Analysis of E2F3 gene variants, expression and clinical significance in melanoma based on multiple databases
2024-01-19HANLiminYEGangDENGChuanDAIJiao4ZHAOHailong
HAN Li-min, YE Gang, DENG Chuan, DAI Jiao4, ZHAO Hai-long✉
1. Department of Endocrine, People’s Hospital of Changshou District, Chongqing 401220, China
2. Department of Hepatobiliary Surgery, People’s Hospital of Changshou District, Chongqing 401220, China
3. Department of Neurology, People’s Hospital of Changshou District, Chongqing 401220, China
4. Department of Pharmacy, People’s Hospital of Changshou District, Chongqing 401220, China
5. Department of Pathophysiology, School of Basic Medical Sciences, Zunyi Medical University, Zunyi 563000, China
Keywords:
ABSTRACT Objective: To investigate the variation, expression and clinical significance of E2F3 gene in melanoma.Methods: Firstly, cBioportal database, Oncomine database and GEO database were used to analyze the variation and expression level of E2F3 gene in melanoma.OSskcm database and TISIDB database were used to analyze the relationship between E2F3 and melanoma prognosis and tumor immune infiltrating cells.Then, the LinkedOmics database was used to identify the differential genes related to E2F3 expression in melanoma and analyze their biological functions.Finally, small molecule compounds for the treatment of melanoma were screened through the CMap database.Results: The mutation rate of E2F3 gene in melanoma is about 4 %, and there are 21 mutation sites.Compared with normal skin tissues,the expression of E2F3 gene in melanoma was significantly increased (P<0.01).The mutation and increased expression of E2F3 gene were associated with the shortened overall survival(OS) of melanoma patients (P<0.05).The CNA level of E2F3 was negatively correlated with the expression levels of lymphocytes such as pDC, Neutrophil, Act DC and Th17, and negatively correlated with the expression levels of chemokines such as CXCL5, CCL13 and CCR1.The methylation level of E2F3 was positively correlated with the expression levels of Th1, Neutrophil, Act DC and other lymphocytes, and positively correlated with the expression levels of CXCL16, CXCL12, CCR1 and other chemokines.The expression level of E2F3 was negatively correlated with the expression levels of lymphocytes such as Th17, Tcm CD4 and Th1, and negatively correlated with the expression levels of chemokines such as CXCL 16, CCL 22 and CCL 2.The expression of 96 genes such as UHRF1BP1 in melanoma was significantly correlated with the expression of E2F3 (|cor| 0.5, P<0.05).The above genes were mainly related to RNA transport, eukaryotic ribosome biogenesis, cell cycle and other pathways.Among them, WDR12, WDR43, RBM28, UTP18, DKC1, PAK1IP1, DDX31,TEX10, TRUB1 and TRMT61B were the top 10 hub genes.YC-1, simvastatin, cytochalasin-d,Deforolimus and cytochalasin-b may be five small molecule compounds for the treatment of melanoma.Conclusion: The mutation and increased expression level of E2F3 gene are related to the poor prognosis of melanoma and participate in the occurrence and development of melanoma by affecting the expression of different tumor immune infiltrating cell subtypes,which may be a potential diagnostic marker and therapeutic target for melanoma.
1.Introduction
Melanoma is a malignant tumor with high mortality, which is mainly produced by melanocytes in the skin or other organs.According to statistics, the number of new cases of melanoma in the world in 2020 was 324635, and the number of deaths due to melanoma reached 57043[1].At present, the treatment of melanoma mainly includes the following four kinds: traditional surgical resection, radiotherapy, chemotherapy, and targeted therapy.There is increasing evidence that adjuvant therapy with targeted drugs or immune drugs significantly improves the overall survival rate of patients with melanoma and is a new way to treat melanoma[2-3].However, the results of several clinical trials have shown that some melanoma patients do not respond effectively to the above drugs,even in people with high load of gene mutation or high expression of PD-1/PD-L1[4].Studies have shown that the 5-year survival rate of patients with melanoma in the early stage is as high as 95%, but the 5-year survival rate of patients with advanced metastatic melanoma is only 5%~10%[5,6].Therefore, it is of great significance to explore more potential markers and targets for the diagnosis and treatment of melanoma.
E2F3 is one of the transcription factors in the E2F family, which is mainly involved in the regulation of cell cycle.In recent years, a few evidence shows that the expression of E2F3 in lung cancer, gastric cancer, breast cancer, liver cancer and other tumors are significantly increased, and can promote the occurrence and development of tumors[7-10].However, at present, the variation, expression, and clinical significance of E2F3 in melanoma are not completely clear.Therefore, this paper aims to analyze the diagnostic value and clinical significance of E2F3 in melanoma through a variety of databases.
2.Materials and methods
2.1 cBioportal database
cBioportal database is a commonly used database for analyzing cancer genomics and visualization, which can analyze the variation of single gene or multi-gene in different tumors.Seven clinical sequencing data of non-metastatic melanoma (a total of 1905 samples) were selected and analyzed after entering E2F3 in the EnterGenes column.The total mutation rate and subtypes of E2F3 gene in melanoma were obtained by Oncoprint, Cancer Types Summary and Mutations menu, and the total survival time curves of E2F3 mutation group and non-mutation group were obtained by Comparison/Survive menu.
2.2 Oncomine database
Oncomine database can be mined to analyze the differential expression data of target genes in different tumor types and their corresponding normal tissues.Cancer type is set as malenoma,Gene is set as E2F3, Analysis Type is set as Cancer VS Normal Analysis, Data Type is set as mRNA, P value<0.05, Fold Change>2,Gene Rank=Top 10%.Finally, click submit to obtain the expression difference of E2F3 gene in melanoma and normal skin tissues.
2.3 GEO database
Four melanoma-related datasets, GSE3189, GSE114445,GSE100050 and GSE46517, were selected.All differentially expressed genes were obtained by online GEO2R analysis tool(P<0.05, Log |FC| 1), and further compared the E2F3 gene expression in melanoma and normal skin.melanoma and normal skin expression levels.
2.4 OSskcm database
The OSskcm database is a survival analysis database dedicated to melanoma developed by Chinese scholars.Its data includes TCGA and some GEO datasets.Gene symbol input E2F3, Data Source TCGA was selected, Split patients by Upper 50%, OS and PFI were selected for Survival, respectively.Finally, KM survival curve was obtained by clicking Kaplan-Meier to compare E2F3 high expression group and low expression group.
2.5 TISIDB database
TISIDB database is a comprehensive database based on TCGA data to analyze the interaction between tumor and immune system.It can analyze the relationship between target genes and immunotherapy,lymphocytes, immunomodulators, chemokines, immune cell subtypes in different tumors.The E2F3 gene in melanoma was obtained from the Lymphocyte, Chemokines and Subtype menus by inputting E2F3 in Gene Symbol.
2.6 LinkedOmics database
LinkedOmics database is a multi-omics online analysis database containing 32 cancer types, which can be used for single gene association analysis and functional enrichment analysis.The RNAseq in the TCGA _ SKCM dataset was selected as the data, and E2F3 was used as the target gene.The Spearman method was used for analysis, and finally the E2F3 gene in melanoma was obtained.
2.7 CMap database
CMap database is a chemical genomics database for screening small molecule compounds.By clicking the Query tool, Query parameters select Gene expression ( L1000 ), with P<0.05, |cor| 0.5 as the cut-off point, the genes that are positively correlated with the expression level of E2F3 ( 61 ) and negatively correlated ( 35 ) are input into the “ UP regulated genes ” and “ DOWN regulated genes” boxes for analysis, and finally the top five potential small molecule compounds are obtained.
3.Results
3.1 Analysis of E2F3 gene variation in melanoma
The results of cBioportal database analysis showed that E2F3 gene still had low variation in melanoma.In the 1905 melanoma population, about 4% of patients had CNA changes or mutations in the E2F3 gene, which mainly included gene amplification (40 samples) and deletion (25 samples), as shown in figure 1.Through the analysis of mutation sites, it was found that there were 21 mutation sites of E2F3 in melanoma, including E20K, G77S, P89S,R134Q, E140D and other sites.The specific location and coding protein are shown in figure 2.
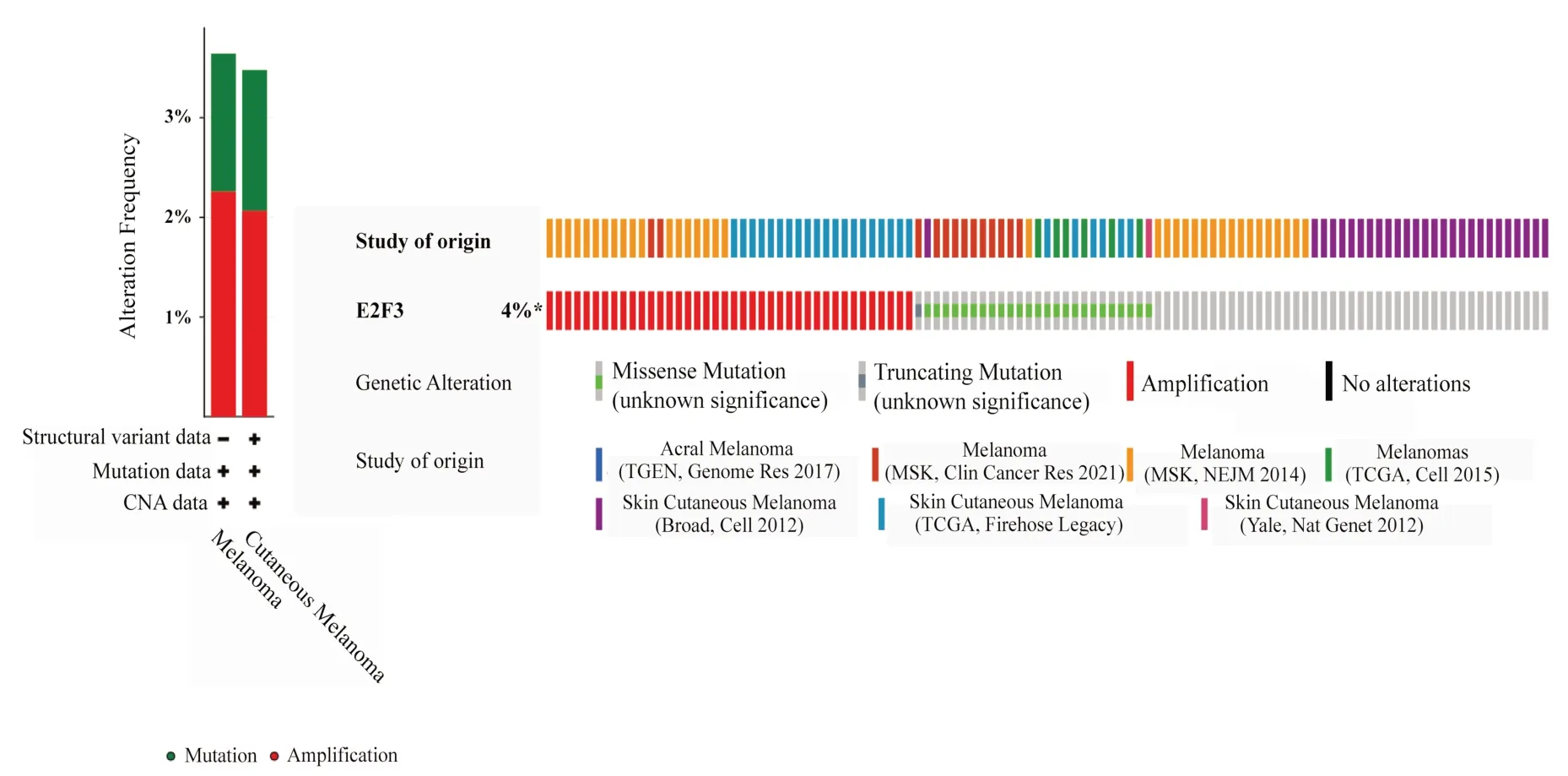
Fig 1 Variation analysis of E2F3 gene in melanoma
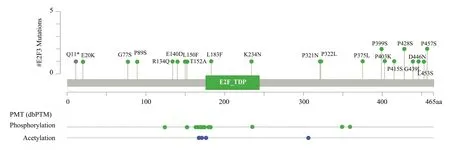
Fig 2 The gene mutation sites of E2F3 in melanoma
3.2 Expression of E2F3 gene in melanoma and normal tissues
The results of Oncomine database showed that the expression of E2F3 gene in various tumors was higher than that in normal tissues.Among them, two studies showed that the expression of E2F3 in melanoma was also significantly increased, as shown in figure 3.The results showed that the expression level of E2F3 gene in melanoma was significantly higher than that in normal skin tissue (t=5.06 or 9.61,P<0.01), and the Fold change values were 3.146 and 3.687,respectively, as shown in Figure 3.Further analysis of GSE3189,GSE114445, GSE100050 and GSE465174 melanoma-related GEO datasets showed that the expression level of E2F3 gene in melanoma was significantly higher than that in normal skin tissues (P<0.01).The specific results are shown in table 1.
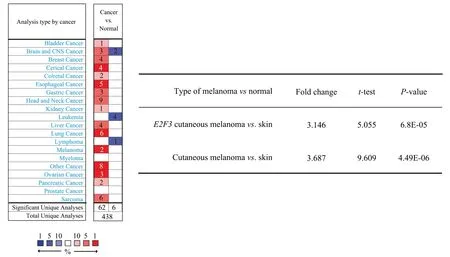
Fig 3 Expression of E2F3 gene in melanoma and normal skin tissues (based on Oncomine database)

Tab 1 Expression of E2F3 gene in melanoma and normal skin tissues (based on GEO database)
3.3 Variation and expression of E2F3 gene and prognosis of melanoma
The results of the cBioportal database showed that the overall survival (OS) of melanoma patients in the E2F3 mutation group was significantly shorter than that in the non-mutation group (P<0.01), as shown in Figure 4.The results of OSskcm database survival analysis also showed that the OS and Progression Free Interval (PFI)of patients with high expression of E2F3 gene in melanoma were significantly lower than those of low expression of E2F3 gene.group(P<0.05), as shown in figure 4.
3.4 The correlation between the variation and expression of E2F3 gene and immune infiltrating cells or chemokines in melanoma
The results of TISIDB database showed that the CNA, methylation and expression of E2F3 were significantly correlated with various immune infiltrating cells and chemokines in melanoma (Sperman method), as shown in figure 5.The CNA level ofE2F3was negatively correlated with the expression levels of lymphocytes such as pDC (rho: -0.374,P<0.01), Neutrophil (rho: -0.361,P< 0.01),Th17 (rho: -0.337, P<0.01), and CXCL5 (rho: -0.293, P < 0.01),CCL13 (rho: -0.265,P<0.01), as shown in figure 5.The methylation level of E2F3 was positively correlated with the expression levels of Th1 (rho: 0.302,P<0.01), Neutrophil (rho: 0.294,P<0.01) and Act DC (rho: 0.273, P<0.01).It was positively correlated with the expression levels of chemokines such as CXCL16 (rho: -0.248,P <0.01), CXCL12 (rho: -0.21, P<0.01) and CCR1 (rho: -0.24, P< 0.01), as shown in Figure 5.The expression level of E2F3 was negatively correlated with the expression levels of Th17 (rho: -0.375, P<0.01), Tcm CD4 (rho: -0.369, P<0.01), Th1 (rho: -0.36,P<0.01), and CXCL 16 (rho: - 0.462,P<0.01), CCL 22 (rho: -0.296,P<0.01), CCL 2 (rho: -0.288, P<0.01), and other chemokines, as shown in Figure 5.
Through further analysis of the immune subtypes and molecular subtypes of melanoma, the results showed that the expression level of E2F3 was also different in different immune subtypes and molecular subtypes (P< 0.05), as shown in figure 6.The expression of E2F3 was the highest in the wound healing group and RAS_Hotspot mutation group, respectively, while the expression was the lowest in the TGF-b dominant and BRAF_Hotspot mutation groups,as shown in Figure 6.
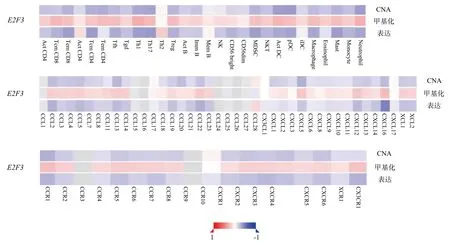
Fig 5 Correlation heat map of E2F3 gene mutation and expression with immune infiltrating cells and chemokines in melanoma
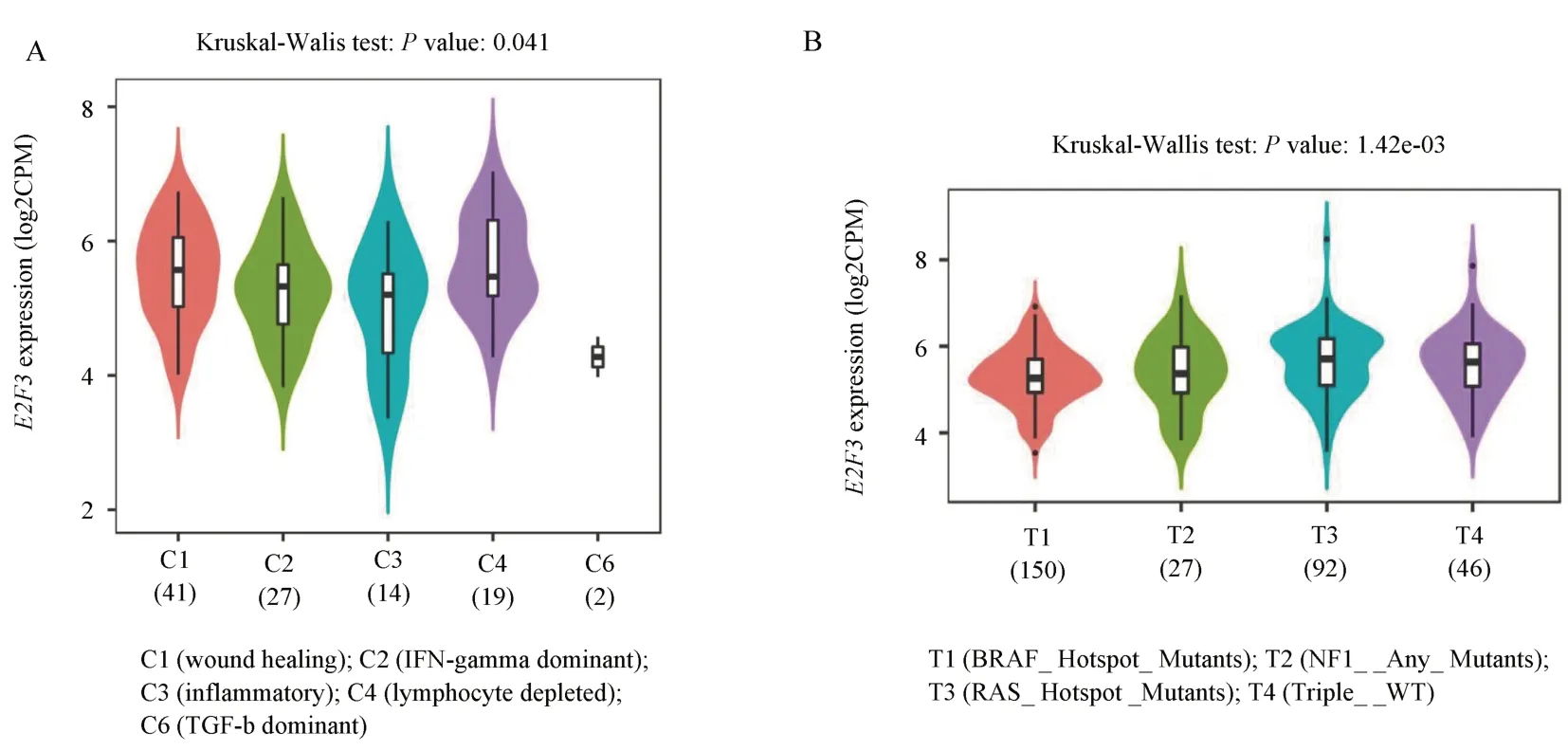
Fig 6 Expression of E2F3 gene in different subtypes of melanoma
3.5 Analysis of E2F3 related genes and their biological functions
The results of LinkedOmics database showed that there were 5369 differentially expressed genes related to E2F3 expression in melanoma under the condition of FDR < 0.05, of which 2856 genes were positively correlated with E2F3, and 2513 genes were negatively correlated withE2F3, indicating that although the expression directions of related genes were inconsistent, these genes andE2F3jointly promoted the occurrence and development of melanoma.The mechanism may be related to changes in biological processes such as apoptosis, cell proliferation and cell cycle.BLM,GLO1,HCG18and other genes were significantly positively correlated with E2F3 gene expression, while IFITM3, IGFBP6,KCTD11and other genes were significantly negatively correlated with E2F3 gene expression, as shown in figure 7.
The biological functions of GO and KEGG were analyzed by GSEA method.The results showed that BP was mainly concentrated in the biological processes such as Ribonucleoprotein complex biogenesis, DNA recombination, and RNA 3‘-end processing.CC is mainly concentrated in chromosomal region, tertiary granule,PcG protein complex and other cellular components.MF is mainly concentrated in helicase activity, DNA secondary structure binding,histone binding and other molecular functions, as shown in table 2.and figure 8.
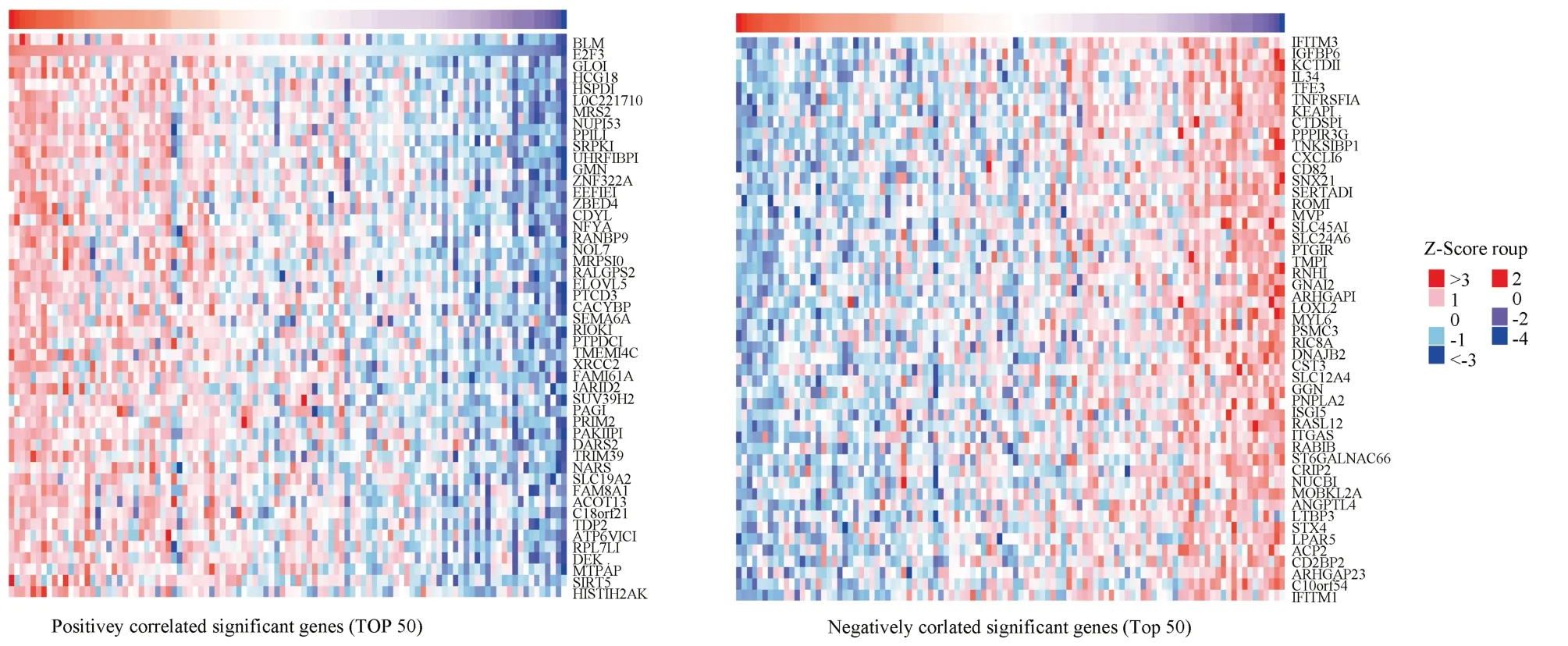
Fig 7 Heat map of the first 50 genes related to E2F3 gene expression in melanoma
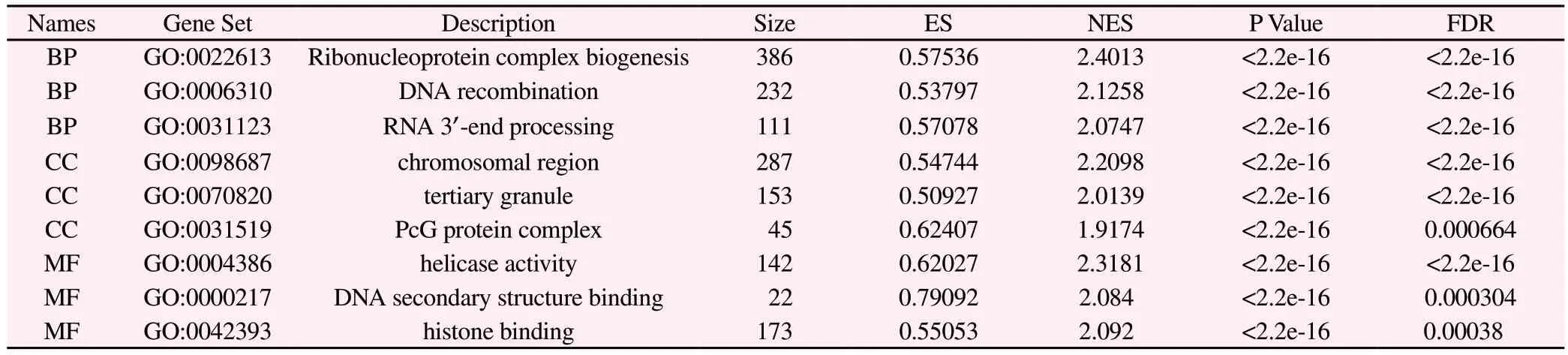
Tab 2 GO biological analysis results of E2F3 related genes
KEGG results showed that it was mainly enriched in RNA transport, ribosome biogenesis of eukaryotes, cell cycle and other pathways, as shown in figure 9.
3.6 Screening of hub genes based on E2F3 related genes
With P<0.05, | cor | 0.45 as the condition, 770 genes related to E2F3 were screened out, and STRING ( https://cn.stringdb.org/) )was used for online PPI network mapping, as shown in figure 10.The results were imported into Cytoscpae 3.8.0, and further analysis was performed using the MCODE and cyto-Hubba modules.A total of 9 modules were obtained from the MCODE analysis results, and the modules with the highest score were as follows, score = 8.667,as shown in figure 11.A total of 10 hub genes were obtained by Hubba (MCC calculation method), which wereWDR12,WDR43,RBM28, UTP18, DKC1, PAK1IP1, DDX31, EX10, TRUB1, TRMT61B, respectively, as shown in figure 11.
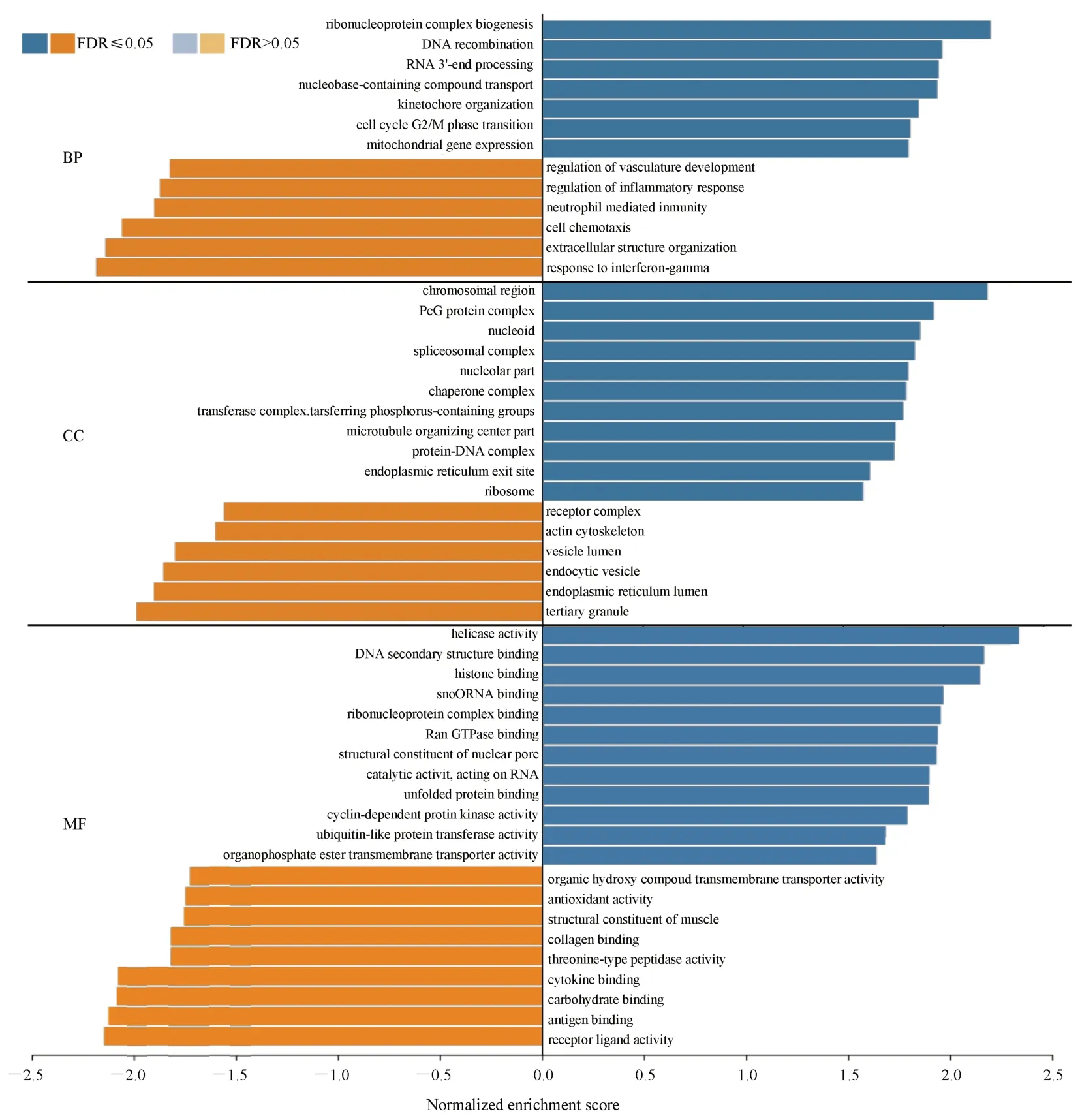
Fig 8 GO analysis results of E2F3 related differential genes
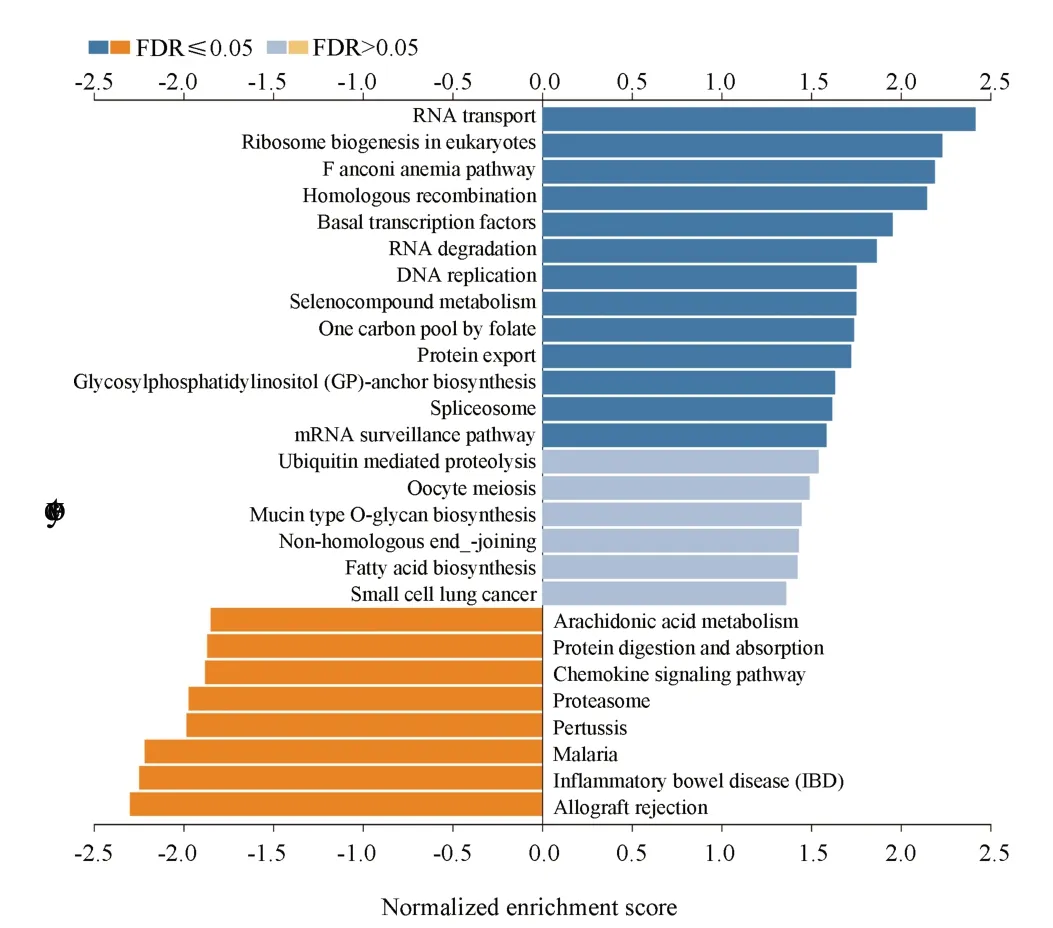
Fig 9 KEGG analysis results of E2F3 related differential
The GEPIA online database (http://gepia.cancerpku.cn/detail.php) was used to identify the table of the above genes in melanoma.The results showed that WDR12 and PAK1IP1 were expressed in melanoma.The level was significantly higher than that of normal tissue (P<0.01), as shown in figure 12.
PAK1IP1 was associated with the stage of melanoma (F= 2.58,P<0.05), as shown in Figure 13.The above results indicate that WDR12 and PAK1IP1 have potential diagnostic value in melanoma.
Fig 10 PPT network enrichment map of E2F3 related genes
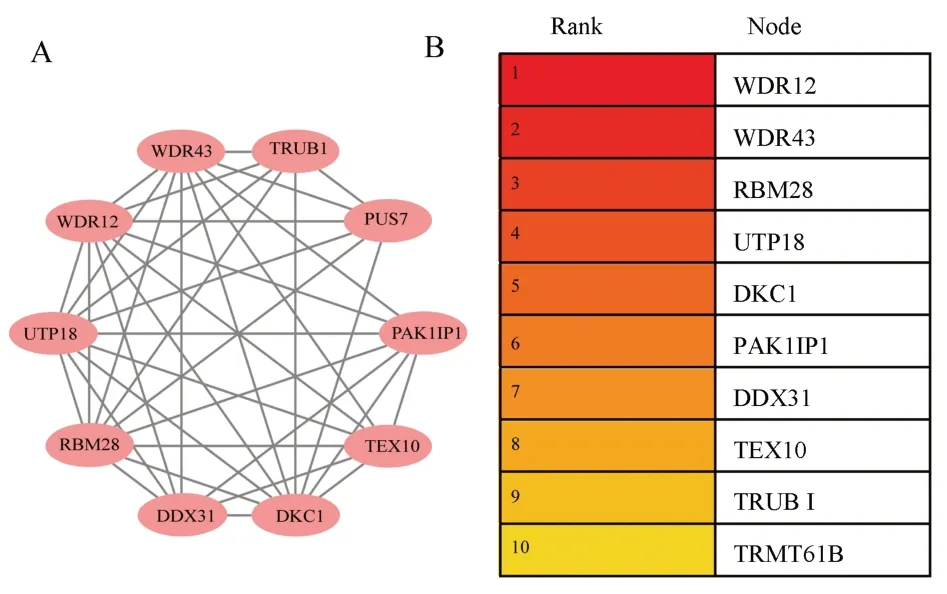
Fig 11 MCODE (Top1) and hub genes (Top10) analysis results of E2F3 related genes
3.7 Screening of small molecular compounds for treatment of melanoma based on E2F3-related genes
Through the CMap database, a total of 2837 related small molecule compounds were screened out.The top five with the lowest scores were YC-1 (Score =-99.37), simvastatin (Score =-99.19), cytochalasin d (Score =-99.01), Deforolimus (Score =-98.94) and cytochalasin b (Score =-98.34), as shown in Figure 14.
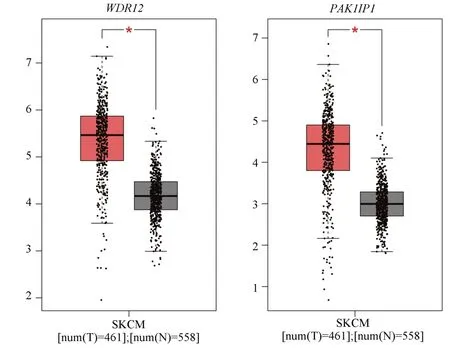
Fig 12 Expression levels of WDR12 and PAK1IP1 in melanoma and normal tissues
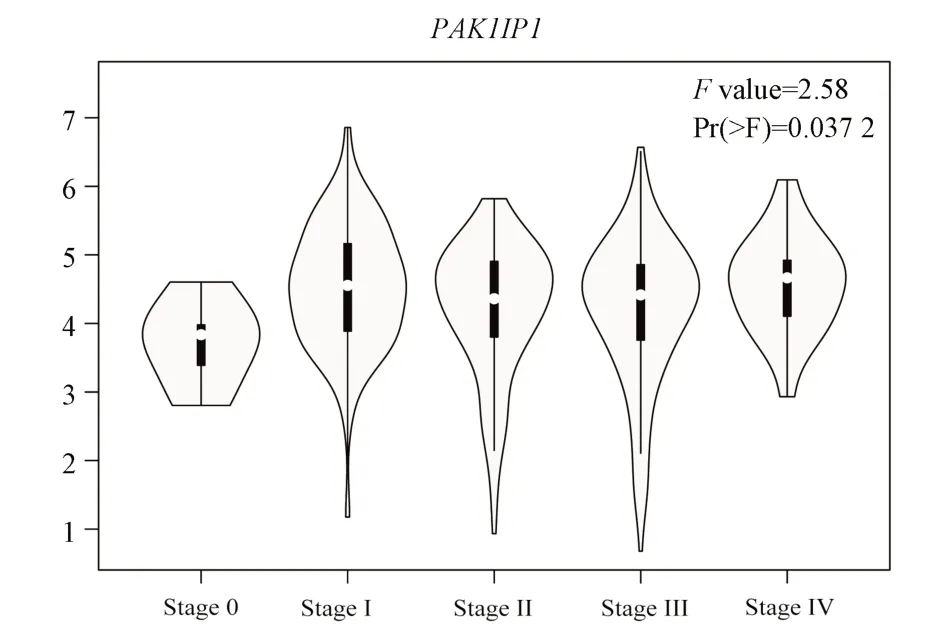
Fig 13 PAK1IP1 and clinical stage of melanoma
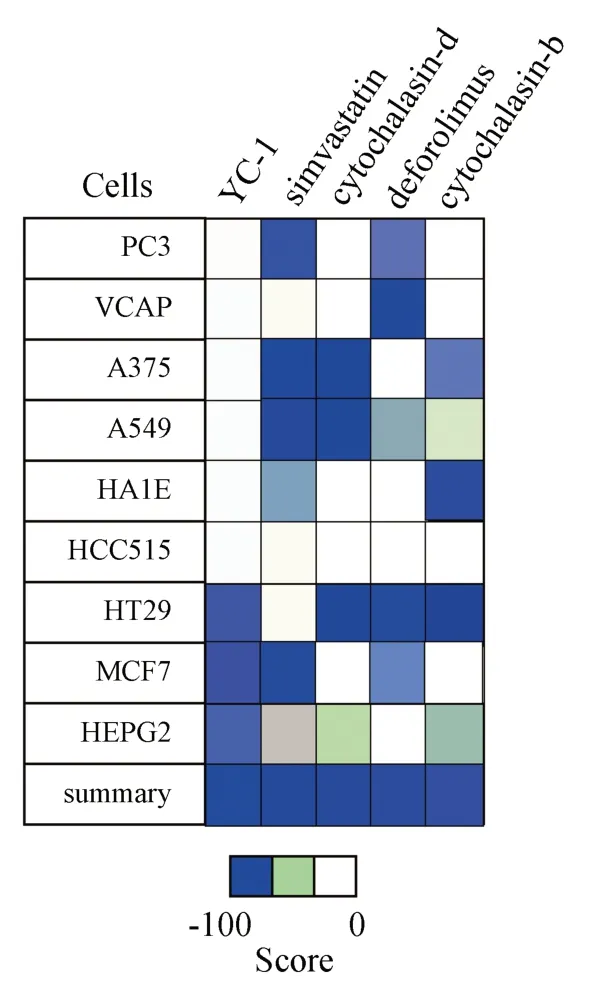
Fig 14 Fractional heat maps of the small molecule compounds for melanoma treatment (Top 5)
4.Discussion
E2F3is a key transcription factor in the E2F family, which has been confirmed to be highly expressed in many tumors and is closely related to the prognosis of tumors.E2F3 is mainly involved in cell cycle progression, DNA replication, DNA repair and apoptosis.It can interact with histone acetyltransferase and promote cell cycle from G1 phase to S phase.Specifically, E2F3 is divided into two isomers:E2F3aandE2F3b, both of which contribute to the activation of E2F3 target gene transcription.However, E2F3a belongs to the activator of gene transcription, while E2F3b belongs to the typical repressor of gene transcription.Therefore, in different studies,E2F3may play different roles in regulating biological functions such as cell proliferation.At present, the variation, expression, and clinical significance ofE2F3in melanoma are not completely clear.
Through a variety of database analysis, this study found thatE2F3gene had low variation in melanoma, and its expression level was significantly higher than that in normal skin group (P<0.01).Specifically, the mutation rate ofE2F3gene in melanoma is about 4 %, mainly CNA changes or mutations, and there are 21 mutation sites.The expression level of E2F3 gene in melanoma is about 2 ~3 times higher than that in normal skin tissue, which indicates that the expression level ofE2F3can assist in the early diagnosis of melanoma.In the prognostic analysis of E2F3 and melanoma, it was found that the OS of melanoma patients in theE2F3mutation group was significantly lower than that in the non-E2F3mutation group(P<0.01).The increase of E2F3 expression level was related to the shortening of OS and PFI in melanoma patients (P<0.05).The above results are mentioned.
Since immune-infiltrating cells in the tumor microenvironment are also another key driver of tumorigenesis and progression, the present study further analyzed the correlation between the variation and expression of E2F3 and immune-infiltrating cells or chemokines in melanoma.The results showed that CNA, methylation and expression of E2F3 were associated with a variety of immune infiltrating cells and chemokines in melanoma (P<0.05), and the CNA and expression levels ofE2F3were negatively correlated with the expression levels of neutrophils, DCs, T-cell subpopulations and a variety of chemokines, which suggests that the elevated levels ofE2F3mutation and expression can affect the subpopulation of immune infiltrating cells in the tumor microenvironment, and thus promote tumorigenesis, and thus promote tumor development.This suggests thatE2F3mutations and elevated expression levels can affect the subpopulations of immune infiltrating cells in the tumor microenvironment, which in turn promotes tumorigenesis, but the specific mechanism is not clear.
Based on the above, the present study also explored the potential mechanism by whichE2F2promotes the occurrence and development of melanoma.Through the analysis of relevant databases, we found that there were 7702 differential genes associated withE2F3expression in melanoma (P<0.05), of which 3956 genes were positively associated with E2F3 and 3746 genes were negatively associated with E2F3.Through KEGG enrichment analysis, it was found that the above genes were mainly enriched in RNA transporter, Ribosome biogenesis in eukaryotes, and E2F3 in eukaryotes.This suggests that E2F3 may be involved in melanoma development through the above pathways.A total of 10 hub genes were obtained by MCODE and cyto-Hubba analyses of the differential genes, among which the expression levels of WDR12 andPAK1IP1were significantly higher in melanoma than in normal tissues (P<0.01), and PAK1IP1 was associated with the stage of melanoma (P<0.05).The above results imply thatWDR12andPAK1IP1have potential value in melanoma.Through GeneCards website, we found that both WDR12 and PAK1IP1 are ribosomal proteins, which are mainly expressed in the nucleus and involved in cell cycle, signaling and other cellular processes.In previous studies, the expression of WDR12 has been shown to be significantly elevated in glioblastoma and hepatocellular carcinoma, and negatively correlated with patient prognosis.In vitro experiments have shown that knockdown of WDR12 or interference with WDR12 expression can inhibit tumor cell proliferation and invasion, and the mechanism is related to the inhibition of cell cycle and Akt-mTORS6K1 signaling pathway[11, 12].PAK1IP1 has been shown to inhibit tumor cell proliferation in HEK293T, A549, H1299 and other tumor cell lines, and overexpression of PAK1IP1 induces the p53 pathway to block the cell cycle and inhibit cell proliferation[13].However,there is no relevant study on the relationship between PAK1IP1 and melanoma, and the relationship between PAK1IP1 and melanoma is still unclear.
Finally, this study explored the potential drugs for the treatment of melanoma through CMap database, and found that YC-1,simvastatin, cytochalasin D, Deforolimus and cytochalasin B may be the first five small molecular compounds in the treatment of melanoma.Among them, simvastatin, cytochalasin D and cytochalasin B have been verified in A375 melanoma cell line (figure 12), while YC-1 and Deforolimus have not been associated with melanoma.YC-1 is an activator of guanylate cyclase and an inhibitor of hypoxia-inducible factor.Its target is mainly HIF1A, GUCY1A2 and other proteins, which can effectively inhibit the expression of HIF1α[14].In vitro studies have found that YC-1 can inhibit the proliferation of hepatocellular carcinoma and breast cancer cells[15-17].Deforolimus (MK 8669) is a selective mTOR inhibitor.Several clinical trials have shown that it has good anticancer effects on sarcomas, malignant hematological diseases, and other solid tumors[18,19].These results suggest that YC-1, simvastatin, cytochalasin D,deforolimus and cytochalasin B have potential application prospects in the treatment of melanoma.
To sum up, this study analyzed the role and value ofE2F3in melanoma from multiple dimensions through a variety of databases,and finally found that the mutation and expression of E2F3 gene were related to the poor prognosis of melanoma and participated in the occurrence and development of melanoma by affecting different tumor immune infiltrating cell subtypes, which may be a potential diagnostic marker and therapeutic target for melanoma.
Authors’ Contribution
Li-Min Han: responsible for the design and writing of the manuscript; Gang Ye, Chuan Deng, and Jiao Dai: responsible for the article-related database analysis; Hailong Zhao: responsible for manuscript revision and proofreading.All authors declare no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- The prevention of high-fat diet-induced non-alcoholic fatty liver disease in mice by Fucoidan
- Mechanism of Qishen Decoction inhibition of macrophage M1 type polarization by targeting TGR5-mediated NLRP3 inflammasome
- Epidemiological characteristics of hyperuricemia in metabolic syndrome and its different components in the physical examination population
- Meta analysis and data mining of the method of yishenhuoxue in the treatment of nonproliferative diabetic retinopathy
- Evaluation of the diagnostic efficacy of noninvasive diagnosis in patients with chronic viral hepatitis B complicated with nonalcoholic fatty liver disease and significant liver fibrosis
- Discover the key genes for glomerular inflammation in patients with type II diabetic nephropathy based on bioinformatics and network pharmacology
