Conformational dynamics as an intrinsic determinant of prion protein misfolding and neurotoxicity
2024-01-24AlessandroCembranPedroFernandezFunez
Alessandro Cembran ,Pedro Fernandez-Funez
The prion protein (PrP) is the key molecular and pathological mediator of prion diseases,a heterogeneous group of brain disorders with fatal outcomes.Prion diseases are rare but deserve special attention because of their unique familial,sporadic,and transmissible etiologies,all caused by a single agent: misfolded conformations of PrP.The novel transmission of prion diseases captured the imagination of generations of scientists set on uncovering the molecular mechanisms underlying protein misfolding: seeded polymerization.This novel mechanism now appears to underlie not only prion diseases but also a group of highly prevalent brain disorders that include Alzheimer’s and Parkinson’s diseases among others,making the study of PrP misfolding highly significant.PrP is a small,secreted protein attached to the extracellular aspect of the membrane.It contains an unstructured N-terminus and a small globular domain with three α-helices and a short β-sheet.Misfolded and aggregated PrP into macromolecular assemblies is assumed to underlie both neuropathology and transmissibility of prion diseases.Recent models propose that different PrP species,including soluble,insoluble,and proteaseresistant assemblies,are responsible for different aspects of prion disease pathology.Regardless of the identity of the PrP species responsible for pathology,the key pathological event is the misfolding of this abundant protein.The overall structural differences between physiological PrPC(cellular) and pathogenic PrPres(protease resistant)are well documented,including an increase in β-sheet content (from 3% to >40%) that promotes self-assembly (Pan et al.,1993;Perez et al.,2010;Christen et al.,2013).Despite the extensive resources generated to examine PrP structure,we still have a limited knowledge about the molecular mechanism regulating PrP conformational dynamics and the initiation of misfolding.
In humans,the most prevalent form of prion diseases has sporadic etiology,meaning that no mutation or infectious seeds are required to initiate PrP misfolding.Unfortunately,the details about how the intrinsic dynamics of human PrP modulates misfolding and disease are currently poorly understood.The PrP structure is highly conserved among mammals,hinting at a critical physiological function.Despite this conservation,few animals suffer endemic prion diseases,including scrapie in sheep and goats,and chronic wasting disease in deer,moose,and elk.Yet,rodents,cattle,felines,and mustelids demonstrate susceptivity to prion transmission in the laboratory or in captivity (farms,zoos).On the other end of the spectrum,some animals show high resistance to prion diseases,including rabbits,canids (dogs and wolves),horses,and pigs.This wide spectrum in susceptibility provides an opportunity to unravel the intrinsic (i.e.,sequence-dependent) determinants of PrP toxicity by identifying differences in the sequence that impact PrP misfolding.In essence,replacing the amino acids with non-conservative substitutions across species with different susceptibility to prion diseases should have an impact on PrP toxicity.Unfortunately,decades of experimentation have revealed complex rules regulating the structure and dynamics of this small protein.For instance,human and dog PrP carry a significant difference in position 159 (human numbering): N (Asn)in human and most animals,D (Asp)/E (Glu) in dogs and other canids.Intriguingly,159N/D only shows local effects by increasing the negative charge on the surface (Lysek et al.,2005).We know that D/E159 is protective because it increases susceptibility to prion transmission in mice expressing dog PrP-D159N (Vidal et al.,2020).Similarly,Drosophila-expressing dog PrP-D159N is toxic in brain neurons (Sanchez-Garcia and Fernandez-Funez,2018).Yet,the reverse substitution (N159D) is not protective in human PrP (Myers et al.,2022).These seemingly contradictory results expose a significant gap in our current understanding of the rules governing the intrinsic mechanisms governing the conformational dynamics of the globular domain regulating the access to soluble and misfolded conformations.This example highlights the importance of better understanding how sequence differences between human and dog PrP combine to create local environments that either stabilize or destabilize N/D/E159.The challenge is to identify how multiple amino acid combinations cooperate to encode the dynamics of entire subdomains that either prevent or promote PrP misfolding.High sequence and conformational variability identify the 3D domain created by the β2-α2 loop and the C-terminal part of helix 3,which we termed the C-terminal 3-dimensional domain (CT3DD),as a subdomain of high interest(Cembran and Fernandez-Funez,2023;Myers et al.,2023).We are concentrating our research on deciphering how the interplay within the CT3DD impacts its dynamics.
With a plethora of mammalian PrP sequences and structures at our disposal,the pressing challenge lies in deciphering the sequenceencoded dynamics governing the subdomains,which in turn modulate PrP’s conformational states.Addressing this knowledge gap,we recently employed temperature replica exchange molecular dynamics (MD) simulations to scrutinize the behavior of the globular domain of human PrP.It is well known that the loops connecting the helices are regions of high conformational entropy (variability) (Cembran and Fernandez-Funez,2023).To examine in more detail the conformational dynamics of the β2-α2 loop,we analyzed the dihedral angles of amino acids 165–175,reduced the dimensionality by principal component analysis,and plotted 3D-Gibbs energy profiles of the trajectories.We found that the β2-α2 loop in human-wild type (WT) explored a large conformational space indicative of a highly dynamic domain.The energy minima from this landscape identified 5 different β-turns for 91% of populations and a 310-turn in 9% of the populations(Myers et al.,2023).This innovative visualization of the conformational landscape provides a reference to analyze the impact of amino acid substitutions on human PrP or as a comparison against animal PrPs.To perturb the dynamics of the CT3DD in human PrP,we introduced a substitution observed in helix 3 of rabbit PrP,an animal highly resistant to prion disease.We selected a unique substitution in rabbit PrP (Y (Tyr)→A (Ala) at 225)as a potential contributor to the stability of the CT3DD.We introduced Y225A in human PrP and performed MD simulations followed by analysis of the trajectories as described above.Remarkably,human-Y225A occupies a significantly smaller conformational space than WT with different loop dynamics: 82% of the populations is in the 310-turn conformation,while the rest occupies the β-turns(Myers et al.,2023).The shift in Y225A dynamics also resulted in a lower solvent exposure and mobility than in human-WT.
These observations offer a new understanding of the heightened dynamic state of human-WT:the high conformational diversity can explain its ability to misfold spontaneously.Introducing substitutions that increase the interactions (bonds)within the CT3DD contribute to stabilizing it,with a preference for the 310-turn.To determine the functional relevance of these observations,we created transgenic flies carrying human PrPWT or Y225A and expressed the constructs at comparable levels.Flies expressing PrPY225A showed lower toxicity in the eyes and in the mushroom body neurons,a higher order brain center in insects,than those expressing PrP-WT.Moreover,flies expressing Y225A accumulated lower levels of insoluble (pathogenic)conformations than flies expressing WT (Myers et al.,2023).Overall,expressing WT and Y225A in flies allowed to efficiently exame the predictions of the simulations in relevant toxicity and aggregation assaysin vivo.This opens the door for further analyses combining MD sims predictions within vivovalidation in flies of other relevant residues in the CT3DD.
Our recent work implies that the misfolding and toxicity of human PrP is encoded,at least in part,in the high conformational dynamics of the CT3DD.In turn,the dynamics of the CT3DD is governed by different amino acids that provide alternative bond configurations that stabilize multiple conformations (310-turn and β-turns) with low energy barriers that facilitate their exchange.This is a testable hypothesis that requires the manipulation of those residues suspected to play relevant roles in the dynamics of the CT3DD followed by analyses of dynamics,bonds,solvent exposure,toxicity,and aggregation.Fortunately,we have developed the assays to carry out these experiments: MD simulations to examine the impact of substitutions on dynamics and stability and transgenic fruit flies to determine thein vivoeffect on aggregation and toxicity.We have started to examine the impact of several candidate residues starting with those unique in rabbit,dog,and horse PrP.Although “humanizing” horse and dog PrP (D159N and S (Ser)167D,respectively)conferred toxicityin vivo,the corresponding changes in human PrP (N159D and D167S) had little effects on toxicity (Sanchez-Garcia and Fernandez-Funez,2018;Myers et al.,2022).The asymmetric effect of these substitutions highlights the relevance of local context,which in this case is the high conformational dynamics of human PrP.Thus,there are still lots of work to do to fully understand the rules governing the conformational dynamics of the CT3DD in human PrP.
For additional context,we conducted a similar analysis of dihedral angles for the entire globular domain for all the PrP structures available in the Protein Data Bank (PDB),including human,animal,WT,and mutants.Each PDB from NMR contains 20 representative structures,providing a readout for the dynamic state of PrP in solution.We recently showed that the β2-α2 loop is the region with the highest conformational entropy of the globular domain (Cembran and Fernandez-Funez,2023).Visualizing the dynamics of the β2-α2 loop for all PrP PDBs as a Gibbs energy plot revealed that the conformational diversity fits in the space occupied by human PrP-WT from MD simulations (Cembran and Fernandez-Funez,2023).Thus,human-WT appears to explore a broad set of conformations present in a wide range of PrPs,including PrP from animals susceptible and resistant to prion diseases,as well as PrP carrying pathogenic and artificial mutations.This overlap further supports the extreme conformational diversity of human-WT.A caveat to these observations is that the conformational space for human PrP was calculated from large sampling in MD simulations and all others from only 20 PDBS from NMR studies.It is fair to assume that the diversity of human PrP may be overrepresented from having more data points.If we believe that the high conformational dynamics of human PrP is real,it likely explains the limited success of introducing single amino acid substitutions using candidate protective residues.This provides a good argument for introducing double and triple substitutions that may work cooperatively to stabilize the CT3DD by creating more stable bonds that increase the energy barriers between conformations.The alignment of the CT3DD for humans and animals resistant to prion diseases identifies several good candidates that can be tested in pairs or triplets.We have already started this process with robust initial results in both MD simulations and fruit flies.Step by step,we are starting to gain a better understanding how PrP sequence encodes misfolding and toxicity thanks to a big contribution from describing the dynamics of the CT3DD.
In conclusion,we have only begun to unravel the rules governing PrP conformation and dynamics.As we discussed above,there are multiple combinations of substitutions that can stabilize the CT3DD in human PrP.We have a good idea about which ones are the best candidates based on sequence and structure comparisons with animals resistant to prion transmission.A systematic approach can allow us to understand the role each substitution plays in CT3DD dynamics.The goal is to identify the combination of substitutions that completely stabilizes the CT3DD in a single conformation that eliminates toxicity and prion conversion.This approach will ultimately allow us to unravel the intrinsic determinants of PrP toxicity by integrating knowledge along this axis:Sequence→Conformation→Dynamics→(dys)Function (Figure 1).At this time,domain conformation (secondary and tertiary structures)can be directly predicted from protein sequences.In most cases,protein structure and function are easy to correlate,and the impact of substitutions or mutations can also be predicted on the structural or catalytic domains.In contrast,domain or subdomain dynamics are harder to predict and need to be examined in detail through biophysical or computational studies.We pose here that conformational dynamics is the key to understanding PrP misfolding and likely to also play a role in developing therapeutics that stabilize PrP and prevent misfolding.This principle may apply to a class of amyloid proteins prone to spontaneous misfolding known to trigger several prevalent disorders,expanding the relevance of these studies.
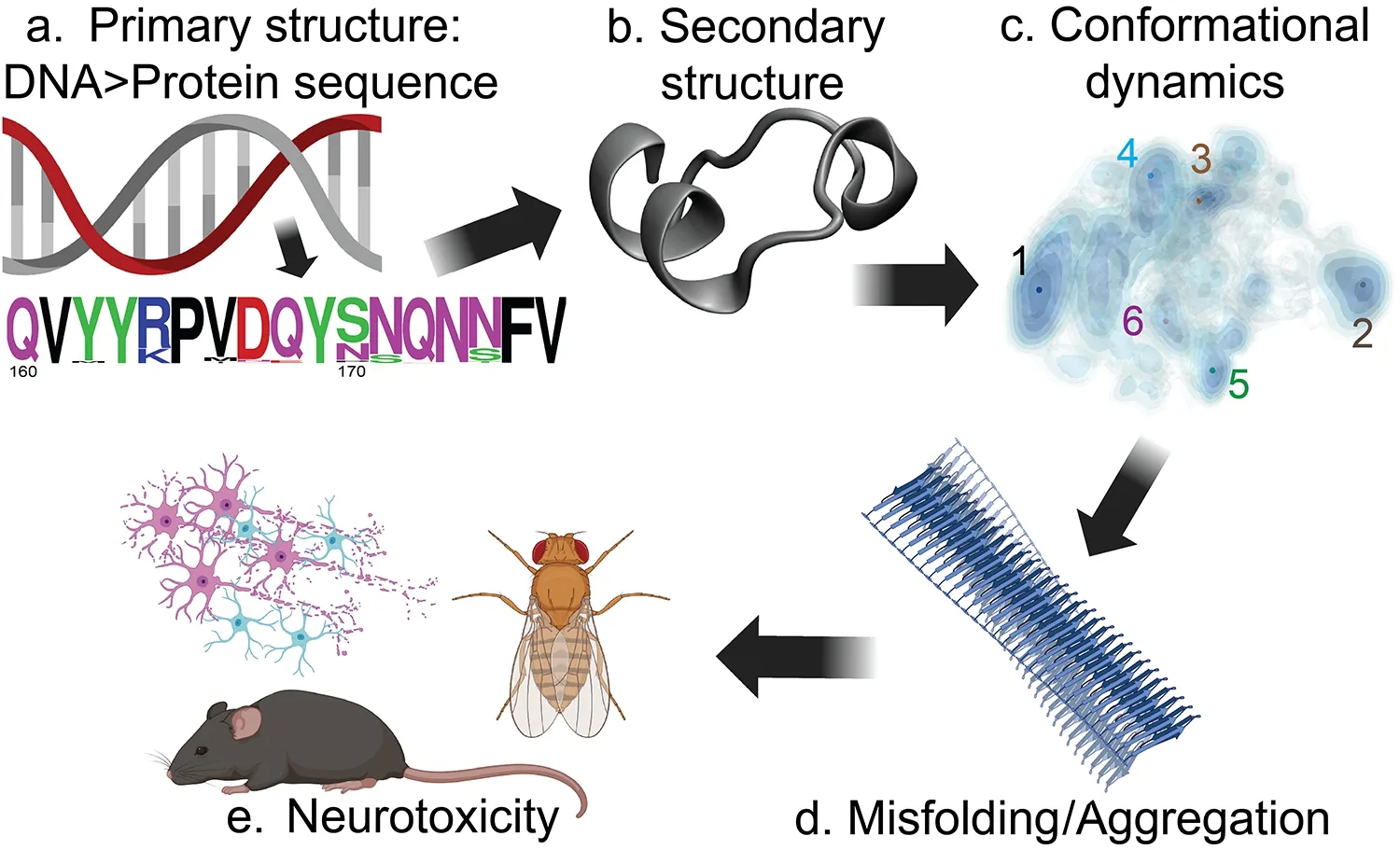
Figure 1 |Understanding structure/function correlation for PrP.
We thank the University of Minnesota Information Technology Support Services for institutional copies of PyMOL and the Minnesota Supercomputing Institute(MSI)at the University of Minnesota.
This work was supported by the NIH grant 7R21NS096627-02 to PFF and the Winston and Maxine Wallin Neuroscience Discovery Fund award CON000000083928 to PFF and AC.
Alessandro Cembran#,Pedro Fernandez-Funez*, #
Department of Chemistry and Biochemistry,University of Minnesota Duluth,Duluth,MN,USA(Cembran A)
Department of Biomedical Sciences,University of Minnesota Medical School,Duluth Campus,Duluth,MN,USA (Fernandez-Funez P)
*Correspondence to:Pedro Fernandez-Funez,PhD,pfernand@d.umn.edu.
https://orcid.org/0000-0002-0103-5593(Pedro Fernandez-Funez)
#Both authors contributed equally to this work.
Date of submission:September 8,2023
Date of decision:November 2,2023
Date of acceptance:November 24,2023
Date of web publication:December 21,2023
https://doi.org/10.4103/1673-5374.391332
How to cite this article:Cembran A,
Fernandez-Funez P(2024)Conformational dynamics as an intrinsic determinant of prion protein misfolding and neurotoxicity.Neural Regen Res 19(10):2095-2096.
Open access statement:This is an open access journal,and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix,tweak,and build upon the work non-commercially,as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Eva Žerovnik,Jožef Stefan Institute,Slovenia.
Additional file:Open peer review report 1.
杂志排行
中国神经再生研究(英文版)的其它文章
- Modulation of p75 neurotrophin receptor mitigates brain damage following ischemic stroke in mice
- Exploring the synergy of the eyebrain connection: neuromodulation approaches for neurodegenerative disorders through transcorneal electrical stimulation
- Pathogenic contribution of cholesteryl ester accumulation in the brain to neurodegenerative disorders
- Cognition and movement in neurodegenerative disorders:a dynamic duo
- Probing the endoplasmic reticulummitochondria interaction in Alzheimer’s disease: searching far and wide
- Music as medicine for traumatic brain injury: a perspective on future research directions
