines Huh-7 and Hep3B were purchased from the Shanghai Institute of Biological Sciences.Under in vitro hypoxic conditions, human hepatocellular carcinoma Huh-7 was cultured with DMEM medium (HyClone, SH30243.01B)supplemented with 10% fetal bovine serum (Gibco, 42F7180K),while Hep3B cells were cultured with MEM medium (HyClone,SH30024.01B) plus 10% fetal bovine serum.Subsequently, these cells continued to be cultured under serum-free conditions.To simulate a hypoxic environment, the cells were each placed in an environment set to 1% O2, 5% CO2and 94% N2balanced gas, and were cultured in an incubator at 37 ℃ for a continuous duration of 20 hours.Upon completion of culture, samples were collected for subsequent experiments.For the negative control, cells were treated with an equivalent amount of phosphate-buffered saline (PBS) for 12 h.
2.2 Analysis of the TCGA database
Using Xiantao Academic(https://www.xiantaozi.com/), the expression of SLC6A8 in normal human tissues and hepatocellular carcinoma tissues was analyzed through the TCGA database, as well as the correlation of SLC6A8 with BNIP3L expression, and the correlation analysis of SLC6A8 with mitochondria autophagy-related genes in hepatocellular carcinoma tissues was also performed.
2.3 RT-PCR to detect SLC6A8 and BNIP3L expression
First, total RNA from cells was extracted using a Trizol reagent kit (Invitrogen, GT1402) according to the instruction manual.The extracted total RNA was then purified using the RNeasy Mini kit(Qiagen, 74104).Next, a reverse transcription reaction was carried out with the SuperScript IV Reverse Transcriptase kit (Invitrogen,18090010) to transcribe RNA into cDNA.The GoScript Reverse Transcription System kit (Promega, A5000) was used for RT-PCR amplification.Expression data were all normalized to the internal reference GAPDH.The 2-ΔΔCtmethod was used to assess relative expression levels.Primers involved in the experimental process are as follows: SLC6A8-F: CATCTCCAAGGTGGCAGAGT; SLC6A8-R: GATGAAGCCCTCCACACCTA; BNIP3L-F: CAGCAGGGACC ATAGCTCTC; BNIP3L-R: TCATGGCTCCACTTTTCCTC;GAPDH-F: CCAGGTGGTCTCCTCTGA; GAPDH-R:GCTGTAGCCAAATCGTTGT.
2.4 Western blot to detect SLC6A8 and BNIP3L protein expression
RIPA lysis buffer (Biyuntian Technology, P0013B) 50 mL was used to lyse Huh-7 and Hep3B cells, and protein samples were collected.The protein concentration was measured using a BCA protein assay kit (Thermo Fisher Scientific, 23225) according to the instruction manual.Protein samples were mixed with SDS sample buffer, electrophoresed on an SDS-PAGE gel (Bio-Rad, 456-1094), and transferred onto a PVDF protein blotting membrane(Merck Millipore, IPVH00010).To prevent non-specific binding,the membrane was blocked with 5% non-fat milk in TBST buffer for 1 h.Next, primary antibodies against SLC6A8 (ab151315) and BNIP3L (ab10433) (Abcam) were added and incubated at a 1:1 000 dilution, at 5 μL per sample, overnight.The membrane was washed three times with TBST buffer, and incubated with HRPlabeled secondary antibodies (Santa Cruz Biotechnology, sc-2004)that matched the primary antibodies at a 1:2 000 dilution, at 5 μL per sample, for 1 hour.The membrane was washed three more times with TBST buffer.Protein bands were visualized using an ECL chemiluminescent substrate kit (GENVIEW, GE2301).Band densities were quantified with Image J software.Relative protein levels were normalized to GAPDH.
2.5 Immunofluorescence observation of mitochondrial autophagy-related proteins
Huh-7 and Hep3B cell lines were cultured on suitable slides for immunofluorescence.SLC6A8 overexpression plasmids and negative control plasmids were transfected into cells using Lipofectamine 2 000 (Thermo Fisher Scientific, 11668019).After 48 h of transfection, cells were fixed with 4% formaldehyde for 15 min.Cells were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich,T9284) in PBS for 10 min.To prevent non-specific binding, cells were blocked with 5% BSA (Sigma-Aldrich, A9418) in PBS for 1 h.Primary antibodies against TOM20 (ab56783) and LC3 (ab128025)(Abcam) were then added and incubated at a 1:200 dilution, at 5 μL per sample, overnight at 4 ℃.Cells were washed three times with PBS, for 5 minutes each time.Fluorescently-labeled goat antirabbit secondary antibodies (Jackson ImmunoResearch, 111-545-144) were added at a 1:500 dilution, at 5 μL per sample, and cells were incubated for 1 hour in a dark room at room temperature.Cells were washed again three times with PBS, for 5 minutes each time.Nuclei were stained with DAPI (Sigma-Aldrich, D9542) at a 1:1000 dilution for 10 min.Cells were washed twice with PBS and mounted with Antifade Mounting Medium (Vector Laboratories, H-1000).Fluorescence was visualized under a fluorescence microscope and images were captured.
2.6 CCK-8 Assessment of Cellular Proliferation
Huh-7 and Hep3B cell cultures were each plated at 5×103cells per well in 96-well plates, with three replicates per group.Transfections of SLC6A8 overexpression plasmid, negative control plasmid, and BNIP3L silencing plasmid were conducted to the cells, employing Lipofectamine 2 000 transfection reagent.At predetermined times(1, 2, 3, 4, 5 d), 10 μlL of CCK-8 reagent (Dojindo, CK04) was added to each well, followed by 3-hour incubation at 37 ℃ in a 5% CO2incubator until the color properly developed.Absorbance(OD values) was read at 450 nm with a microplate reader.Data was normalized to the negative control group, to calculate cell proliferation rates.
2.7 Statistical Analysis
Measurements are expressed as mean ± SD (Standard Deviation).Data were analyzed and plotted using GraphPad Prism 9.0.0 software.The comparisons between two groups were conducted with Student’s t-test, whereas one-way ANOVA was used for multiple group comparisons.Statistical significance was defined as P<0.05.
3.Results
3.1 Significant overexpression and positive correlation of SLC6A8 with BNIP3L in hepatic cancer tissue was observed
Analysis of the SLC6A8 gene through TCGA database demonstrated its significant overexpression in hepatic cancer tissues(t=0.94,P<0.001), refer to Figure 1A; also, SLC6A8 expression positively correlated with BNIP3L expression in hepatic cancer tissues (R=0.327, P<0.001), see Figure 1B.The association analysis of SLC6A8 with autophagy-related genes in hepatic cancer tissues,including BNIP3L, PINK1, ATG5, ATG7, ULK1, and MAP1LC3B,indicated meaningful correlations, as shown in Figure 1C.
3.2 Overexpression of SLC6A8 Promotes Mitochondrial Autophagy
Using RT-PCR technique, we investigated the mRNA expression of SLC6A8 in human hepatoma Huh-7 and Hep3B cells overexpressing SLC6A8.The results revealed that the relative expression of SLC6A8 in the OE-SLC6A8 group (SLC6A8 overexpressed group) was 4.29±0.34, significantly higher than the NC group (negative control)SLC6A8 relative expression of 1.01±0.02 (t=16.63, P<0.001), see Figure 2A.The Western blot assay for SLC6A8 protein level in both groups showed that, the OE-SLC6A8 group had significantly higher SLC6A8 protein relative expression (0.81±0.02) compared to the NC group with 0.36±0.02 (t=24.32,P<0.001), see Figure 2B.Mitochondrial autophagy was observed via immunofluorescence by staining the mitochondrial protein TOM20 and the autophagy protein LC-3.The result indicated notable increase of mitochondrial autophagy (yellow part) in the OE-SLC6A8 group compared to the NC group, see Figure 2C.A CCK-8 assay was performed to analyze the proliferation rate of human hepatoma Huh-7 and Hep3B cells overexpressing SLC6A8, and the proliferation rate of the OESLC6A8 group on the fifth day (1.69±0.15) was significantly higher than the NC group 2.85±0.13 (t=22.20, P<0.001), see Figure 2D.
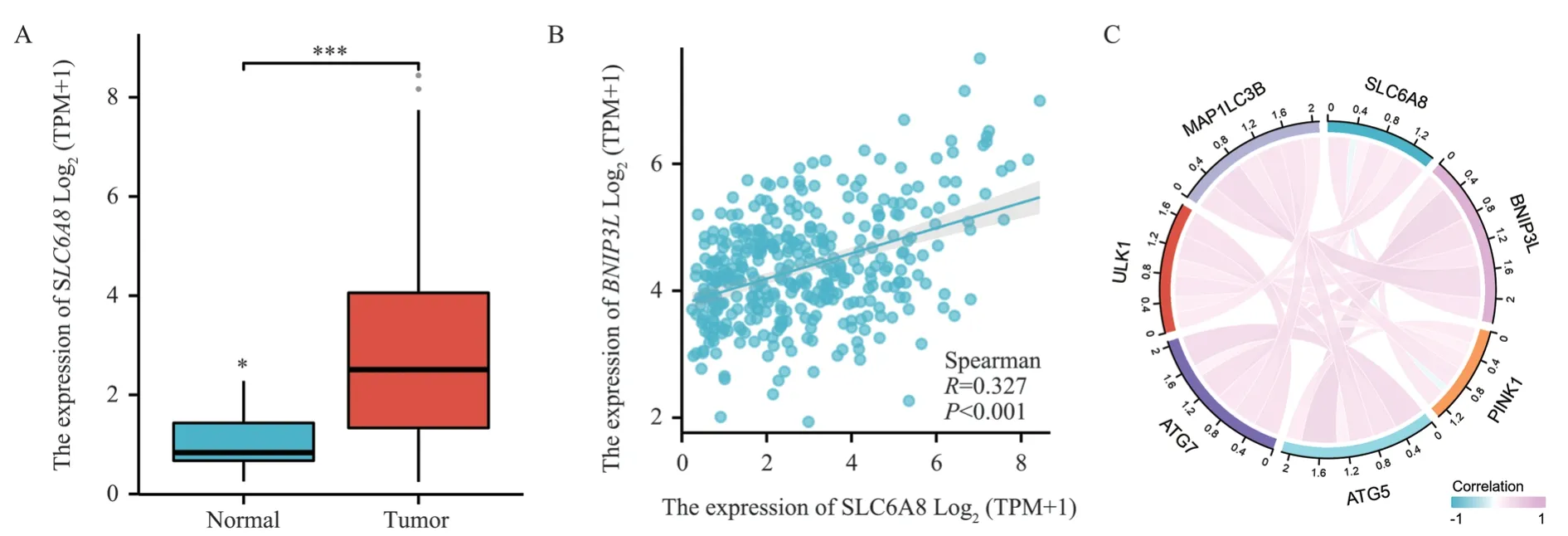
Fig 1 SLC6A8 is highly expressed in liver cancer tissue and positively correlated with BNIP3L expression.
3.3 Overexpression of SLC6A8 Enhances BNIP3L Expression
RT-PCR was conducted to detect the expression of BNIP3L mRNA in human hepatoma Huh-7 and Hep3B cells overexpressing SLC6A8.According to the results, the relative expression of BNIP3L mRNA in the OE-SLC6A8 group was significantly higher(1.40±0.13) compared to the NC group with 1.03±0.07 (t=4.34,P<0.05), see Figure 3A.Utilizing Western blot for BNIP3L protein level assessment revealed that the OE-SLC6A8 group had significantly higher BNIP3L protein relative expression (0.82±0.03)than the NC group with 0.34±0.03 (t=19.60,P<0.001), see Figure 3B.
3.4 Silencing of BNIP3L Reversed the Stimulatory Effect of SLC6A8 Overexpression on Hepatic Cancer Cell
Proliferation A CCK-8 assay was used to check the proliferation rate of human hepatoma Huh-7 and Hep3B cells with silenced BNIP3L following overexpression of SLC6A8.The data showed a lower cell proliferation rate (2.01±0.14) in the OE-SLC6A8+KDBNIP3L group (overexpressed SLC6A8 + silenced BNIP3L) than the OE-SLC6A8 group with 2.60±0.23 (q=8.945,P<0.001) on day 5.The proliferation rate of the OE-SLC6A8 group was significantly higher than the NC group with 1.77±0.13 (q=12.54,P<0.001), see Figure 4.
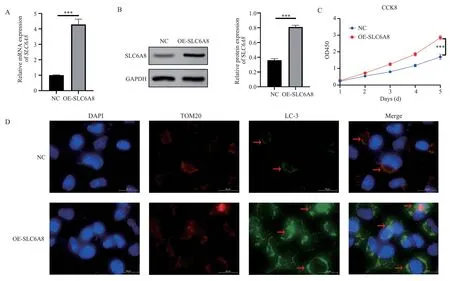
Fig 2 Overexpression of SLC6A8 promotes mitochondrial autophagy and promotes liver cancer cells proliferation.

Fig 3 Overexpression of SLC6A8 promotes BNIP3L expression.

Fig 4 Silencing BNIP3L can reverse the promoting effect of SLC6A8 overexpression on liver cancer cells proliferation.*** means P<0.001
4.Discussion
Hepatocellular carcinoma (HCC) is a primary liver tumor,accounting for over 90% of all primary liver tumors[3].Despite advancements in treatments for liver cancer in recent years[3,19],the complex pathogenesis and late-stage diagnosis of the disease contribute to persistently high mortality rates[20,21].Studies have discovered aberrant gene expression and regulation to be among the key factors contributing to the onset of liver cancer[22].Overexpression or silencing of certain genes may induce cellular malignant transformation and ultimately provoke the initiation of liver cancer[23,24].
The SLC6A8 gene, which encodes a creatine transporter[25] ,has been discovered in recent research to be associated with the malignant progression of a variety of tumors, such as colorectal cancer and non-small cell lung cancer[26,27].Additionally, SLC6A8 has notably been up-regulated in several cancer types[28].Our preliminary research revealed that knocking down the SLC6A8 gene can suppress the invasion and migration of human liver cancer Huh-7 and Hep3B cells[17].In this study, our further experimental results demonstrate that the overexpression of SLC6A8 significantly promotes not only mitochondrial autophagy but also proliferation of human liver cancer Huh-7 and Hep3B cells, which is in line with previous research[17].
According to reports, mitochondrial dysfunction is related to the development and progression of many chronic liver diseases[29].Mitochondrial autophagy is a critical biological process maintaining cellular mitochondrial quality through selective degradation of damaged and aged mitochondria[30] , which has a significant impact on the initiation and progression of hepatocellular carcinoma[31].Studies have found that mitochondrial autophagy is associated with metabolic reprogramming, resistance to anticancer therapies and cancer stem cells, providing new strategies for cancer treatment[32,33].BNIP3L is a critical protein that regulates mitochondrial autophagy[34], and its dysfunction may lead to the dysregulation of mitochondrial autophagy, thereby affecting the initiation and progression of HCC[31].This research demonstrates the most significant positive correlation between SLC6A8 and the mitochondrial autophagy-related gene BNIP3L in liver cancer tissues.Research results indicate that overexpression of SLC6A8 can notably promote the expression of BNIP3L mRNA and protein,and that silencing BNIP3L can reverse the proliferative effects of SLC6A8 overexpression on liver cancer cells.The above results suggest that SLC6A8 regulates the expression of BNIP3L, impacting mitochondrial autophagy and influencing the initiation and progression of HCC.
This study, while it has yielded meaningful results, does possess limitations.Our research is primarily based on cellular experiments,with in vivo animal experiments and clinical trials currently unperformed; therefore, the specific roles and mechanisms of SLC6A8 and BNIP3L require further exploration.Additionally,we only studied the relationship between SLC6A8 and BNIP3L;however, SLC6A8 may have interactive effects with other genes or pathways, which also provides directions for future research.In summary, our research found high expression of SLC6A8 in liver cancer tissues and its positive correlation with BNIP3L.Overexpression of SLC6A8 can promote mitochondrial autophagy and proliferation of liver cancer cells, and silencing BNIP3L can reverse the proliferative effects of SLC6A8 overexpression.These discoveries not only provide new insights for understanding the pathogenesis of HCC but also present potential targets for developing novel therapeutic strategies for hepatocellular carcinoma.
Conflict of Interest Statement
All authors have no conflicts of interest and agree to submit this manuscript.
Author’s contribution description
Author Contribution Degree of contribution LU Yuan experiments, statistical analysis,and manuscript writing mainly Tang Jun cell culture assistance Li Wen-chuan RT-PCR testing assistance Xu Zuo-ming Western blot testing assistance Luo Zong-jiang CCK8 testing assistance Ma Jia-sheng immunofluorescence observation of autophagy assistance Pu Jian article review and funding support mainly
杂志排行
Journal of Hainan Medical College的其它文章
- Effect of honokiol regulating SIRT3 on chronic hypoxia-induced microglia and astrocyte polarization
- Clinical characteristics of patients with early-and late-onset optic neuromyelitis optica spectrum disease
- Distribution of gene polymorphisms associated with aspirin antiplatelet in the Han NSTEMI population
- Predictive value of systemic immunity index for sepsis in low-medium risk community-acquired pneumonia
- To analyze the differentially expressed genes in chronic rejection after renal transplantation by bioinformatics
- Effect of different interventions on orthodontic tooth movement acceleration: A network meta-analysis
