Eugenol targeting CrtM inhibits the biosynthesis of staphyloxanthin in Staphylococcus aureus
2024-01-24JingChngBoChenZeqinDuBowenZhoJihuiLiZiyiLiKnnppnArunchlmTingShiDongqingWeiChunleiShi
Jing Chng, Bo Chen, Zeqin Du, Bowen Zho, Jihui Li, Ziyi Li,Knnppn Arunchlm, Ting Shi, Dongqing Wei, Chunlei Shi,
a MOST-USDA Joint Research Center for Food Safety, School of Agriculture and Biology, State Key Laboratory of Microbial Metabolism,Shanghai Jiao Tong University, Shanghai 200240, China
b School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China
Keywords: Staphylococcus aureus Staphyloxanthin Eugenol Virtual screening 4,4’-Diapophytoene synthase (CrtM)
ABSTRACT Staphylococcus aureus is a serious foodborne pathogen threatening food safety and public health. Especially the emergence of methicillin-resistant Staphylococcus aureus (MRSA) increased the diff iculty of S. aureus treatment. Staphyloxanthin is a crucial virulence factor of S. aureus. Blocking staphyloxanthin production could help the host immune system counteract the invading S. aureus cells. In this study, we f irst screened for staphyloxanthin inhibitors using a virtual screening method. The outcome of the virtual screening method resulted in the identification of eugenol (300 μg/mL), which significantly inhibits the staphyloxanthin production in S. aureus ATCC 29213, S. aureus Newman, MRSA ATCC 43300 and MRSA ATCC BAA1717 by 84.2%, 63.5%, 68.1%, and 79.5 %, respectively. The outcome of the growth curve assay, f ield-emission scanning electron, and confocal laser scanning microscopy analyses confirmed that eugenol at the test concentration did not affect the morphology and growth of S. aureus. Moreover, the survival rate of S. aureus ATCC 29213 and MRSA ATCC 43300 under H2O2 pressure decreased to 51.9% and 45.5% in the presence of eugenol, respectively. The quantitative RT-PCR and molecular simulation studies revealed that eugenol targets staphyloxanthin biosynthesis by downregulating the transcription of the crtM gene and inhibiting the activity of the CrtM enzyme. Taken together, we f irst determined that eugenol was a prominent compound for staphyloxanthin inhibitor to combat S. aureus especially MRSA infections.
1. Introduction
Staphylococcus aureusis one of the most important foodborne pathogens across the world, accounting for a significant number of food safety incidents caused by Gram-positive pathogens[1-3]. In recent years, the improper use of “Antimicrobial Therapy” againstS. aureushas led to the emergence of antimicrobial-resistantS. aureusstrains[2,4-5]. It was reported that methicillin-resistantS. aureus(MRSA) was responsible for about 20 000 estimated deaths per year in the United States than acquired immune def iciency syndrome (AIDS), which indicates a serious public health threat[6-7]. In addition, the increasing development diff iculty and cost further slowed the development of new antimicrobial agents. Given the persistence of the current situation, very few antimicrobial agents could be used inS. aureusclinical treatments in the “Post-Antimicrobial Era”[8-9].Hence, there is an urgent demand to study and develop new treatment strategies that aid the potential of “Antimicrobial Therapy”.
“Antivirulence Therapy” has recently gained a lot of interest in treating infectious diseases[10-11]. In brief, antivirulence treatment aims to increase pathogens’ susceptibility to host immune systems and antimicrobial agents without leaving any selection pressure for developing resistance. Staphyloxanthin, a yellow pigment, is reported as an essential virulence factor ofS. aureus. It is proved that staphyloxanthin shieldsS. aureusfrom reactive oxygen species(ROS)-mediated killing by host immune systems. Therefore,staphyloxanthin is regarded as an important target of antivirulence therapy for treatingS. aureusinfections[12-13].
TheS. aureusutilizes different enzymes, such as 4,4’-diapophytoene synthase (CrtM), 4,4’-diapophytoene desaturase (CrtN),4,4’-diaponeurosporene oxidase (CrtP), glycosyltransferase (CrtQ)and acyltransferase (CrtO) for the production of staphyloxanthin[14].Targeting any of these enzymes in this biosynthetic pathway could lead to the loss of staphyloxanthin production[12,15]. For instance,the compound BPH-652 was found to inhibit the CrtM and thus blocked staphyloxanthin biosynthesis[16]. Similarly, naftifine blocked staphyloxanthin biosynthesis by inhibiting CrtN activity and madeS. aureusmore susceptible to H2O2and human blood[12]. Hence,identifying compounds that inhibit staphyloxanthin synthesis might be a novel antivirulence strategy for treating complicatedS. aureusinfections. It could offer novel approaches to treat complicatedS. aureusinfections by developing anti-staphyloxanthin compounds.In particular, the protein structure of CrtM (PDB ID: 3ACX) had been determined by X-ray diffraction method and deposited in the RCSB protein databank (https://www.rcsb.org/)[17], which provided much convenience for CrtM-inhibitor screening using molecular simulation method.
This study used a virtual screening method to screen natural compounds with anti-staphyloxanthin activity. The outcome of the virtual screening method yielded eugenol, a natural food additive from cloves, as a staphyloxanthin inhibitor targeting CrtM. Moreover,the mode of action of eugenol against CrtM was determined by quantitative RT-PCR and molecular simulation analyses. These results indicated the potential of eugenol as a staphyloxanthin inhibitor and offered evidence for future research into eugenol’s potential as an antivirulence agent.
2. Materials and methods
2.1 Reagents
All reagents were purchased from Sigma-Aldrich, Shanghai,China, unless specified. The working concentration of auraptene,naringenin, morin, osthole, menthol, cinnamaldehyde, thymoquinone,and linalool solutions were prepared in dimethyl sulfoxide (DMSO),arachidic acid and terpineol solutions were in chloroform and eugenol in ethanol. The final concentrations of DMSO, chloroform, and ethanol in all sample solutions were less than 1% (V/V), which had no apparent adverse effect on the growth ofS. aureus.
2.2 Bacterial strains and growth conditions
Four strains ofS. aureus(S. aureusATCC 29213,S. aureusNewman, MRSA ATCC 43300 and MRSA ATCC BAA1717) were used in this study. All 4 strains were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). All the strains were maintained in Baird-Parker (BP) agar plates supplemented with yolk potassium citrate potassium enrichment solution (Land Bridge Technology Ltd., Beijing, China). For assay purposes, the strains were subcultured in tryptic soy broth (TSB) (Land Bridge Technology Ltd.,Beijing, China) at 37 °C to grow up to an appropriate cell density for further experiments.
2.3 Virtual screening
The molecular structures of 11 092 natural compounds retrieved from the Chinese traditional medicine ingredients library, organic molecules database (both developed by Prof. Dongqing Wei, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University,Shanghai, China), and ZinC (http://zinc.docking.org/)[18]were used as ligand library in this study. Subsequently, the interaction between CrtM (PDB ID: 3ACX) and the ligand library was analyzed using AutoDock 4.2[19]. Root mean square deviation (RMSD) was used to evaluate the binding force, docking score, and ligand efficiency.
2.4 Growth curve assay
The growth curves ofS. aureusin the presence and absence of test compounds were determined[20]. In brief,S. aureusstrains were cultured in TSB till they reached an optical density (OD600nm) value of 0.1. Subsequently, the culture was mixed with TSB at the ratio of 1:19(V/V) in the well of a 100-well honeycomb plate, respectively. Then,the specific compound was added to the cultures as required. The growth ofS. aureusstrains was monitored at 600 nm using BioScreen(Bioscreen C MBR automated turbidometric analyzer, Growth Curves Ltd, Finland).
2.5 Quantification of staphyloxanthin
The quantification of staphyloxanthin production inS. aureuswas performed using the methanol extraction method[12]. In brief,1% of the test strains (OD600nm= 0.5) were allowed to grow in TSB supplemented with and without the test compounds at specified concentrations. Following incubation at 37 °C for 24 h, the cells were harvested by centrifugation (8 000 r/min, 5 min, 4 °C) and washed with phosphate buffered solution (PBS). Methanol was used to extract staphyloxanthin from the test strains, and this procedure was performed thrice. Finally, the absorbance of the methanol was read at OD462nmusing a spectrophotometer (Tecan Sunrise, Tecan Inc.,Mannedorf, Switzerland).
Further, the effect of eugenol on the characteristic peaks of staphyloxanthin (1 161 and 1 525 cm–1) was investigated using Raman spectroscopy[21]. In brief, under default settings, methanol extract containing staphyloxanthin was analyzed using a laser confocal Raman spectroscopy (Senterra R200-L, Bruker Optics., Germany).The samples were excited using 532 nm laser (10 mW) for 30 s. All the IR spectra were plotted as absorbance.
2.6 Eugenol susceptibility test
The agar dilution method was used to determine the susceptibility ofS. aureusstrains to eugenol[20]. In brief,S. aureusstrains (OD600nm= 0.1)were serially diluted. Then, 10 μL of cultures at each dilution was spotted on the Mueller Hinton agar (MHA; Land Bridge Technology Ltd., Beijing, China) plate supplemented with different concentrations of eugenol. The MHA plates were further incubated at 37 °C for 16 h and imaged.
2.7 Field-emission scanning electron microscopy (FESEM)
In brief, eugenol-treated and untreatedS. aureuscells were prepared, harvested, and washed with PBS, as mentioned in section 2.5. The cell pellets were resuspended in sterile water containing 2.5% glutaraldehyde and incubated overnight at 4 °C to fix the cells.After centrifugation, the cells were dehydrated with a water-ethanol gradient (30%, 50%, 70%, 90%, 95%, and 100%) for 10 min with shaking at each concentration[22]. Following ethanol dehydration, the cells were fixed on SEM supports. The samples were critical point dried and sputter coated with gold before being examined under FESEM (Sirion 200, FEI Co., Ltd., Hillsboro, OR, USA).
2.8 Confocal laser scanning microscopy (CLSM)
The change in cell membrane integrity ofS. aureuswas studied with CLSM using membrane permeable and impermeable fluorescent dyes[23]. In brief,eugenol-treated and untreatedS. aureuscells were prepared, harvested, and washed with PBS, as mentioned in section 2.5. The equally blended SYTO 9 (membrane impermeable dye) and propidium iodide (PI, membrane permeable dye) mixture was mixed with cell suspension (3:1 000,V/V) and stood for 15 min in the dark.Finally, the samples placed on a glass slide were observed and imaged using a CLSM (Leica TCS SP8 STED 3X, Leica Microsystems,Nanterre, France). Green fluorescence would be present inS. aureuswith a healthy cell membrane, whereas red fluorescence would be present inS. aureuswith a damaged cell membrane.
2.9 Hydrogen peroxide (H2O2) sensitivity assay
The impact of eugenol on the sensitivity ofS. aureusto H2O2was studied[24]. Eugenol-treated and untreatedS. aureuscells were prepared, harvested, and washed with PBS, as mentioned in section 2.5. Hydrogen peroxide (20 mmol/L) was added to the cell suspensions (OD600nm= 1.0) and incubated at 37 °C for 1 h. Following H2O2treatment, the number of viable cells in the cell suspension was enumerated by incubating at 37 °C for 18 h.
2.10 Reverse-transcription quantitative PCR
Briefly, 1% of the test strains (OD600nm= 0.5) were allowed to grow in TSB supplemented with and without eugenol at specified concentrations. Following incubation at 37 °C for 8 h, the cells were harvested by centrifugation (10 000 r/min, 3 min, 4 °C) and washed thrice with PBS. Following the supplier’s protocol, total RNA was extracted and reverse transcribed into cDNA using a SPARKeasy Improved Bacteria RNA kit and SPARKscript II RT Plus kit(SparkJade, Shandong, China), respectively. Finally, quantitative PCR was conducted with the Mastercycler ep realplex system (Eppendorf,Hamburg, Germany). The 2−ΔΔCtmethod was used to estimate the relative expression levels of the target genes[25]. The data set of the target genes was normalized with the expression of the housekeeping gene,gyrB. The detailed sequences of primers used in this study are listed in Table 1.

Table 1 The sequence of primers used for RT-qPCR analysis.
2.11 Molecular docking and molecular dynamics
The chemical structure of eugenol (CID: 3314) was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and the molecular structure of CrtM (PDB ID: 3ACX) was retrieved from the RCSB protein databank (https://www.rcsb.org/). Molecular docking analysis was carried out using Autodock 4.2. The interactions and their 3D/2D structures were visualized through PyMOL v2.0[19,26].Molecular dynamics of the CrtM-eugenol complex was studied using Amber v16[27]. The protein-ligand binding affinity between CrtM and eugenol was computed using molecular mechanics generalized born surface area (MM-GBSA) methods implemented in Amber v16[28].
2.12 Statistical analysis
The results were analyzed byt-test using SPSS software (version 26.0; SPSS, Inc., Chicago, USA). Differences were defined as significant atP< 0.05.
3. Results
3.1 The virtual screening of potential staphyloxanthin inhibitors
Based on the virtual screening results and the availability of natural products, a total of 11 potential staphyloxanthin inhibitors were identified with a stronger binding force toward CrtM. The identified compounds were auraptene, naringenin, morin, osthole,menthol, arachidic acid, eugenol, cinnamaldehyde, thymoquinone,terpineol, and linalool (Table 2).

Table 2 The virtual screening results of staphyloxanthin inhibitors.
3.2 The staphyloxanthin inhibitory effect of potential staphyloxanthin inhibitors
Based on the “Antivirulence Therapy”, the ideal staphyloxanthin inhibitor should be able to block staphyloxanthin biosynthesis without affecting the growth ofS. aureus. In order to establish the reference testing concentration for the subsequent staphyloxanthin inhibition experiment, we first evaluated the effects of the 11 putative staphyloxanthin inhibitors listed above on the growth ofS. aureusATCC 29213 using a growth curve assay. As depicted in Fig. 1,the concentrations above 1 μg/mL of thymoquinone, 16 μg/mL of auraptene, 100 μg/mL of naringenin, morin, menthol, and cinnamaldehyde, 200 μg/mL of osthole, 300 μg/mL of linalool,400 μg/mL of terpineol, and 500 μg/mL of arachidic acid and eugenol showed a significant influence onS. aureusgrowth.
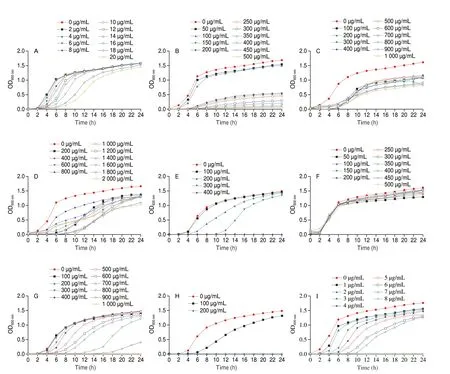
Fig. 1 Growth curve of S. aureus ATCC 29213 cultured in the presence and absence of natural compounds identified through virtual screening. (A) Auraptene;(B) Naringenin; (C) Morin; (D) Osthole; (E) Menthol; (F) Arachidic acid; (G) Eugenol; (H) Cinnamaldehyde; (I) Thymoquinone; (J) Terpineol; (K) Linalool.
The staphyloxanthin inhibitory effect of these compounds below the growth inhibitory concentration was determined (Fig. 2). As shown in Fig. 2G, eugenol at the test concentrations was found to have a significant inhibitory effect on staphyloxanthin biosynthesis ofS. aureusATCC 29213. In particular, eugenol at 300 μg/mL inhibited the staphyloxanthin production inS. aureusATCC 29213 to a maximum of 84.2%. However, eugenol at higher concentrations showed no significant effect on staphyloxanthin production compared to 300 μg/mL. Therefore, it was determined that eugenol was an effective staphyloxanthin inhibitor with 300 μg/mL as the optimal staphyloxanthin inhibitory concentration (OSIC) that exerted no selection pressure onS. aureusgrowth.

Fig. 2 The staphyloxanthin inhibitory effect of natural compounds against S. aureus ATCC 29213. (A) Auraptene; (B) Naringenin; (C) Morin; (D) Osthole;(E) Menthol; (F) Arachidic acid; (G) Eugenol; (H) Cinnamaldehyde; (I) Thymoquinone; (J) Terpineol; (K) Linalool.* Significantly different from the control (0 μg/mL) group (P < 0.05).
3.3 Eugenol alters staphyloxanthin production in S. aureus
In addition toS. aureusATCC 29231, the impact of eugenol on staphyloxanthin production was assessed in otherS. aureusstrains,such asS. aureusNewman, MRSA ATCC 43300 and MRSA ATCC BAA1717. As shown in Fig. S1, eugenol at 300 μg/mL reduced the staphyloxanthin pigment production inS. aureusNewman by 63.5%(Fig. S1A), MRSA ATCC 43300 by 68.1% (Fig. S1B), and MRSA ATCC BAA1717 by 79.5% (Fig. S1C). Furthermore,S. aureusATCC 29213 and MRSA ATCC 43300 were used in the following experiment.
A series of experiments were performed to confirm the effect of eugenol at OSIC on the growth ofS. aureusATCC 29213 and MRSA ATCC 43300. As shown in Fig. 3, it was apparent that bothS. aureusATCC 29213 and MRSA ATCC 43300 had an excellent growth state under the treatment of eugenol below 300 μg/mL compared with the control. In addition, FESEM and CLSM were used to check the effect of eugenol at OSIC on the structural integrity ofS. aureusATCC 29213 and MRSA ATCC 43300. As shown in Fig. 4, the FESEM results showed that untreated cells had a typical cell membrane structure ofS. aureus(smooth and globular-shaped). At the same time, the structure of the eugenol-treated cells was also similar to untreated cells. The results of CLSM analysis also validated the FESEM results (Fig. 5).
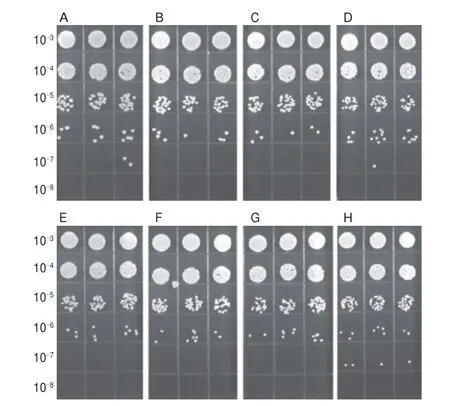
Fig. 3 Growth status of S. aureus ATCC 29213 and MRSA ATCC 43300 on MHA medium supplemented with eugenol at different concentrations.(A) S. aureus ATCC 29213, 0 μg/mL; (B) S. aureus ATCC 29213, 100 μg/mL;(C) S. aureus ATCC 29213, 200 μg/mL; (D) S. aureus ATCC 29213, 300 μg/mL;(E) MRSA ATCC 43300, 0 μg/mL; (F) MRSA ATCC 43300, 100 μg/mL;(G) MRSA ATCC 43300, 200 μg/mL; (H) MRSA ATCC 43300, 300 μg/mL).The 10–3–10–8 on the left side represents the dilution ratio.

Fig. 4 FESEM images of S. aureus ATCC 29213 and MRSA ATCC 43300 treated with and without eugenol. (A) S. aureus ATCC 29213, 0 μg/mL;(B) S. aureus ATCC 29213, 300 μg/mL; (C) MRSA ATCC 43300, 0 μg/mL;(D) MRSA ATCC 43300, 300 μg/mL.
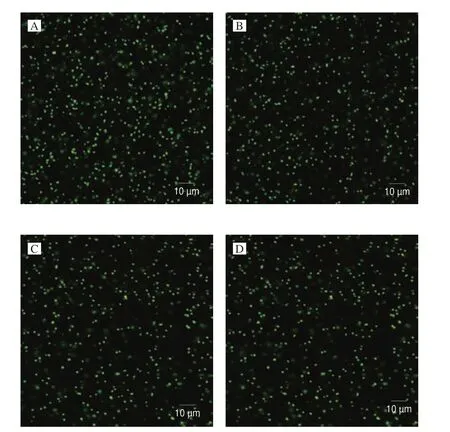
Fig. 5 CLSM images of S. aureus ATCC 29213 and MRSA ATCC 43300 treated with and without eugenol. (A) S. aureus ATCC 29213, 0 μg/mL;(B) S. aureus ATCC 29213, 300 μg/mL; (C) MRSA ATCC 43300, 0 μg/mL;(D) MRSA ATCC 43300, 300 μg/mL.
The effect of eugenol on staphyloxanthin production was qualitatively analyzed by spottingS. aureusATCC 29213 and MRSA ATCC 43300 cultures on the MHA plates supplemented with eugenol at different concentrations (100, 200, and 300 μg/mL).The obtained result was consistent with the result of the staphyloxanthin quantification assay (Figs. 2G and S1B). The results showed that the eugenol at increasing concentration gradual changed the color ofS. aureuscolonies from yellow to white(Fig. S2). Further, the Raman spectroscopy results demonstrated that the signal strength of staphyloxanthin characteristic peaks(1 161 and 1 525 cm–1) were inversely proportional to the eugenol’s concentration. In particular, there were nearly no staphyloxanthin characteristic peaks detected when theS. aureuscells were treated with 300 μg/mL eugenol (Fig. 6).
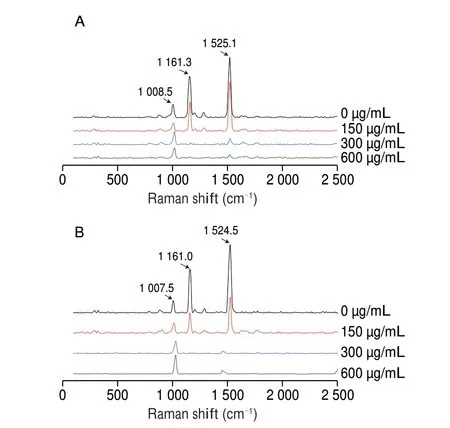
Fig. 6 The Raman spectra of staphyloxanthin from (A) S. aureus ATCC 29213 and (B) MRSA ATCC 43300 treated with eugenol (0, 150, 300, and 600 μg/mL).
3.4 The effect of eugenol on the H2O2 sensitivity of S. aureus
The inhibition of staphyloxanthin production would increase the susceptibility ofS. aureusto ROS produced by the host immune response. To substantiate this, the survival assay ofS. aureusATCC 29213 and MRSA ATCC 43300 under H2O2pressure (20 mmol/L)was performed. As shown in Fig. 7, eugenol at OSIC reduced the survival rate ofS. aureusATCC 29213 and MRSA ATCC 43300 by 51.9% and 45.5%, respectively.
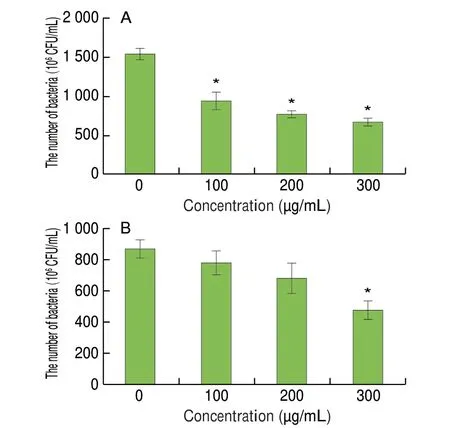
Fig. 7 Effect of eugenol at different concentrations on the survival of(A) S. aureus ATCC 29213 and (B) MRSA ATCC 43300 under H2O2 pressure(20 mmol/L). * Significantly different from the control (0 μg/mL) group (P < 0.05).
3.5 Impact of eugenol on the expression of crt operon
The transcription of genescrtP,crtQ,crtM, andcrtNinvolved in staphyloxanthin biosynthesis upon eugenol treatment was determined using RT-qPCR. In Fig. 8, the transcription ofcrtMandcrtQwas significantly downregulated by eugenol treatment compared to the untreated control (P< 0.05). In particular, the transcription ofcrtMwas significantly downregulated by 0.22-fold (P< 0.05).

Fig. 8 Effect of eugenol (300 μg/mL) on the expression of staphyloxanthin biosynthesis genes in S. aureus ATCC 29213. * Significantly different from the control (0 μg/mL) group (P < 0.05).
3.6 Properties of the interaction between eugenol and CrtM
Molecular docking analysis was performed to elucidate the interaction of eugenol with CrtM. As shown in Figs. 9A and 9B,eugenol had hydrophobic interaction with the amino acid residues of Phe22, Phe26, Val37, Tyr41, Val133, Val137, Leu141, Leu164 and Gln165. Moreover, the hydroxyl groups of eugenol had conventional hydrogen bonding interaction with Ala134 with a bond length of 1.9 Å.
The molecular dynamics simulation between eugenol and CrtM(20 ns,n= 3) showed that the RMSD could reach a steady state,which suggested that eugenol could stably bind with CrtM (Fig. 9C).The binding free energy between eugenol and CrtM calculated by MM-PBSA was –79.91 kJ/mol, which suggested that there was a strong interaction between eugenol and CrtM. The most populated structure and the initial structure of eugenol-CrtM complex were quite comparable, indicating that the interaction between eugenol and the corresponding active site of CrtM was stable (Fig. 9D). To sum up,the molecular simulation studies showed that there was a strong and stable interaction between eugenol and CrtM.
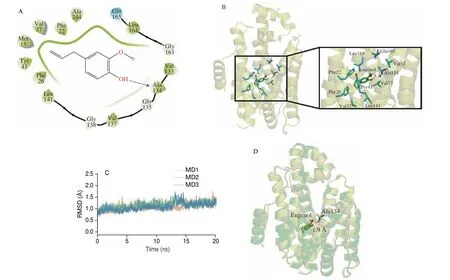
Fig. 9 In silico studies showing the molecular interaction between eugenol and CrtM. (A) Representative molecular docking image depicting the interaction of eugenol with CrtM. (B) 3D image showing the binding pattern of eugenol with CrtM. (C) The RMSD value of the protein structure during molecular dynamics simulation (eugenol and CrtM). (D) Comparison of the most clustered structure and the initial structure of eugenol and CrtM in molecular dynamics simulation.
4. Discussion
In recent years, the improper use of antimicrobial agents posed a severe threat to food safety and public health[28]. As a novel treatment strategy, “Antivirulence Therapy” has lured global attention and showed enormous development potential[29]. In “Antivirulence Therapy”, recent studies have shown that natural compounds targeting staphyloxanthin production inS. aureusmay be used to treatS. aureusinfections[12,30-31]. In this study, we tried to screen the potential staphyloxanthin inhibitors from natural products using virtual screening. As a result, 11 potential compounds were identified from the ligand library containing 11 092 natural compounds with good docking scores and ligand efficiency. Among the identified compounds, eugenol at 300 μg/mL was found to have an excellent inhibitory effect on the staphyloxanthin production on bothS. aureusand MRSA. Further, the potential therapeutic target of eugenol was determined as CrtM using RT-qPCR and molecular simulation studies. To the best of our knowledge, this is the first report revealing the staphyloxanthin inhibitory nature of eugenol. These results further substantiated the potential of natural products as staphyloxanthin inhibitors and provided important data support for further application of eugenol as an antivirulence agent.
In recent years, virtual screening has become a novel strategy in the drug development process by increasing drug candidate selection efficiency[32-33]. It was reported that virtual screening was successfully used in a series of drug developments, such as COVID-19[34],cancer[35], diabetes[36], Parkinson’s[37], and so on. In particular, with the development of cryo-electron microscopy technology and Alphafold 2,the structure information of target protein is very accurate and increases the reliability of virtual screening[38-39]. In this study, we used the virtual screening method to mine staphyloxanthin inhibitors from natural products for the first time. Through virtual screening,we successfully identified eugenol as an efficient staphyloxanthin inhibitor, which suggests the competence and reliability of computerbased screening to identify potential natural products against different human diseases.
The continuous selection pressure of compounds onS. aureususually resulted in the emergence of resistant strains[2,5]. Therefore,it is imperative to reduce the threat ofS. aureusto the host without exerting selection pressure. This study demonstrated that 300 μg/mL of eugenol (without influencingS. aureusgrowth) could increase the susceptibility ofS. aureus(including MRSA) to host immune response, which suggests eugenol as a potent antivirulence agent.These results might provide a new solution for treatingS. aureusinfections and increase our understanding of eugenol as food and medicine. Comparing the earlier research on small molecules, the staphyloxanthin inhibitory effect of OSIC-eugenol was superior to OSIC-carvacrol[40], OSIC-citral[41]and OSIC-ZY-214-4[42]. More,the structural alteration and synergistic drug combination might further lower the OSIC of eugenol, which would expand its potential applications.
The potential target of eugenol on staphyloxanthin biosynthesis was determined as CrtM by RT-qPCR and molecular simulation studies. It was suggested that eugenol might inhibit the staphyloxanthin biosynthesis by downregulating the transcription ofcrtMand inhibiting CrtM enzyme activity simultaneously.CrtM was the first key enzyme to catalyze farnesyl diphosphate to 4,4’-diapophytoene in the staphyloxanthin biosynthesis procedure.Blocking the function or the synthesis of CrtM has resulted in the deficiency of staphyloxanthin[43]. Some other reports indicated that CrtM might be an attractive target for natural products, which suggested the general applicability of CrtM as a staphyloxanthin inhibitory target[44-46]. Overall, the current study reiterates the importance of targeting staphyloxanthin in the antivirulence treatment againstS. aureusinfections and the role of CrtM in developing novel staphyloxanthin inhibitors.
5. Conclusion
This study found eugenol as a potential staphyloxanthin inhibitor using a virtual screening method. Subsequently, it was identified that 300 μg/mL of eugenol could significantly inhibit the staphyloxanthin biosynthesis of bothS. aureusand MRSA without influencing their growth and cell morphology. At the same time, the susceptibility ofS. aureusand MRSA to H2O2increased significantly upon eugenol treatment. Furthermore, it was revealed that eugenol mainly targeted the expression ofcrtMand the enzyme activity of CrtM simultaneously to block staphyloxanthin production. In sum,as an antivirulence agent, eugenol has a strong ability to inhibit the staphyloxanthin biosynthesis of bothS. aureusand MRSA.These findings open up a new area of research for investigation,optimization, and application of eugenol as an antivirulence agent againstS. aureuspathogenesis and subsequent infections.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (31972169 and 32001798).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://doi.org/10.26599/FSHW.2022.9250115.
杂志排行
食品科学与人类健康(英文)的其它文章
- Betalains protect various body organs through antioxidant and anti-inf lammatory pathways
- Effects of Maillard reaction and its product AGEs on aging and age-related diseases
- Characterization of physicochemical and immunogenic properties of allergenic proteins altered by food processing: a review
- Polyphenol components in black chokeberry (Aronia melanocarpa)as clinically proven diseases control factors—an overview
- Food-derived protein hydrolysates and peptides: anxiolytic and antidepressant activities, characteristics, and mechanisms
- Recent advances in the study of epitopes, allergens and immunologic cross-reactivity of edible mango