Association of DNA methylation/demethylation with the functional outcome of stroke in a hyperinflammatory state
2024-01-24YuboWangLingZhangTianjieLyuLuCuiShunyingZhaoXuechunWangMengWangYongjunWangZixiaoLi
Yubo Wang ,Ling Zhang ,Tianjie Lyu ,Lu CuiShunying ZhaoXuechun WangMeng Wang,Yongjun Wang,,Zixiao Li,
Abstract Inflammation is closely related to stroke prognosis,and high inflammation status leads to poor functional outcome in stroke.DNA methylation is involved in the pathogenesis and prognosis of stroke.However,the effect of DNA methylation on stroke at high levels of inflammation is unclear.In this study,we constructed a hyperinflammatory cerebral ischemia mouse model and investigated the effect of hypomethylation and hypermethylation on the functional outcome.We constructed a mouse model of transient middle cerebral artery occlusion and treated the mice with lipopolysaccharide to induce a hyperinflammatory state.To investigate the effect of DNA methylation on stroke,we used small molecule inhibitors to restrain the function of key DNA methylation and demethylation enzymes.2,3,5-Triphenyltetrazolium chloride staining,neurological function scores,neurobehavioral tests,enzyme-linked immunosorbent assay,quantitative reverse transcription PCR and western blot assay were used to evaluate the effects after stroke in mice.We assessed changes in the global methylation status by measuring DNA 5-mc and DNA 5-hmc levels in peripheral blood after the use of the inhibitor.In the group treated with the DNA methylation inhibitor,brain tissue 2,3,5-triphenyltetrazolium chloride staining showed an increase in infarct volume,which was accompanied by a decrease in neurological scores and worsening of neurobehavioral performance.The levels of inflammatory factors interleukin 6 and interleukin-1 beta in ischemic brain tissue and plasma were elevated,indicating increased inflammation.Related inflammatory pathway exploration showed significant overactivation of nuclear factor kappa B.These results suggested that inhibiting DNA methylation led to poor functional outcome in mice with high inflammation following stroke.Further,the effects were reversed by inhibition of DNA demethylation.Our findings suggest that DNA methylation regulates the inflammatory response in stroke and has an important role in the functional outcome of hyperinflammatory stroke.
Key Words:DNA demethylation;DNA methylation;DNMT3A;functional outcome;hyperinflammatory state;interleukin;neuroinflammation;stroke;TET2
Introduction
Currently,there is no ideal treatment to repair the impaired neurological function after stroke.Targeting inflammation following ischemic stroke may be a solution for this problem(Jayaraj et al.,2019).Suppressing neuroinflammation in animal models has been shown to ameliorate ischemic stroke injury (Wang et al.,2018;Jayaraj et al.,2019;Han et al.,2021).And limiting the systemic inflammatory response following inflammatory agents improved the prognosis of stroke to some extent (Anrather and Iadecola,2016).To better understand the underlying connections of these studies,the possible mechanisms of stroke and inflammation need to be further investigated.
Epigenetics may provide unique insights to this problem(Qureshi and Mehler,2010;Kassis et al.,2017;Ng et al.,2018).DNA methylation is one of the most intensively studied epigenetic mechanisms involved in the pathogenesis and prognosis of stroke.Previous studies have shown that epigenetic alterations following transient middle cerebral artery occlusion (tMCAO) in mice upregulated overall DNA methylation in the brain after ischemia and reperfusion (Jhelum et al.,2017;Endres et al.,2000;Stanzione et al.,2020;Choi et al.,2022).Downregulation of DNA methylation by genetic or pharmacological inhibition of DNA methyltransferases (DNMTs)appears to contribute to better outcomes after the injury(Endres et al.,2000;Dock et al.,2015;Choi et al.,2018).In addition,inhibition of ten-eleven translocation methylcytosine dioxygenases 2 (TET2),which plays an important role in DNA demethylation,increased infarct volume after tMCAO(Miao et al.,2015).DNA methylation is associated with neuroinflammation during stroke and immunosuppression in poststroke processes.Studies suggested that DNA methylation adjusts microglia/macrophage polarization,B-and T-cell development and differentiation (Morales-Nebreda et al.,2019),as well as the expression of inflammatory cytokines such as interferon-γ (Chen et al.,2007).
On the basis of previous research,it seems that the simple downregulation of DNA methylation and poststroke inflammation can improve prognosis.However,the situation may be somewhat complicated when stroke is advanced or accompanied by hyperinflammation.A number of stroke patients also have other diseases that affect the systemic inflammatory status at baseline,such as sepsis or dental infections.Stroke activates a strong inflammatory response,and a hyperinflammatory status at baseline elevates the risk of stroke (Lindsberg and Grau,2003;Anrather and Iadecola,2016;Shao et al.,2019).Approximately 8.5% of patients with ischemic stroke have had sepsis in the previous 1-year risk period before stroke,and the risk of stroke is immensely elevated after sepsis for the subsequent year (Boehme et al.,2017).However,there is currently a lack of relevant studies on the regulation of inflammation after stroke for these patients.
The inflammatory response impacts DNA methylation(Stenvinkel et al.,2007;Niwa et al.,2010;Myte et al.,2019).For example,inflammatory factor interleukin (IL)-6 can interfere with the expression pattern of DNMTs (Hegde et al.,2020).However,the epigenetic alterations and detailed biochemical pathways of stroke combined with hyperinflammation remain unclear.Further,the incidence of stroke combined with hyperinflammation is not negligible and deserves further investigation.To gain a deeper understanding and to provide new insights into the treatment of stroke patients in a highinflammatory state,in this study,we hypothesized that altering DNA methylation and demethylation processes would have varying effects on stroke in a high-inflammatory state.Our aim was to investigate the role of DNA methylation abnormalities in the prognosis of high-inflammatory stroke.
Methods
Animals
Adult male C57BL/6 mice (8–10 weeks old,22–25 g;Kang et al.,2013;Cui et al.,2016;Liu et al.,2019) were used for the experimentation.Only male mice were used here due to the consideration that female mice had menstrual cycles,which may influence the results.Animal experiments were conducted in the laboratories of Beijing Neurosurgical Institute.Animals were group housed at 22 ± 2°C under a 12-hour light/dark cycle.Food and water were available ad libitum for the duration of the experiment.The Animal Welfare Ethics Committee of the Beijing Neurosurgical Institute approved the animal procedures (approval No.202003004) on January 4,2021,which were conducted following the ethical principles outlined in the NIH Guide for the Care and Use of Laboratory Animals (8thed.,National Research Council,2011).This study is reported in accordance with the ARRIVE 2.0 guidelines (Animal Research: Reporting ofIn VivoExperiments;Percie du Sert et al.,2020) .
Study design
Using a random number table,we randomly allocated mice into four groups (Figure 1): (1) Sham group,in which mice underwent tail vein injection of vehicle (4% dimethylsulfoxide dissolved in saline) plus surgery without tMCAO;(2) Vehicle group,in which mice received vehicle administration plus tMCAO;(3) DNA methylation inhibition group,in which mice received RG108 (DNA methyltransferase inhibitor,Selleck Chemicals,Houston,TX,USA,Cat# S2821) plus tMCAO;(4)DNA demethylation inhibition group,in which mice received Bobcat339 (DNA demethyltransferase inhibitor,Selleck Chemicals,Cat# S6682).

Figure 1 | The experimental design.
Mice in each group were anesthetized and subjected to either sham surgery or tMCAO after drug administration for 3 consecutive days.Intraperitoneal injection of high-dose lipopolysaccharide (LPS) was conducted at the same time to induce a hyperinflammatory state.Reperfusion was achieved after 60 minutes in groups that received tMCAO,and the drug was injected again through the tail vein.All mice underwent behavioral testing at 24 hours after surgery.Functional deficits were measured by the Modified Garcia Scale,followed bytissue collection for infarct volume estimation and histological analysis as detailed below.
Drug administration
RG108 and Bobcat339 were each dissolve din dimethylsulfoxide and diluted in saline at a concentration of 1 mg/mL.RG108 (200 µL,10 mg/kg),or the same volume of Bobcat339 or vehicle,was administrated by tail vein injection 3 days before surgery,followed by daily injection once a day until the date of surgery.All mice were subjected to intraperitoneal injections of LPS (10 mg/kg) to induce hyperinflammation at the onset of tMCAO (Li et al.,2005).Reagents were administered again at the onset of reperfusion to ensure the efficacy.
tMCAO model
The tMCAO procedure was performed as previously described (Moisse et al.,2008;Dong et al.,2020).Anesthesia was induced with 4% vaporized isoflurane (RWD Life Science,Shenzhen,China,Cat# R510-22) and then maintained through a mask with 1.5% isoflurane.Under an operating microscope,the right common carotid artery and external carotid artery were dissected.Then,tMCAO was induced by a monofilament (Doccol Corporation,Sharon,MA,USA,Cat#602056PK10),which was guided from the external carotid artery into the internal carotid artery and the origin of the MCA.The filament was then retreated to achieve reperfusion after 60 minutes.For sham surgery,the operating field was exposed and the relevant arteries were isolated as was performed for tMCAO,but the arteries were not dissected and the filament was not inserted.Wounds were stitched up and mice were monitored carefully after surgery for signs of discomfort for 24 hours.
Neurological function assessment
The Modified Garcia Score was applied to assess sensorimotor function of the mice at approximately 24 hours after tMCAO,as previously described (Garcia et al.,1995).We test five demensions of neurological deficts.First,body proprioception was tested by touching the mouse with a blunt stick on each side of the body.Scores indicate the following: 3,mouse reacted by turning head and was equally startled by the stimulus on both sides;2,bilateral weak response or brisk ipsi and weak contra response;1,unilateral response;and 0,no response.Second,response to vibrissae touch was tested by brushing a blunt stick against the vibrissae on each side.Scores indicate the following: 3,mouse reacted by turning head or was equally startled by the stimulus on both sides;2,bilateral weak response or brisk ipsi and weak contra response;1,unilateral response;and 0,no response.Third,limb symmetry was tested by holding the mouse in the air by the tail to observe symmetry in the movement of the four limbs.Scores indicate the following: 3,all four limbs extended symmetrically;2,mild flexion of forelimb and/or mid flexion of hindlimb;1,contra forelimb flexed but hindlimb extended or forelimb extended and hindlimb flexed;and 0,contra forelimb and hindlimb completely flexed.Fourth,Lateral turning was tested by suspending the mouse 10 cm above the tabletop.Scores indicate the following: 3,bilateral turning equal and>45 degrees;2,bilateral turning equal but <45 degrees;1,unequal turning;and 0,no turning.
Finally,forelimb walking was tested by holding the mouse by the tail and make mouse walk on forelimbs.Scores indicate the following: 3,briskly walks forward in symmetry;2,moves to one side;1,moves in circle;and 0,cannot move on forelimbs.Scores for each dimension range from 0–3,and higher scores indicate better sensorimotor functional performance.The experiments were performed by investigators blind to the groupings.
Behavioral testing
Behavioral testing was performed after neurological function assessments.The investigators were blinded to allocation during experiments and outcome assessments.
The foot misplacement test assesses locomotor and sensory function in mice with central nervous system disorders(Chavez et al.,2019).Mice were placed in the testing room for 60 minutes before the experiment.An Automatic Foot Misplacement Apparatus (BiosebIn VivoResearch Instruments,Pinellas Park,FL,USA,Cat# BIO-FMA) was used for the testing.Mice were placed on the elevated horizontal ladder and trained to cross the device.Sensors recorded paws slipping through the steps of the ladder.Locotronic software(BiosebIn VivoResearch Instruments) was used to measure the paw misplacements and the time to cross a distance of 50 cm.Each mouse was tested three times and the average number of missed steps was recorded.
The open field test was performed to evaluate locomotion,hyperactivity,and anxiety-related behaviors (Yoshizaki et al.,2020).Each mouse was placed in the center of a 50 cm × 50 cm × 40 cm acrylic black box.Total distance,mean velocity,path length and other information were recorded by an overhead camera during a 30-minute period.Data were analyzed using Smart 3.0 software (Panlab,S.L.U.,Barcelona,Spain).
The rotarod test was used to measure motor coordination and balance (Schaar et al.,2010).In this test,the mice had to walk on the rotating rod to prevent falling (XR-YLS-4C,XinRuan,China).Mice were pretrained for 3 days at 4 r/min to ensure that each mouse could stay on for at least 3 minutes.The formal experiment was started 30 minutes after the mice were readapted to the training.The speed of rod rotation was gradually accelerated from 4 r/min to 40 r/min over 5 minutes,and the holding time of mice was recorded.Before and after the experiment,the instrument was fully cleaned with 75% ethanol and wiped dry.The experiment was repeated three times with an interval of 30 minutes.The average holding time of the three experiments was calculated as the final result.
The hanging test was used to measure the locomotion ability of the limbs and was performed according to the wire hanging test method (Feng et al.,2020).The mice were suspended on a barbed wire grid for 3 days of pretraining,and those that hung suspended for less than 5 minutes were excluded.One pretraining session was conducted before the formal experiment,and then the experiment was conducted threetimes with an interval of 30 minutes each time.The average holding time was recorded as the final result.
Measurement of infarction volume by 2,3,5-triphenyltetrazolium chloride staining
Mice were anesthetized and sacrificed after neurological function assessments and behavioral testing.Brains were removed quickly and sliced into 1-mm-thick coronal sections on ice using a mouse brain slicing mold (RWD Life Science,Cat# 68707).Sections were then stained with 2%2,3,5-triphenyltetrazolium chloride (TTC;Solarbio,Beijing,China,Cat# T8170) for 10 minutes,followed by fixation in 4%paraformaldehyde (Solarbio,Cat# P1110) for approximately 20 minutes.Infarction volumes were determined using ImageJ 1.53f51 software (NIH,Bethesda,MD,USA;Schneider et al.,2012).Contralateral and ipsilateral hemisphere and infarct areas were traced using ImageJ software.The total infarct size was calculated as a percentage by dividing the infarct size by the total size of the bilateral hemisphere in six consecutive coronal sections.To correct for tissue swelling,the infarct size was calculated using the equation described by Boyko et al.(2011).Corrected infarct size=infarct size × contralateral hemisphere size/ipsilateral hemisphere size.
Splenectomy and spleen weighing
Splenectomy of mice was performed under the isoflurane anesthesia,as previously described.A longitudinal incision approximately 0.5 cm long was made on the left side of the abdomen.The attachments and blood vessels of the spleen were dissected and cauterized using a cautery pen with a heated tip.The spleen was then separated and weighed immediately (Guan et al.,2023).
Western blotting
Brain samples collected at 24 hours after tMCAO were lysed using RIPA buffer (RIPA:PMSF=100:1).Cells were broken up in a grinder and the resultant liquid was then centrifuged.After centrifugation at 13,400 ×gat 4°C for 30 minutes,the supernatant was collected as a protein extract.The concentrations of proteins were determined using a BCA Protein Assay kit (Thermo Fisher Scientific,Waltham,MA,USA,Cat# 23225).The supernatant was added into 1× SDS loading buffer (APPLYGEN,Beijing,China,Cat# B1012-5)with a final concentration of 0.1%.The SDS samples were boiled for 10 minutes before loading.Proteins (25 µg) were fractionated by electrophoresis on 10% Nupage Bis-Tris gels (APPLYGEN,Cat# T202110) and then transferred onto a polyvinylidene fluoride membrane (Merck Millipore,Billerica,MA,USA,Cat# IPFL00010).Then,membranes were incubated with primary antibodies against GADPH (mouse,1:2000,ZSGB-BIO,Beijing,China,Cat# TA-08,RRID: AB_2747414),DNMT3A (mouse,1:1000,Abcam,Cambridge,UK,Cat#ab2850,RRID: AB_303355),DNMT3B (mouse,1:500,Santa Cruz Biotechnology,Santa Cruz,CA,USA,Cat# sc-81252,RRID: AB_1122311),DNMT1 (mouse,1:500,Santa Cruz Biotechnology,Cat# sc-271729,RRID: AB_10710384),TET1(mouse,1:500,Santa Cruz Biotechnology,Cat# sc-293186),TET2 (rabbit,1:1000,Cell Signaling Technology,Danvers,MA,USA,Cat# 36449,RRID: AB_2799102),IL-1β (mouse,1:500,Cell Signaling Technology,Cat# 12242,RRID: AB_2715503),IL-6 (mouse,1:500,Santa Cruz Biotechnology,Cat# sc-28343,RRID: AB_627805),NF-kβ (mouse,1:500,Santa Cruz Biotechnology,Cat# sc-8008,RRID: AB_628017) and p-NF-kβ (mouse,1:500,Santa Cruz Biotechnology,Cat# sc-136548,RRID: AB_10610391) at 4°C overnight.After being washed three times with Tris-buffered saline with Tween 20,the membranes were incubated for 1–2 hours at room temperature with horseradish peroxidase (HRP)-conjugated donkey anti-mouse IgG (1:5000,Jackson Immunoresearch Laboratories,West Grove,PA,USA,Cat# 711-035-151,RRID:AB 2340771) or HRP-conjugated donkey anti-rabbit IgG(1:5000,Jackson Immunoresearch Laboratories,Cat# 711-035-152,RRID: AB 10015282).Protein levels were determined using enhanced chemiluminescence HRP detection reagents(Solarbio,Cat# PE0010) and Image Reader (Syngene,Cambridge,UK,G:BOX Chemi XX9).
Quantitative reverse transcription-polymerase chain reaction
For cDNA synthesis,total RNA was extracted from the ischemic tissue using the EASYspin tissue/cell total RNA extraction kit (Aidlab Biotechnologies,Ltd.,Beijing,China,Cat# RN2802) and was reverse transcribed into cDNA using 5× All-In-One RT Master Mix (Applied Biological Materials Inc.,Richmond,BC,Canada,Cat# G490).The real-time monitoring of polymerase chain reaction (PCR) amplification reaction was performed on a Step-One Plus PCR instrument (Applied Biosystems,Waltham,MA,USA) using a miScript SYBR Green PCR Kit (Thermo Fisher Scientific,Cat# A25741).The primers used for quantitative reverse transcription-PCR (qRT-PCR) are listed inTable 1.The qRT-PCR conditions were as follows: 95°C for 5 minutes denaturation,followed by 40 cycles of 95°C for 15 seconds,and 60°C for 60 seconds.The fold change in the expression of each gene was calculated using the ΔΔCt method.Relative protein expression was normalized to glyceraldehyde-3-phosphate dehydrogenase.

Table 1 |Quantitative reverse transcription-polymerase chain reaction primers targeting the related genes
Enzyme linked immunosorbent assay
A peripheral blood sample was collected from the orbital venous sinus of each mouse under isoflurane anesthesia,as previously described.A capillary tube was inserted at the medial canthus under the nictitating membrane at a 45° angle,aiming for the back of the orbit to collect the peripheral blood.Cytokine levels (5-mc,5-hmc,and IL-6) in the peripheral blood were detected using 5-mc (Epigenteck,Farmingdale,NY,USA,Cat# P1030),5-hmc (Epigenteck,Cat# P1032) and IL-6 (Abcam,Cat# ab222503) enzyme linked immunosorbent assay (ELISA)kits according to the manufacturer’s instructions.Absorbance at 450 nm was measured using a microplate reader (Tecan Trading AG,Männedorf,Switzerland,# SPARK).Each sample was assayed in triplicate.
Statistical analysis
No statistical methods were used to predetermine sample sizes;however,our sample sizes were similar to those reported in previous publications (Kalappa et al.,2013;Jin et al.,2014).Statistical analysis data are presented as scatter plots combined with mean ± standard error of the mean.Data analysis was performed using GraphPad Prism 8.0 (GraphPad,San Diego,CA,USA,www.graphpad.com).Between-group comparisons were made using one-way or two-way analysis of variance followed by Tukey’spost hoctest.P<0.05 was considered statistically significant.
Results
Hypomethylation status increases infarction volume in hyperinflammatory brain
First,we investigated the effects of abnormal DNA methylation and demethylation on the mouse tMCAO model under a hyperinflammatory state.In the LPS-induced hyperinflammatory state,TTC staining measurement at 24 hours after tMCAO showed that,compared with the vehicle group,the infarct volume in the RG108 group was increased approximately 14% (P<0.01),whereas that in the Bobcat339 group had a slight reduction (Figure 2AandB).When we compared the two treatment groups,there was a reduction in infarct volume in the Bobcat339 group compared with the RG108 group (approximately 17%,P<0.001;Figure 2AandB).There was no significant difference in the weight of mice from the beginning of injection to the time of surgery (Figure 2C).There was no significant difference in spleen weight between the groups (Figure 2D).

Figure 2 |In a hyperinflammatory state,inhibition of DNA methyltransferase(DNMT3A) led to increased infarction volume,and inhibition of translocation methylcytosine dioxygenases 2 (TET2) showed the opposite result.
Hypomethylation lowers neurofunctional scores in ischemic mice in a hyperinflammatory state
Next,we evaluated the neurological deficits caused by cerebral ischemia after RG108 or Bobcat339 intervention at high inflammatory levels.The Modified Garcia Test scores were used to evaluate sensorimotor function after 24 hours of tMCAO.Compared with the Sham group,all ischemic mice had significantly lower neurological function scores (P<0.001;Figure 3F).Compared with the Vehicle group,the RG108 group had lower scores in body proprioception (P<0.05;Figure 3A),vibrissae touch (Figure 3B),limb symmetry (Figure 3C),lateral turning (Figure 3D),and forelimb walking (P<0.05;Figure 3E),and in total neurological assessment scores (P<0.01;Figure 3F),whereas the Bobcat339 group showed the opposite trend.The Bobcat339 group had significantly better individual scores than the RG108 group (P<0.001;Figure 3F).These results indicated that altering DNA methylation or demethylation may affect sensorimotor function at 24 hours after tMCAO in a hyperinflammatory state.

Figure 3 |In a hyperinflammatory state,inhibition of DNMT3A resulted in poorer neurological function scores,and inhibition of TET2 elevated the scores.
Hyperinflammatory hypomethylation status causes motor behavioral deficits after cerebral ischemia
We next performed four commonly used motor behavioral tests to detect motor deficits.Before the experiments,we trained all of the mice and excluded those that did not perform well before surgery or because of the drug injection.The foot misplacement test was used to evaluate the coordination of mice during marching.At 24 hours after stroke,the foot misplacement test showed that compared with that in the vehicle group,the average number of missed steps in the RG108 group was significantly increased (P<0.05),whereas that in the Bobcat339 group was decreased(Figure 4A).
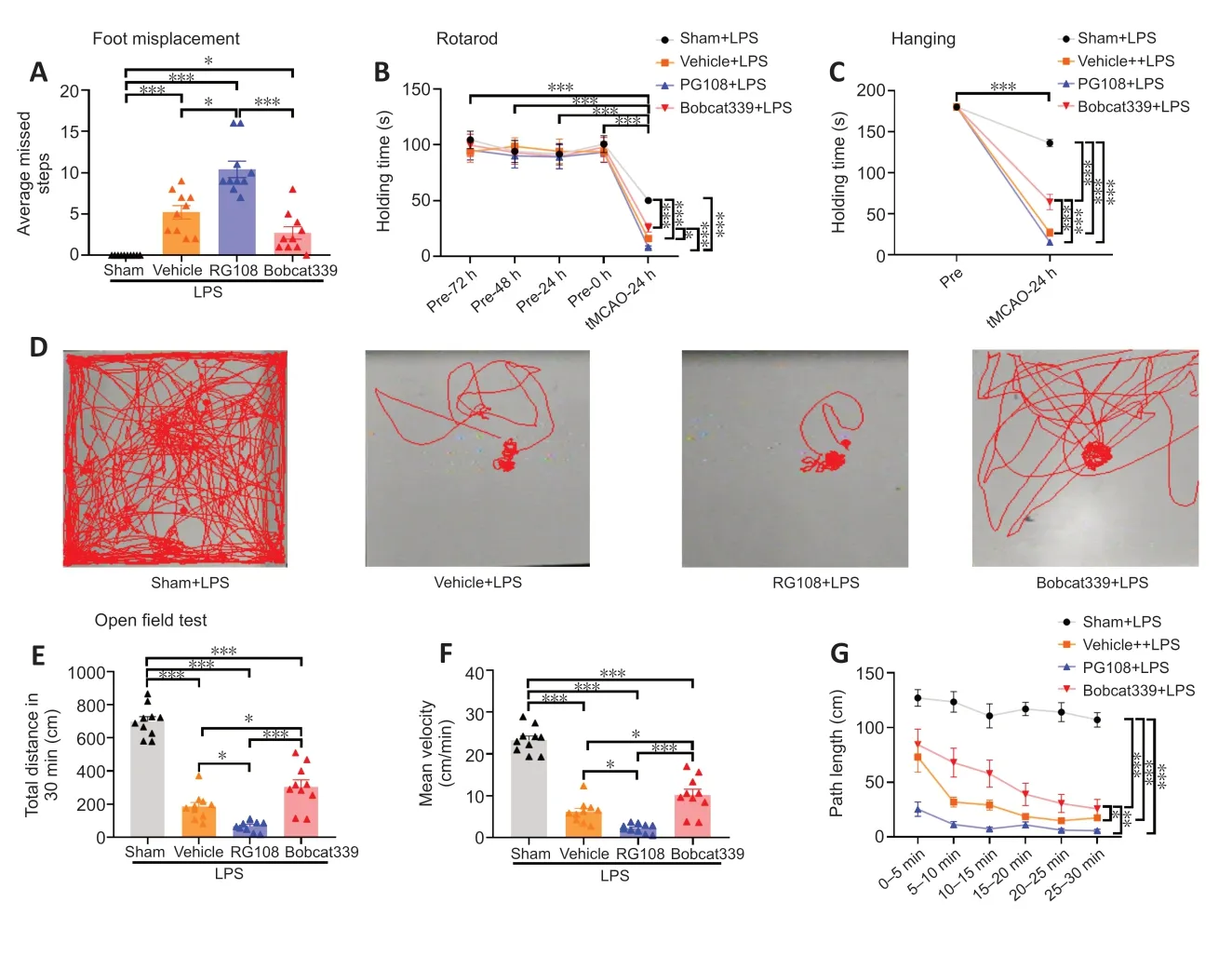
Figure 4 |Motor behavioral deficits after cerebral ischemia in different intervention groups under a high inflammatory state.
The rotarod test was used to evaluate motor balance in mice.Mice in a hyperinflammatory state had significantly reduced performance compared with pre-experimental normal levels of inflammation (P<0.001).In the hyperinflammatory state,the tMCAO model group had a shorter retention time than the Sham group (P<0.001),and the RG108 group had the shortest time of all of the groups (Figure 4B).
The hanging test was used to assess motor dysfunction in the limbs of mice.Motor performance in mice under hyperinflammatory conditions was decreased compared with that at pre-experimental levels,and mice in the tMCAO group performed worse than the Sham group (P<0.001).When all tMCAO groups were compared with each other,the RG108 group had the shortest hanging time and the Bobcat330 group was improved (RG108vs.Bobcat339,P<0.001;Figure 4C).
The open field test was used to evaluate the general exploratory behavior of mice in each group (Figure 4D).Total distance (Figure 4E),mean velocity (Figure 4F),and path length (Figure 4G) were markedly diminished in all tMCAO groups compared with the Sham group (P<0.001).This might be due to the combination of high inflammation and stroke,which led to a significant drop in the motor ability of mice.The performance of the RG108 group and Vehicle group were poor,and there was statistical difference between them,whereas the Bobcat339 group had a better performance compared with these two groups.
These findings suggest that excessive inflammation impaired mobility of the mice and showed that mobility was visibly impaired after stroke combined with hyperinflammation.Inhibition of DNA methylation may exacerbate this deficit,and inhibition of DNA demethylation may alleviate it.
RG108 affects the levels of DNMT3A
To investigate the changes of DNMT3A protein levels in mice after RG108 intervention,qRT-PCR and western blot analysis were performed on brain tissue after ischemia.As shown inFigure 5AandB,the mRNA expression level ofDnmt3awas highest in the RG108 group,whereas the protein expression level was the lowest in this group.We simultaneously detected DNMT1 andDnmt3band found that their mRNA expression levels followed the same trend as Dnmt3a.However,the changes in protein levels were not significant(Figure 5C–F).

Figure 5 |In a hyperinflammatory state,inhibition of DNMT3A function results in changes of DNA methylation level.
Bobcat339 affects the levels of TET2
Similarly,to investigate the effect of Bobcat339 on TET2 function,we used western blot and qRT-PCR to detect changes in TET2 after Bobcat339 treatment.The results showed thatTet2mRNA expression was upregulated in the Bobcat339 group (P<0.01;Figure 6A),whereas protein levels were decreased when compared with those in the vehicle group (P<0.001;Figure 6B).Next,we used the same methods to detect TET1 in the TET family and found that the trend of changes inTet1mRNA and protein was similar to that of TET2,but the changes in TET2 were more significant(Figure 6CandD).

Figure 6 |In a hyperinflammatory state,inhibition of TET2 function results in changes of DNA demethylation level.
Hypomethylation status elevates ischemic brain tissue inflammation levels in a state of high inflammatory stroke
To investigate the inflammation levels in different treatment groups after hyperinflammatory stroke,we used western blot and qRT-PCR to detect mRNA and protein expression levels of IL-1β and IL-6 in ischemic brain tissue.The mRNA and protein levels of IL-6 were obviously raised in the RG108 group.In contrast,the Bobcat339 group had diminishedIL-6mRNA and protein levels.The trend of IL-1β was similar to that of IL-6,but the variations were more pronounced in IL-6(Figure 7).

Figure 7 |Changes in levels of inflammatory markers in ischemic braintissue.
After high inflammatory stroke,RG108 and Bobcat339 lead to systemic changes in DNA methylation and demethylation levels and inflammation levels
Next,to investigate changes in the systemic methylation status induced by continuous administration of RG108 and Bobcat339,we measured the levels of DNA 5-mc and DNA 5-hmc in peripheral blood by ELISA.Compared with the Vehicle group,the RG108 group had a significant decrease in DNA 5-mc (P<0.01;Figure 8A),and the Bobcat339 group had a significant decrease in DNA 5-hmc (P<0.05;Figure 8B).Additionally,to investigate the systemic inflammation level,we measured IL-6 levels in peripheral blood.Consistent with the ischemic brain tissue,the IL-6 level was significantly increased in the RG108 group (RG108vs.Vehicle,P<0.001;RG108vs.Bobcat339,P<0.01;Figure 8C).
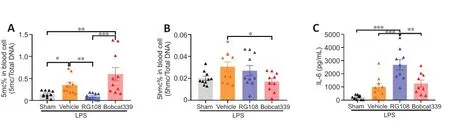
Figure 8 |Changes in DNA methylation and demethylation and inflammation levels in peripheral blood.
After high inflammatory stroke,alteration of DNA methylation and demethylation leads to changes in related inflammatory pathways,including the NF-κB pathway
To gain insight into the mechanisms underlying the observed effects,we detected the key factors of the five classic inflammatory pathways after stroke.The change in NF-κB was the most significant.Specifically,NF-κBmRNA expression was significantly upregulated in the RG108 group compared with that in the other groups (RG108vs.vehicle,P<0.05;RG108vs.Bobcat339,P<0.05;Figure 9A).mTORmRNA expression was also significantly upregulated in the vehicle and RG108 group,but there was no statistical difference between them(vehiclevs.sham,P<0.05;RG108vs.sham,P<0.05;Figure 9A).Other inflammatory pathways including AKT,Nrf2 and MAPK did not present statistical difference between vehicle group and sham group.Subsequently,we examined the key complex protein NF-κB p65 of the NF-κB pathway by western blot and found that the expression level of phosphorylated NF-κB p65 (p-NF-κB,representing the activation of NF-κB)was highest in the RG108 group and lowest in the Bobcat339 group (RG108vs.Bobcat339,P<0.05;Figure 9BandC).
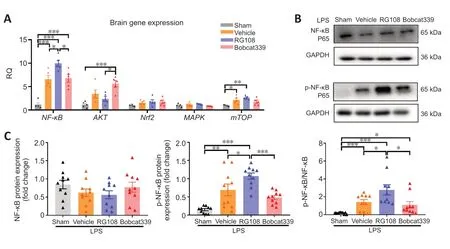
Figure 9 |In a hyperinflammatory state,DNA methylation and demethylation changes affect the NF-κB pathway.
Discussion
Our study aimed to explore the influence of DNA methylation on the prognosis of stroke in an LPS-induced hyperinflammatory state.The findings contribute to a deeper understanding of the interplay between DNA methylation,stroke and inflammation.Our results showed that in an LPS-induced systemic inflammatory state,inhibiting DNA methylation led to an increase in infarct volume,a decrease in neurological scores and behavioral performance,and worse functional outcome in mice after stroke.Inhibiting DNA methylation also led to higher levels of ischemic braintissue and systemic inflammation.In contrast,inhibiting DNA demethylation improved these measures.
In this experiment,RG108 and Bobcat339 were selected as the DNA methylation and demethylation inhibitors,respectively.We selected RG108 as the DNA methylation inhibitor rather than alternatives,such as decitabine,5-azacytidine or zebularine,because it is a non-nucleoside catalytic DNMT inhibitor that blocks the activity of DNMTs directly.Further,RG108 lacks cytotoxicity (Lyko and Brown,2005;Siedlecki et al.,2006) and does not induce gene derepression or alter the DNA methylation state on monosomic chromosomes as decitabine does (Greve et al.,2021).Compared with other DNA demethylation inhibitors,Bobcat339 has higher selectivity and potency,and can effectively inhibit the DNA demethylation process.Additionally,our data on long-term monitoring of body weight and behavior in mice during continuous administration demonstrated the safety of the inhibitor (Chua et al.,2019;Dusadeemeelap et al.,2022).
In the present study,we found that DNMT3A inhibition resulted in increased infarction volume and worse functional outcome under hyperinflammatory conditions.Conversely,inhibition of TET2 may have a beneficial outcome.Previous studies showed that increased total DNA methylation levels after ischemia may worsen the prognosis of stroke (Endres et al.,2000;Jhelum et al.,2017;Choi et al.,2022).DNMT inhibitor decitabine has been shown to reduce inflammation and improve outcomes in sepsis by inhibiting the NF-κB signaling pathway (Hegde et al.,2020) and has also been reported to reduce the severity of ischemic injury after tMCAO(Endres et al.,2000).DNMT inhibitor zebularine has also been shown to improve LPS-induced neuroinflammation (Matt et al.,2018).Further,some previous studies indicated that TET may play a neuroprotective role and inhibition of TET2 increases infarct volume (Miao et al.,2015;Gontier et al.,2018;Morris-Blanco et al.,2021).These different outcomes may be due to the differences between the normal poststroke inflammatory response and a hyperinflammatory environment.One possible explanation is that DNA methylation has a dual role in stroke as neuroprotective or damaging,depending on the stage and severity of stroke.Apoptotic mechanisms are engaged in milder forms of ischemia,whereas excitotoxic and free radical-mediated mechanisms play a more dominant role after prolonged periods of ischemia (Choi,1988;Endres et al.,1997,1998a,b).Furthermore,animal experiments showed that DNA methylation inhibition induced by DNMT1 gene deletion had no protective effect in an ischemia model of excitotoxic or necrotic cell death (Endres et al.,2000).This result implied that the neuroprotective effect of DNA hypomethylation might be more prominent in mild stroke driven by apoptotic mechanisms (Kristián and Siesjö,1998;Xing et al.,2012).In our study,LPS application may boost oxygen consumption and radical generation,thus enhancing oxidative stress and excitotoxicity.Hyperinflammation may alter the stroke pattern and its main injury mechanism,limiting the protective effects of DNA hypomethylation.
Our results showed that after application of the inhibitor,Dnmt2aandTet2mRNA levels increased and their protein expression levels decreased.In a previous study,Dnmt3amRNA levels increased in isolated monocytes treated with LPS(Wisler et al.,2022),which is consistent with our results of the mRNA levelsin vivo.RG108 was shown to downregulate DNMT3a protein levels (Shao et al.,2017),which was the opposite trend as the protein levels in our study.This may be due to the inhibitor interfering with the enzymatic activity,leading to reduced functionality and triggering posttranscriptional regulatory responses.In this context,there may be an increase in mRNA transcription to compensate for the decreased protein expression,such that in response to the decrease in DNMT3A and TET2 protein expression,the cells may attempt to increaseDnmt3aandTet2mRNA transcription,resulting in an overall increase in transcriptional mRNA levels after inhibitor treatment.This may involve several complex factors,such as post-transcriptional regulation,protein degradation,and post-translational modifications.These findings underscore the complexity of intracellular regulatory networks and hold significant importance in understanding the roles of DNMT3A and TET2 in the process of high inflammation during stroke and their involvement in related inflammatory responses.This represents a crucial direction for our further research efforts.
We further tested the inflammatory factors after stroke in a high inflammatory state after the alteration of DNA methylation.We selected IL-1β and IL-6 as indicators for neuroinflammation.Our results showed that the change in IL-6 was more significant than that of IL-1β in ischemic braintissue.Higher levels of IL-6 correlate with cerebral perfusion deficits and infarct sizes and an increased risk of poor functional outcome in stroke (Hotter et al.,2019;Li et al.,2022),as well as with the adjusted incidence of the outcome cluster recurrent stroke,myocardial infarction or vascular death after stroke (Whiteley et al.,2011).Studies showed that compared with IL-1β,IL-6 had higher differential gene expression in the acute phase and had better performance in predicting poor functional outcome of stroke (Zhang et al.,2018;Shi et al.,2021;Coveney et al.,2022),which is consistent with our findings.We consider that IL-6 may be a better inflammatory target than IL-1β,and that these findings may provide a new reference for the selection of inflammatory indicators after stroke.
Stroke is commonly considered as a neurological disorder caused by a localized ischemic or hemorrhagic event in the brain (Nguyen and Chauhan,2023;Ugidos et al.,2023).However,mounting evidence suggests that stroke is not merely a local brain injury but a systemic disorder that involves multiple organs and systems.The inflammatory response is one of the key mechanisms underlying the systemic effects of stroke (Taing et al.,2023).Following a stroke,inflammatory mediators are released into circulation,causing a systemic inflammatory response that can affect distant organs and systems beyond the brain (Chamorro et al.,2016).Our results showed that inhibition of DNA methylation and demethylation led to systemic changes in DNA methylation levels and inflammation after high-inflammatory stroke,which were consistent with the ischemic brain tissue.Therefore,it is becoming increasingly clear that stroke is a complex,multiorgan disease that requires a systemic approach to its management and prevention.
The process of stroke involves a variety of pathological changes,including neuroinflammation,apoptosis,autophagy,excitotoxicity,and oxidative stress.These pathological changes are mediated by a complex interplay of various interactive signaling pathways,such as the NF-κB pathway,MAPK pathway,mTOR pathway,PI3K-Akt pathway,and Nrf2 pathway(Qin et al.,2022).The NF-κB pathway is one of the most studied pathways among these and is particularly important in the inflammatory response.The NF-κB pathway plays a pivotal role in microglia-associated neuroinflammation through inflammasomes and also participates in autophagy (Wang et al.,2021;Qin et al.,2022).We screened the expression of inflammatory pathways after stroke and found that under a high inflammatory state,alteration of DNA methylation led to changes in poststroke inflammatory pathways including the NF-κB pathway and the mTOR pathway,especially in the former one.While there was no significant changes in the AKT,Nrf2 and MAPK pathways.These findings point towards our future research direction and may provide a basis for targeted therapy of high inflammatory stroke.
Current findings support that DNA methylation has a significant protective effect against stroke combined with hyperinflammation.However,there are some limitations and future directions that should be considered.Additional experiments need to be conducted to investigate the relationship between DNA methylation,stroke and inflammation.For example,DNA methylation inhibition using knockout mice is a more stable and reproducible approach.Further,our study found different responses of inflammatory pathways after DNA methylation or demethylation inhibition after high-inflammatory stroke,and future studies should explore the molecular mechanisms in more detail.
In conclusion,we investigated the effect of DNA methylation on the functional outcome of stroke in a high inflammatory state.The findings demonstrated that in a state of high inflammation,inhibition of DNMT3A function led to higher levels of inflammation,increased infarct size,and poor prognosis in stroke.Conversely,inhibition of TET2 function produced favorable results.Multiple inflammatory signaling pathways may be affected by DNA methylation,and NF-κB may be one of the key pathways regulating stroke inflammation.This finding provides a novel potential therapeutic target in poststroke recovery.
Author contributions:Study conception and design:YuW,TL,YongW and ZL;experiment implementation:YuW,LZ,TL,LC,SZ and XW;data analyze:YuW,LZ,TL and MW;manuscript draft:YuW and LZ;critical input and study supervision:YongW and ZL.All authors approved the final version of the manuscript.
Conflicts of interest:The authors declare that they have no competing interests.
Data availability statement:No additional data are available.
Open access statement:This is an open access journal,and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix,tweak,and build upon the work non-commercially,as long as appropriate credit is given and the new creations are licensed under the identical terms.
杂志排行
中国神经再生研究(英文版)的其它文章
- Modulation of p75 neurotrophin receptor mitigates brain damage following ischemic stroke in mice
- Conformational dynamics as an intrinsic determinant of prion protein misfolding and neurotoxicity
- Exploring the synergy of the eyebrain connection: neuromodulation approaches for neurodegenerative disorders through transcorneal electrical stimulation
- Pathogenic contribution of cholesteryl ester accumulation in the brain to neurodegenerative disorders
- Cognition and movement in neurodegenerative disorders:a dynamic duo
- Probing the endoplasmic reticulummitochondria interaction in Alzheimer’s disease: searching far and wide
