基于转录组测序筛选与柚裂果相关的基因
2023-11-18卢艳清林燕金王贤达卢新坤
卢艳清,林燕金,王贤达,卢新坤
基于转录组测序筛选与柚裂果相关的基因
卢艳清,林燕金,王贤达,卢新坤
福建省农业科学院果树研究所,福州 350013
【目的】柑橘类果实生长季节内出现的裂果现象是一类生理失调病害,然而目前尚未完全揭示柑橘类果实裂果的分子机理。本研究通过对文旦柚抗裂和易裂品种果皮转录组的比较分析,筛选果实抗裂相关基因。【方法】以抗裂品种(‘度新1号文旦柚’)的正常果(此品种无裂果),以及易裂品种(‘度尾文旦柚’)的正常果和裂果为材料,果实均取自两个时期(时期A:2021年8月3日;时期B:2021年8月20日,裂果敏感期)。取果实果顶部位的果皮(以果顶为中心,30 mm半径范围内的果皮)用于转录组测序。【结果】将易裂品种裂果果皮与两品种正常果果皮转录组进行比较分析,在时期A共筛选到1 660个差异基因,易裂品种裂果果皮与两品种正常果果皮间相同的差异基因104个;在时期B共筛选到1 972个差异基因,易裂品种裂果果皮与两品种正常果果皮间相同的差异基因82个。对易裂品种裂果果皮与两品种正常果果皮间所有差异基因的GO富集分析结果显示:在生物学过程分类中,两个时期均富集到差异基因的主要亚类包括代谢过程、细胞过程、单生物过程、生物调控、刺激响应和信号。对易裂品种裂果果皮与两品种正常果果皮间所有差异基因的代谢通路分析结果显示:两个时期均富集到差异基因的主要代谢途径包括碳代谢、植物MAPK信号途径、植物激素信号转导和内质网蛋白过程,这些代谢途径富集的差异基因也最多。发现了一些与果实抗裂能力相关的重要基因:两品种正常果果皮扩张蛋白-A1基因表达量均显著高于易裂品种裂果果皮,两个时期结果一致;在时期A,两品种正常果果皮类钙调磷酸酶蛋白B亚基基因表达量显著高于易裂品种裂果果皮,但在时期B无显著差异;易裂品种裂果果皮热激转录因子、丝氨酸/苏氨酸蛋白激酶、生长素响应蛋白和脱水响应元件结合蛋白基因表达量显著高于两品种正常果果皮,两个时期结果一致。【结论】与果皮弹性、水分运动、高温和水分亏缺逆境响应相关的基因是调控文旦柚果实抗裂能力的关键基因。
柚(L. Osbeck);裂果;果皮;转录组;抗裂基因
0 引言
【研究意义】裂果是很多类果实(例如:樱桃、番茄、荔枝、柑橘、枣、苹果)一个普遍的生理失调病害。开裂果实易腐烂,商品价值明显降低。研究表明,裂果不仅与植物品种相关[1],而且也与自然环境(例如:高温、高湿)相关[2]。因此,综合分析果实的生长特性及其对自然环境的响应机制,有助于寻找到有效的防治裂果的方法。【前人研究进展】在果实生长季节内,果实快速生长期是多类果实的裂果敏感时期。例如,横径快速生长期内的荔枝果实易裂果[3];柑橘果实快速生长期伴随着果实开裂[4-5]。这些研究促使人们去寻找果实生长季节内果肉与果皮间生长应力存在差异的原因,所以大量的研究集中在果肉的生长势和果皮的机械力度,以寻找抑制裂果的方法。通过对抗裂品种和易裂品种果实[6-7],以及裂果和正常果果实[8]的比较,发现果实可溶性固形物和糖含量与裂果敏感度呈正相关。糖与果实吸水能力相关[9],所以果肉部位的渗透吸水是果实水分吸收的重要途径之一。另外,气孔和果面微小裂隙也是果实吸水的重要途径。樱桃(L.)果实气孔密度与果实水分导度呈正相关[10]。枣果实气孔大小与裂果率呈正相关[11]。研究发现枣果面微小裂隙的形成源自气孔[12]。离体樱桃外果皮的渗透吸水量与角质层微小裂隙数量呈正相关[13]。果皮机械力度高,果实抗裂能力高[14]。果实细胞壁组分影响果皮机械力度。与易裂荔枝品种‘糯米糍’相比,抗裂的荔枝品种‘淮枝’果皮纤维素、半纤维素、不溶性果胶含量高,可能有助于果皮机械力度和果实抗裂能力的提高[15]。易裂品种‘壶瓶枣’果实纤维素含量和裂果率的偏相关系数为-0.971[16]。番茄抗裂资源果实半纤维素含量高于番茄易裂资源果实[17]。水溶性果胶含量低或者原果胶含量高与果实抗裂能力呈正相关[17-19]。扩张蛋白是一类细胞壁蛋白[20]。扩张蛋白通过调控细胞壁聚合物成分的变化影响细胞壁的延展性,例如,纤维素糖化[21]和半纤维素结构修饰[22]。‘富士’(Borkh.)苹果中果皮表达量高于外果皮,引起外果皮生长速率低于中果皮,从而导致果实环裂[23]。扩张蛋白基因表达量也与荔枝和枣果实的抗裂能力呈正相关[24-25]。【本研究切入点】尽管果肉部位糖积累会引起渗透吸水,但不能通过抑制果肉糖积累的方法防止裂果。而通过调控果皮裂隙的形成、细胞壁物质的积累和果皮弹性防治裂果是一条可行的途径。【拟解决的关键问题】‘度尾文旦柚’(L. Osbeck)在中国福建省广泛栽培,该品种裂果严重,尚无有效的防治裂果的方法。‘度新1号文旦柚’是‘度尾文旦柚’的一个抗裂果突变株(果实生长季节内无裂果出现)。因此,本研究以‘度尾文旦柚’裂果和正常果及‘度新1号文旦柚’正常果为材料,通过比较两品种果顶部位果皮转录组的差异,揭示与果实抗裂能力相关的果皮转录组特征。
1 材料与方法
1.1 果实样品
‘度尾文旦柚’是一个果实生长季节内裂果率高的品种。8月初开始出现裂果。该品种果实仅果顶部位果皮开裂。‘度新1号文旦柚’(抗裂品种)源自‘度尾文旦柚’的芽变株,果实生长季节内无裂果出现。两品种的正常果和裂果见图1。两个柚资源栽植在同一个商品果园(地理坐标:118°59,25°44N,位于中国福建省莆田市仙游县),栽培管理制度相同。本试验以6年生抗裂品种的正常果(图1-1)、6年生易裂品种的正常果(图1-2)和裂果(图1-3)为材料。果实样品取自两个时期(时期A:2021年8月3日和时期B:2021年8月20日)和3个生物学重复(单株为一个生物学重复)。以果顶为中心,切取3 cm半径范围内的果皮,-80 ℃条件下保存,用于转录组测序和实时荧光定量分析。

A:‘度新1号文旦柚’正常果;B:‘度尾文旦柚’正常果;C:‘度尾文旦柚’裂果;1:A图中矩形框标注部位的放大图;2:B图中矩形框标注部位的放大图;3:C图中矩形框标注部位的放大图
1.2 cDNA文库构建与测序
每个样品取1 μg RNA用于构建cDNA文库。构建文库的试剂为NEBNext®Ultra™ RNA Library Prep Kit,试验方法参考说明书。
1.3 原始读序质量控制、组装和unigene功能注释
去除raw read中的测序接头和引物序列,过滤低质量序列,从而获得高质量的clean read。计算碱基质量值≥Q30(错误率0.1%)的碱基百分含量和GC碱基含量。序列组装软件为Trinity v2.5.1[26](https:// github.com/trinityrnaseq/trinityrnaseq/wiki)。
使用DIAMOND v2.0.4[27](https://github.com/ bbuchfink/diamond)将Unigene序列与NR[28](ftp://ftp. ncbi.nih.gov/blast/db/)、Swiss-Prot[29](http://www.uniprot. org/)、COG[30](http://www.ncbi.nlm.nih.gov/COG/)、KOG[31](http://www.ncbi.nlm.nih.gov/KOG/)、eggNOG[32](http://eggnogdb.embl.de/)和KEGG[33](http://www. genome.jp/kegg/)数据库比对。使用KOBAS v3.0[34](http://kobas.cbi.pku.edu.cn/kobas3)获得unigene在KEGG中的KEGG Orthology结果。使用InterProScan[35](https://www.ebi.ac.uk/interpro/download/)获得unignene的GO Orthology结果。预测完unigene的氨基酸序列之后使用HMMER[36]与Pfam[37](http://pfam. xfam.org/)数据库比对。以BLAST-value不大于1e-5和HMMER E-value不大于1e-10为标准,获得unigene注释信息。
1.4 差异基因的筛选
FPKM(fragments per kilobase of transcript per million mapped reads)是每百万reads中来自比对到某一基因每千碱基长度的reads数目,据此计算基因表达量。
DESeq2 v1.6.3[38](http://www.bioconductor.org/ packages/release/bioc/html/DESeq.html)用于样品间基因差异表达分析。依据Benjamini-Hochberg方法对原有假设检验得到的显著性值进行校正,以校正后的值(FDR)作为筛选差异表达基因的关键指标。将FDR小于0.01且差异倍数大于等于1.5作为差异表达基因的筛选标准。
1.5 差异基因GO功能富集与KEGG代谢通路富集分析
使用topGO v2.28.0(http://www.bioconductor.org/ packages/release/bioc/html/topGO.html)软件对注释到GO数据库的差异基因进行富集分析。利用富集因子分析差异基因在KEGG代谢通路中的富集程度。
1.6 实时荧光定量分析
实时荧光定量分析用于检测基因表达量。应用CTAB法[39]提取果皮总RNA。第一链cDNA合成试剂为script all-in-one RT mix with dsDNase kit(P710)(Jinbaite, Beijing, China)。以(genne ID:GQ389668.1)为内参基因。应用primer3(http://bioinfo. ut.ee/primer3-0.4.0/primer3/)在线设计正、反向引物(表1)。荧光定量仪为Mastercycler ep realplex (Eppendorf, Hamburg, Germany),定量试剂为2×SYBR green qRT-PCR mix kit(p519)(Jinbaite, Beijing, China)。25 μL反应体系:1 μL cDNA模板,0.5 μL正向引物(10 μmol·L-1),0.5 μL反向引物(10 μmol·L-1),12.5 μL 2×SYBR green qRT-PCR mix,10.5 μL去DNase/ RNase水。实时荧光定量PCR程序:94 ℃ 3 min,45个循环(94 ℃10 s,60 ℃ 34 s)。2-ΔΔCT[40]计算基因相对表达量。

表1 用于qRT-PCR试验的引物
2 结果
2.1 序列的组装和质量控制
每个样品GC碱基含量约为44%,Q30均超过91%。共组装获得305 757个转录本和53 075个unigene,序列的平均长度和N50分别为1 024 bp和2 329 bp(表2)。
2.2 unigene 功能注释
在8个数据库中注释到功能的unigene 33 334个,每个数据库注释到的基因数量见表3。
2.3 差异基因的筛选
通过对两品种裂果和正常果果皮转录组的比较筛选到的差异基因数量见表4。通过对易裂品种裂果果皮与两品种正常果果皮间转录组的差异分析,在时期A,共筛选到1 660个差异基因,从A1 vs A3和A2 vs A3两组转录组的比较中,发现相同的差异基因104个,其中30个基因的表达量在A1与A2果皮样品间也存在显著差异;在时期B,共筛选到1 972个差异基因,从B1 vs B3和B2 vs B3两组转录组的比较中,发现相同的差异基因82个,其中17个基因表达量在B1与B2果皮样品间也存在显著差异。
2.4 差异基因的GO富集分析
对易裂品种裂果果皮与两品种正常果果皮间所有差异基因(两个时期)的GO富集分析结果见图2—4。在生物学过程分类中,两个时期均富集到差异基因的主要亚类包括代谢过程、细胞过程、单生物过程、生物调控、刺激响应和信号(图2)。在分子功能分类中,两个时期均富集到差异基因的主要亚类包括催化活性、结合、转运体活性及与核酸结合的转录因子活性(图3)。在细胞组分分类中,两个时期均富集到差异基因的主要亚类包括细胞、细胞膜和细胞器(图4)。从差异基因富集到的主要亚类可以发现,果皮对自然环境或者细胞内信号的响应和代谢调控与果实的抗裂能力相关。

表2 转录本和unigene序列长度分布

表3 在数据库中得到功能注释的unigene数量
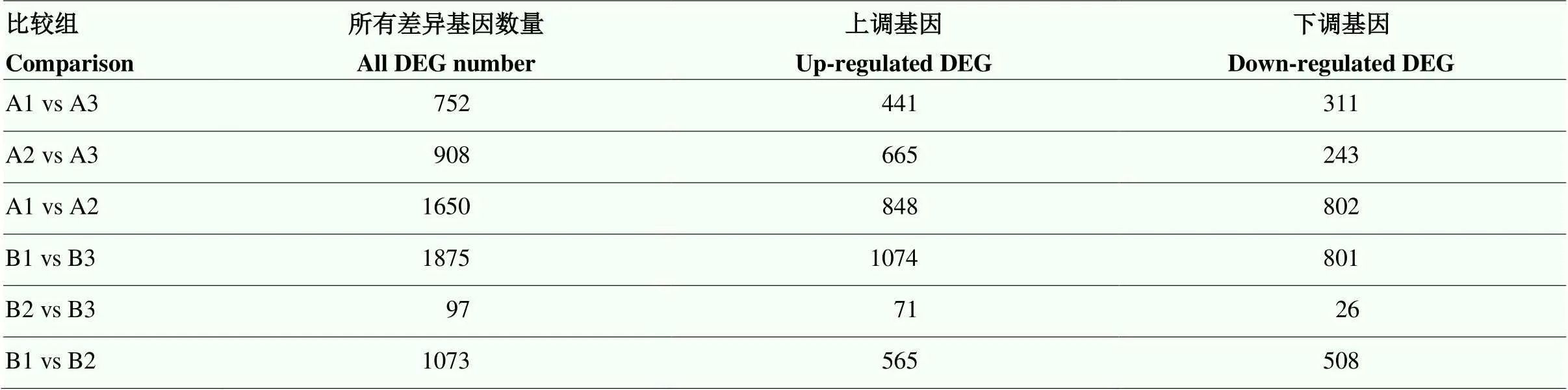
表4 基于两个时期6组果皮样品间的比较筛选到的差异表达基因
A表示果实取样时间(2021年8月3日);B表示果实取样时间(2021年8月20日);1为‘度新1号文旦柚’(抗裂品种)正常果果皮;2为‘度尾文旦柚’(易裂品种)正常果果皮;3为‘度尾文旦柚’(易裂品种)裂果果皮。下同
A, fruit sampling time (August 3, 2021); B, fruit sampling time (August 20, 2021); 1, pericarp of normal fruits from Duxin 1 pomelo (cracking-resistant cultivar); 2, pericarp of normal fruits from Duwei pomelo (cracking-sensitive cultivar); 3, pericarp of cracked fruits from Duwei pomelo (cracking-sensitive cultivar). The same as below
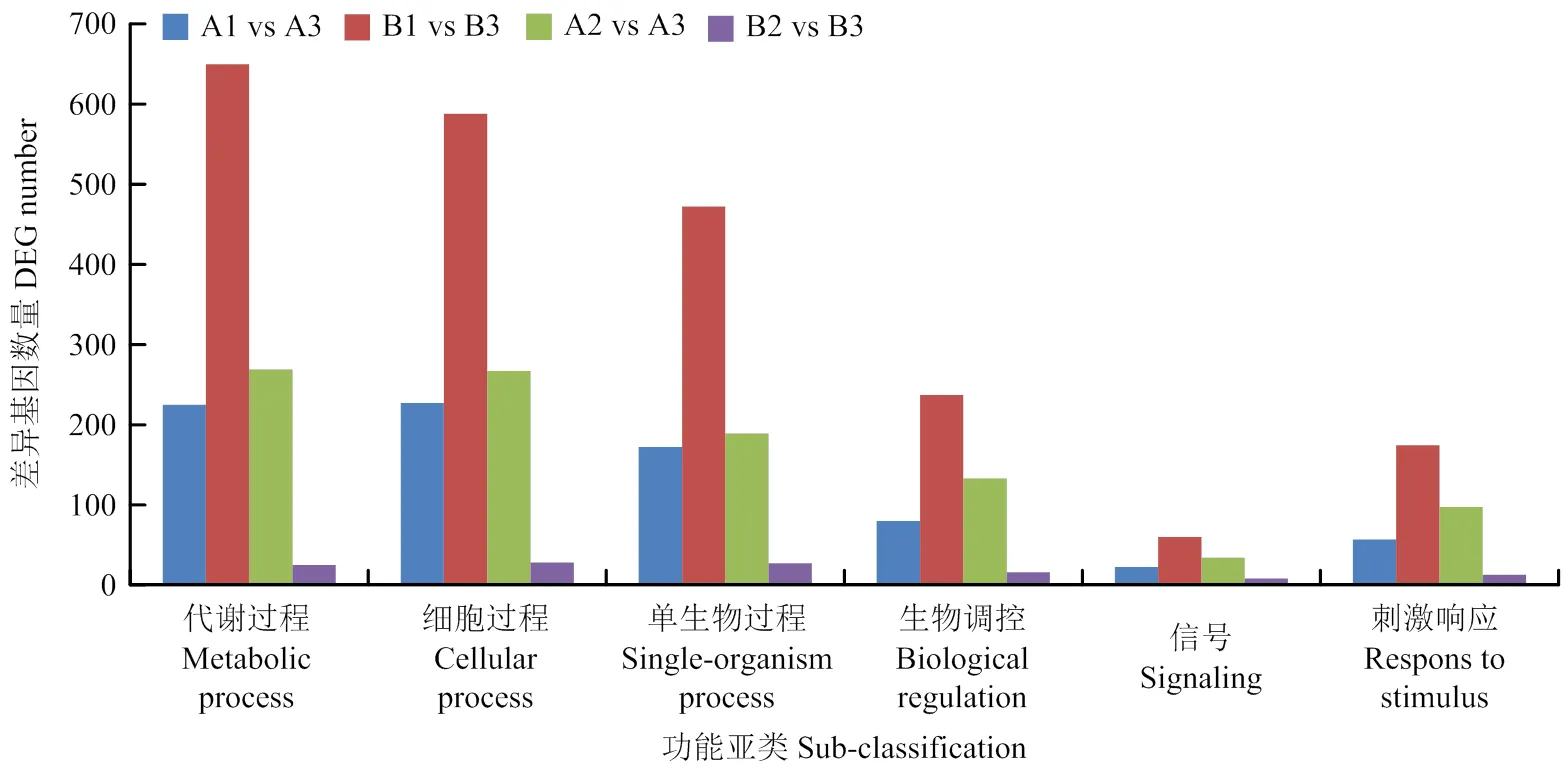
图2 生物学过程中基于转录组比较筛选到的所有差异基因注释到的主要功能亚类

图3 分子功能类别中基于转录组的比较筛选到的所有差异基因注释到的主要功能亚类
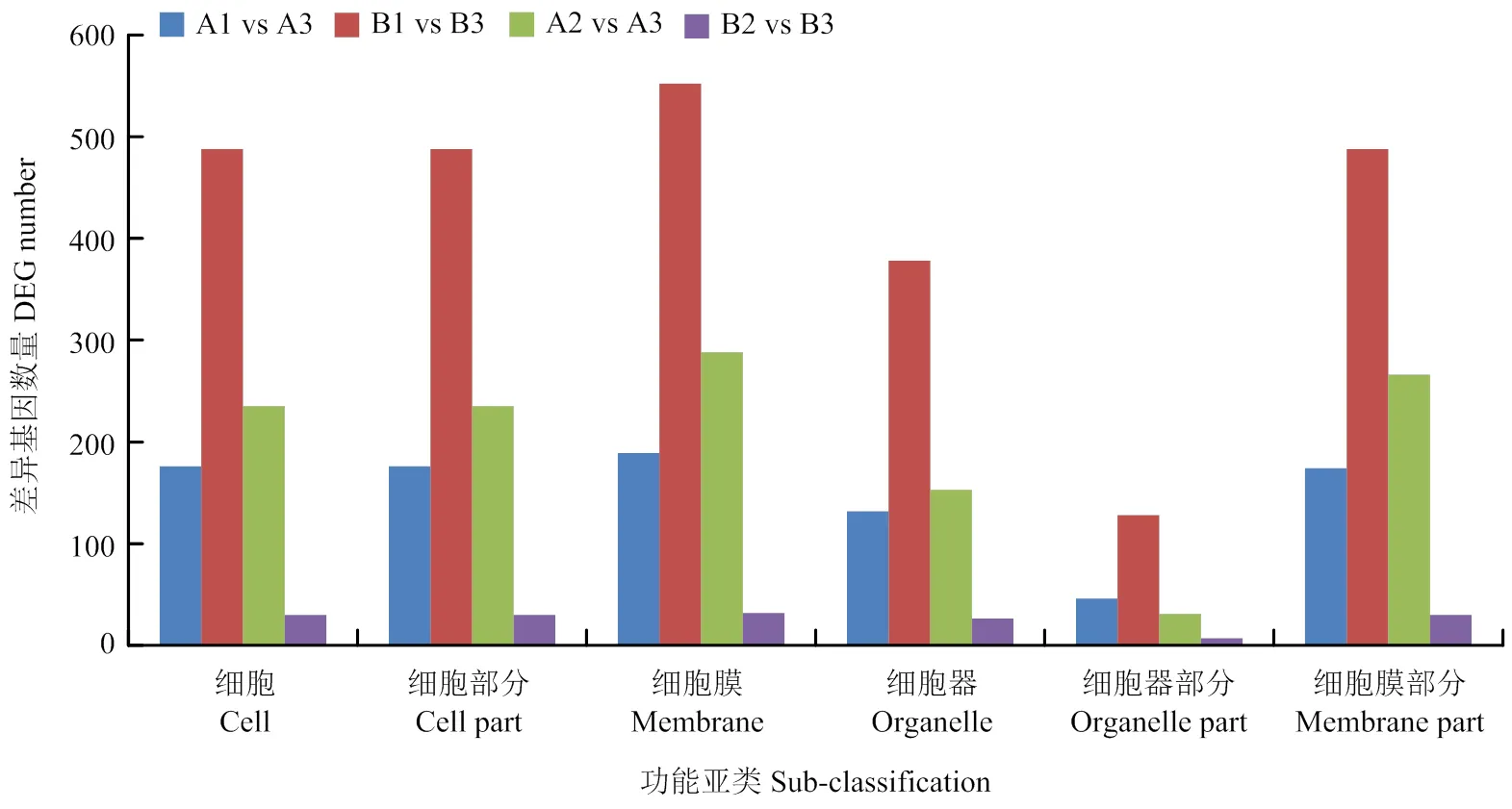
图4 细胞组分类别中基于转录组比较筛选到的所有差异基因注释到的主要功能亚类
2.5 差异基因代谢通路富集分析
基于A1 vs A3、B1 vs B3、A2 vs A3和B2 vs B3 4组转录组的比较筛选到的差异基因分别富集到101、123、89和37个代谢途径。易裂品种裂果果皮与两品种正常果果皮间所有差异基因富集到相同的代谢途径共30个(表5)。这30个代谢途径中碳代谢、植物激素信号转导、植物MAPK信号途径、内质网蛋白过程和植物病原菌互作富集的差异基因较多。从这几个重要的相同代谢途径中也可以发现,裂果与正常果间的差异基因涉及信号转导途径和生物调控。与植物病原菌互作相关的差异基因表达量的变化可能与果皮裂口的形成相关,而不是调控果实抗裂能力的基因。
2.6 裂果相关基因的筛选
以A1 vs A3和A2 vs A3两组样品中104个相同的差异基因,以及B1 vs B3和B2 vs B3两组样品的82个相同差异基因为核心(这些基因的表达量在易裂品种裂果果皮与两品种正常果果皮间均存在显著差异),综合分析这些基因表达量在时期内和时期间的变化趋势,以及GO功能和KEGG代谢通路显著富集分析的结果,筛选与果实抗裂能力相关的基因。在与细胞壁代谢相关的差异基因中,发现2个编码扩张蛋白-A1的基因(c35353.graph_c0、c19677.graph_c0)表达量在两个时期呈现显著差异。时期A,c35353.graph_c0基因在易裂品种裂果果皮的表达量显著低于正常果果皮,也显著低于抗裂品种正常果果皮,而在两品种正常果果皮中的表达量无显著差异。时期B,c19677.graph_c0基因表达量的变化规律与c35353.graph_c0基因完全相同(表6)。
在两个时期均发现生长素信号途径的基因表达量呈现显著变化。时期A,生长素响应蛋白30基因(c17406.graph_c1)表达量在易裂品种裂果果皮中显著高于正常果果皮,也显著高于抗裂品种正常果果皮,但在两品种正常果果皮间无显著差异。时期B,编码生长素响应蛋白26的基因(c37618.graph_c0)表达量变化规律与时期A完全一致(表7)。
在两个时期均筛选到涉及信号转导途径的差异基因。两时期易裂品种裂果果皮丝氨酸/苏氨酸蛋白激酶基因表达量显著高于两品种的正常果果皮,特别是时期B,易裂品种正常果果皮此类基因表达量也显著高于抗裂品种正常果果皮。表明丝氨酸/苏氨酸蛋白激酶基因表达量的提高易引发裂果。尽管受体蛋白激酶基因的表达量在易裂品种裂果果皮与两品种正常果果皮间存在显著差异,但两个时期的变化规律相反。另外,在时期A筛选到钙结合蛋白、钙转运ATP酶、小磷酸化酶样蛋白和类钙调磷酸酶B亚基基因,在时期B筛选到细胞壁受体激酶基因,除类钙调磷酸酶B亚基基因在易裂品种裂果果皮中表达量显著低于两品种正常果果皮外,其余基因在易裂品种裂果果皮中的表达量显著高于两品种正常果果皮,但均仅在一个时期呈现显著差异(表8)。
与两品种正常果果皮相比,易裂品种裂果果皮表达量呈现显著变化的转录因子主要包括热激转录因子、脱水响应元件结合蛋白和其他类转录因子(bHLH、MYC、SRM、BOA、BIM),仅热激转录因子和脱水响应元件结合蛋白基因表达量在两个时期均呈现显著变化。易裂品种裂果果皮热激转录因子和脱水响应元件结合蛋白基因表达量显著高于两品种正常果果皮(表9)。

表5 基于转录组比较筛选到的所有差异基因富集到相同的KEGG代谢途径
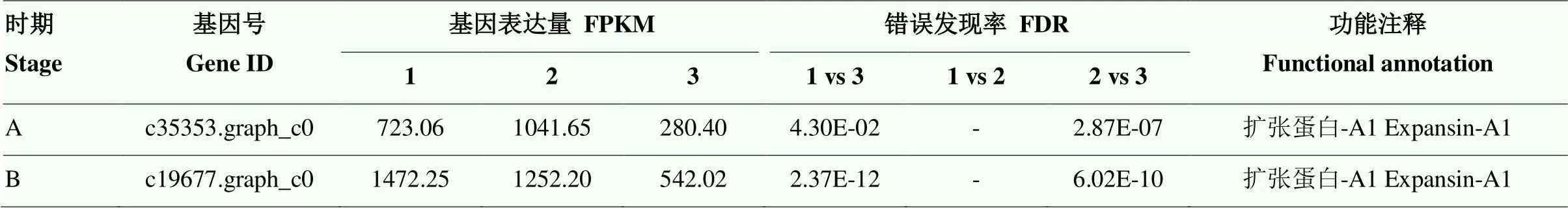
表6 ‘度尾文旦柚’与‘度新1号文旦柚’果皮细胞壁代谢途径的差异基因
“-”表示两样品间基因表达量无显著差异。下同
“-” indicate no significant difference in levels of gene transcripts between the two samples. The same as below
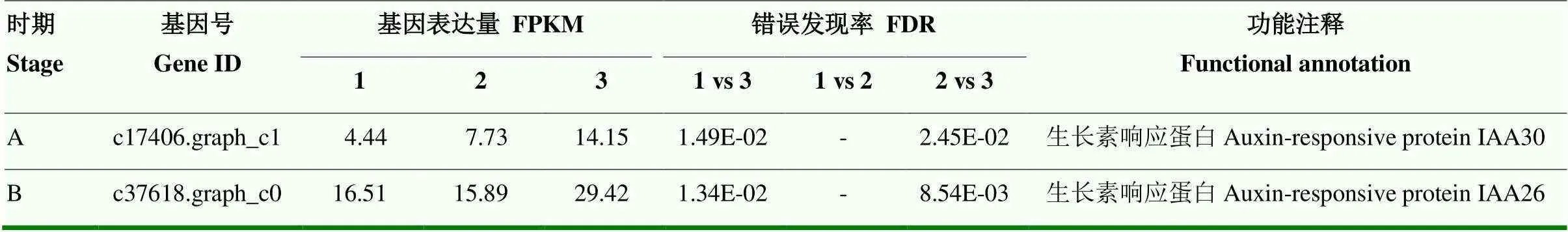
表7 ‘度尾文旦柚’与‘度新1号文旦柚’果皮植物激素信号转导途径差异基因
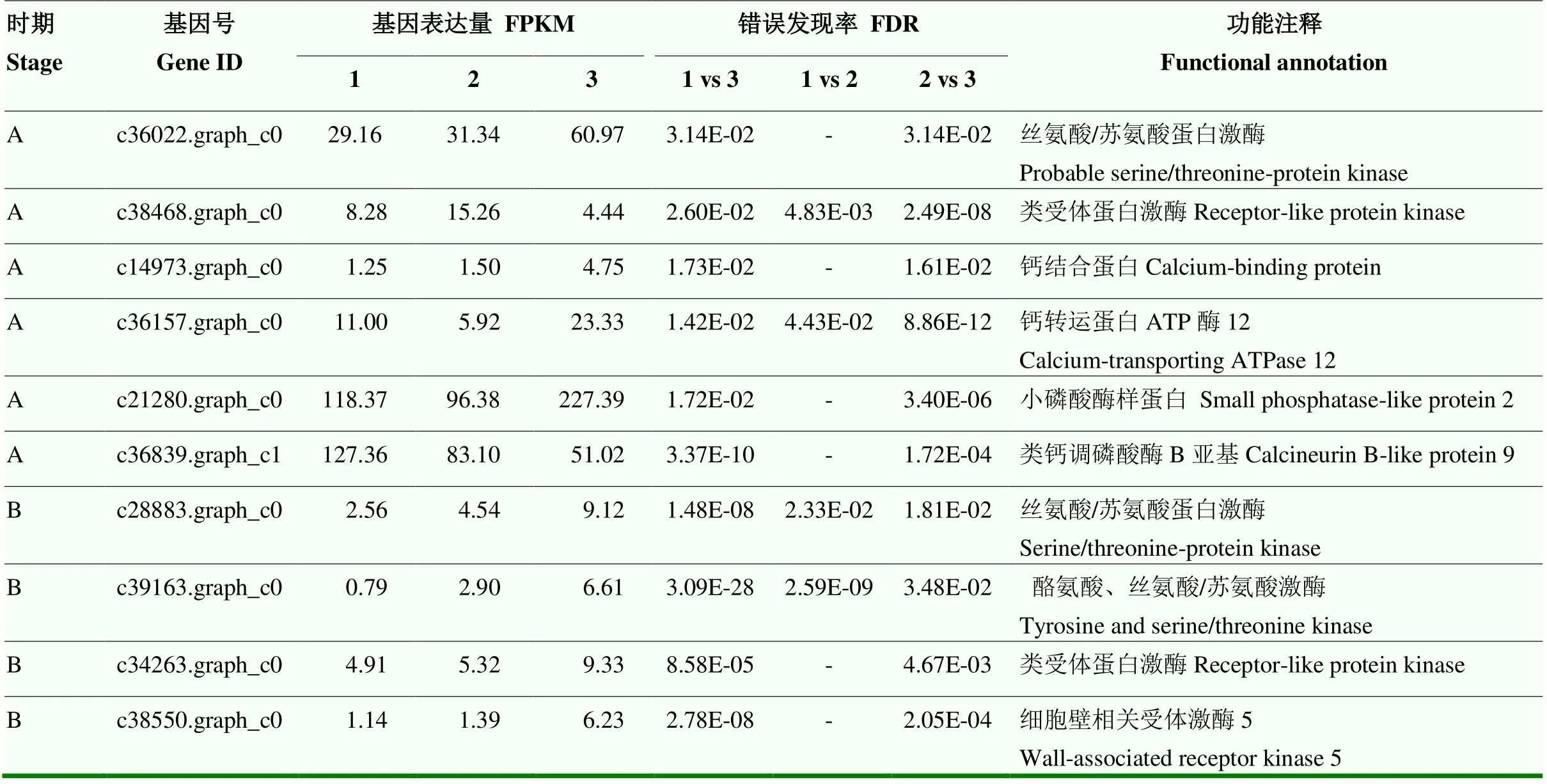
表8 ‘度尾文旦柚’与‘度新1号文旦柚’果皮植物信号途径差异基因
2.7 差异表达基因表达量的qRT-PCR验证
qRT-PCR方法用于验证8个差异基因的表达量。RNA-seq和qRT-PCR两类方法测得的基因表达量皮尔逊相关系数为0.8193。qRT-PCR结果显示,果皮样品间差异基因表达量的变化趋势与RNA-seq结果较为一致(图5)。
3 讨论
3.1 扩张蛋白与裂果间的关系
扩张蛋白是一类细胞壁蛋白超家族,分为4个亚家族:扩张蛋白A、扩张蛋白B、类扩张蛋白A和类扩张蛋白B;该类蛋白主要分布在伸展的细胞壁[20]。扩张蛋白通过提高纤维素酶和果胶酶活性促进纤维素和果胶降解[41],由此引起细胞壁松弛[42]。研究发现番茄扩张蛋白(SlExp1)功能缺失引起番茄果实硬度提高[22]。另外,扩张蛋白表达的变化也与裂果相关。在荔枝(Sonn.)果皮中鉴定到两个扩张蛋白基因(、),在果实快速生长初期检测到抗裂品种‘淮枝’mRNA,之后其表达量提高,在果实快速生长末期达到最高值,尽管在果实快速生长期的易裂品种‘糯米糍’果皮中也检测到,但其表达量保持稳定;另外,仅在‘淮枝’果皮中检测到mRNA[24]。与枣(Mill.)易裂品种果实相比,枣抗裂品种果皮扩张蛋白基因(、、、)的表达量高[25]。本研究也显示扩张蛋白基因表达量与果实抗裂能力呈正相关。由于‘度尾文旦柚’(易裂品种)正常果和裂果果皮间扩张蛋白基因表达量也存在显著差异,推测环境因素能够调控该基因的表达。

*表示4组比较(A3 vs A1、A3 vs A2 B3 vs B1 B3 vs B2)中两个样品间基因表达量(qRT-PCR)差异显著(P<0.05)

表9 ‘度尾文旦柚’与‘度新1号文旦柚’果皮差异转录因子
3.2 高温逆境响应基因与裂果间的关系
本试验发现在每个取样时期,样品间热逆境转录因子的表达量均呈现显著差异。易裂品种裂果果皮热激转录因子表达量显著高于两品种正常果果皮,而两品种正常果果皮间该基因表达量无显著差异。热激转录因子转录水平与植物对高温的适应力呈正相关[43-44]。由于柚果实快速生长期(8—9月)果实易裂果,此期是高温时节,热激转录因子表达量的提高可能是果实对高温逆境的响应。生长素响应蛋白(IAA26)[45]也参与植物对高温逆境的响应。本研究发现,与两品种正常果果皮相比,易裂品种裂果果皮中生长素响应蛋白基因(、)表达量显著提高。这也表明高温可能是引起柚裂果的重要环境因素。
3.3 信号途径和水分运动相关基因与裂果间的关系
在时期A,两品种正常果果皮类钙调磷酸酶B亚基、受体蛋白激酶和脱水响应元件结合蛋白基因表达量显著高于易裂品种裂果果皮。据报道,类钙调磷酸酶B亚基和受体蛋白激酶基因的表达量与植物对干旱或者渗透胁迫的适应力呈正相关[46-48]。本试验样品间这3个基因(编码类钙调磷酸酶B亚基、受体蛋白激酶和脱水响应元件结合蛋白)表达量的差异表明柚果实生长发育过程中果皮可能存在水分亏缺现象。另外,两品种正常果果皮信号转导途径基因(钙结合蛋白、钙转运ATP酶、丝氨酸/苏氨酸蛋白激酶、小磷酸化酶样蛋白)和脱水响应元件结合蛋白(DREB,一类转录因子)基因表达量显著低于易裂品种裂果果皮。尽管这些基因的转录水平均与植物对水分亏缺的适应力呈正相关[49-52],但因无法排除这些基因(丝氨酸/苏氨酸蛋白激酶基因除外)是否由果皮开裂后细胞水分散失所引起,尚难断定其与裂果间的关系。
在时期B,与两品种正常果果皮相比,易裂品种裂果果皮中编码丝氨酸/苏氨酸蛋白激酶、酪氨酸和丝氨酸/苏氨酸激酶、受体蛋白激酶、受体蛋白、细胞壁受体激酶和DREB的基因表达量均显著提高。此期不仅易裂品种裂果果皮丝氨酸/苏氨酸蛋白激酶基因表达量显著高于两品种正常果果皮;而且,易裂品种正常果果皮此基因表达量也显著高于抗裂品种正常果果皮,表明此基因表达量与果实抗裂能力呈负相关。在水稻()植株对金属和机械损伤的响应中,细胞壁受体激酶基因表达量提高[53]。据此推测柚裂果果皮中细胞壁受体激酶的基因表达量提高可能是由果皮开裂引起。此期尽管裂果果皮水通道蛋白TIP2-1基因表达量是两品种正常果果皮的5—10倍,但此基因表达量的变化可能是对果皮裂隙形成后的响应,而不是引起裂果的上游调控基因。
综合分析两个时期样品间果皮信号转导途径和水分运动相关基因表达量的差异,发现在果实快速生长初期(时期A),果皮类钙调磷酸酶B亚基转录水平与柚果实抗裂果能力呈正相关;而脱水响应元件结合蛋白、果皮丝氨酸/苏氨酸蛋白激酶基因表达量与柚果实抗裂能力呈负相关(两个取样时期规律一致)。考虑到高温逆境引起柑橘植株蒸腾量提高[54],所以柚果实生长季节内的高温可能与果皮水分亏缺相关。高温逆境和水分亏缺均能引起植物氧化逆境的形成,主要表现为丙二醛和过氧化氢含量、相对电导率提高,叶绿素和相对含水量降低[52,55-56]。而活性氧的积累易引起细胞凋亡[57]。因此,推测柚果实生长季节内高温引起果皮水分亏缺,继而导致细胞凋亡及微小裂隙的形成。
4 结论
本研究对果实快速生长初期和果实快速生长期的易裂品种(‘度尾文旦柚’)裂果和正常果果皮,以及抗裂品种(‘度新1号文旦柚’)正常果果皮进行转录组测序。综合分析果皮样品间转录组的差异,发现品种间抗裂能力的差异主要与扩张蛋白、高温和水分亏缺逆境响应基因和生长素响应蛋白基因的表达水平相关。扩张蛋白-A1、类钙调磷酸酶B亚基(干旱逆境响应蛋白)基因表达量与柚果实抗裂能力呈正相关。脱水响应元件结合蛋白、热逆境转录因子A-4b、热逆境转录因子A-8、丝氨酸/苏氨酸蛋白激酶(干旱逆境响应蛋白)和生长素响应蛋白(IAA26、IAA30,高温逆境响应蛋白)基因转录水平与柚果实抗裂能力呈负相关。
[1] 芮文婧, 王晓敏, 张倩男, 胡学义, 胡新华, 付金军, 高艳明, 李建设. 番茄353份种质资源表型性状遗传多样性分析. 园艺学报, 2018, 45(3): 561-570.
RUI W J, WANG X M, ZHANG Q N, HU X Y, HU X H, FU J J, GAO Y M, LI J S. Genetic diversity analysis of 353 tomato germplasm resources by phenotypic traits. Acta Horticulturae Sinica, 2018, 45(3): 561-570. (in Chinese)
[2] MITRA S K, PATHAK P K, DEBNATH S, SARKAR A, MONDAL D. Elucidation of the factors responsible for cracking and sunburn in litchi and integrated management to minimize the disorders. Acta Horticulturae, 2010, 863: 225-234.
[3] MUTHOO A K, RAINA B L, NATHU B L. Fruit cracking studies in some commerical cultivars of litchi (Sonn). Advances in Plant Sciences, 1999, 12(2): 543-547.
[4] 叶正文, 叶兰香, 张学英. “朋娜” 等脐橙的裂果规律及赤霉素防裂效果. 上海农业学报, 2002, 18(4): 52-57.
YE Z W, YE L X, ZHANG X Y. The fruit cracking rules of navel orange varieties such as “pengna” and the effect of gibberellin (ga) preventing fruits from cracking. Acta Agriculturae Shanghai, 2002, 18(4): 52-57. (in Chinese)
[5] 李娟, 陈杰忠. 柑桔裂果发生类型、过程及预防对策. 广东农业科学, 2011, 38(10): 32-33, 37.
LI J, CHEN J Z. The style, process and control of cracking fruit in citrus. Guangdong Agricultural Sciences, 2011, 38(10): 32-33, 37. (in Chinese)
[6] 郗鑫, 刘晨筱, 陈虹, 白晋华, 郭红彦. 不同枣品种果实性状与裂果的相关性. 山西农业科学, 2016, 44(10): 1476-1478, 1507.
XI X, LIU C X, CHEN H, BAI J H, GUO H Y. Correlation analysis of fruit characters and cracking in different varieties of jujube. Journal of Shanxi Agricultural Sciences, 2016, 44(10): 1476-1478, 1507. (in Chinese)
[7] 栗现芳, 陈晓龙, 问徐鹏, 赵怡, 马辉. 枣易裂品种和抗裂品种间各糖组分含量与裂果的相关性分析. 分子植物育种, 2020, 18(18): 6180-6186.
LI X F, CHEN X L, WEN X P, ZHAO Y, MA H. Correlation analysis between the sugar components and fruit cracking in easily cracked and resistant jujube. Molecular Plant Breeding, 2020, 18(18): 6180-6186. (in Chinese)
[8] 李建国, 黄辉白. 荔枝果实理化特性及果皮形态学与裂果易感性的关系. 华南农业大学学报, 1995, 16(1): 84-89.
LI J G, HUANG H B. Phybico- chemical properties and peel morphology in relation to fruit-cracking susceptibility in litchi fruit. Journal of South China Agricultural University, 1995, 16(1): 84-89. (in Chinese)
[9] YU J, ZHU M T, WANG M J, TANG W Y, WU S, ZHANG K, YANG G S. Effect of nordihydroguaiaretic acid on grape berry cracking. Scientia Horticulturae, 2020, 261: 108979.
[10] BEYER M, KNOCHE M. Studies on water transport through the sweet cherry fruit surface: V. conductance for water uptake. Journal of the American Society for Horticultural Science, 2002, 127(3): 325-332.
[11] LI N, FU L J, SONG Y Q, LI J, XUE X F, LI S R, LI L L. Water entry in jujube fruit and its relationship with cracking. Acta Physiologiae Plantarum, 2019, 41(9): 162.
[12] SONG Y Q, LI J, FU L J, LI N, LI L L. Change of fruit surface characteristics and its relationship with water absorption and fruit cracking in‘Huping’. Scientia Silvae Sinicae, 2018, 54(12): 52-59.
[13] PESCHEL S, KNOCHE M. Characterization of microcracks in the cuticle of developing sweet cherry fruit. Journal of the American Society for Horticultural Science, 2005, 130(4): 487-495.
[14] HUANG X M, YUAN W Q, WANG H C, LI J G, HUANG H B, SHI L, YIN J H. Linking cracking resistance and fruit desiccation rate to pericarp structure in(Sonn.). The Journal of Horticultural Science and Biotechnology, 2004, 79(6): 897-905.
[15] HUANG X M, WANG H C, GAO F F, HUANG H B. A comparative study of the pericarp of litchi cultivars susceptible and resistant to fruit cracking. The Journal of Horticultural Science and Biotechnology, 1999, 74(3): 351-354.
[16] 张鹏飞, 张燕, 巩磊, 纪薇, 高美英, 刘亚令, 郝晓娟. 植物生长调节剂对枣果皮细胞壁多糖的影响研究. 山西农业大学学报(自然科学版), 2014, 34(2): 174-178.
ZHANG P F, ZHANG Y, GONG L, JI W, GAO M Y, LIU Y L, HAO X J. Effects of plant growth regulators on pericarp cell wall polysaccharide of Chinese jujube. Journal of Shanxi Agricultural University (Natural Science Edition), 2014, 34(2): 174-178. (in Chinese)
[17] YANG Z E, WU Z, ZHANG C, HU E M, ZHOU R, JIANG F L. The composition of pericarp, cell aging, and changes in water absorption in two tomato genotypes: mechanism, factors, and potential role in fruit cracking.Acta Physiologiae Plantarum, 2016, 38(9): 215.
[18] CHOI J H, LEE B, GU M M, LEE U Y, KIM M S, JUNG S K, CHOI H S. Course of fruit cracking in ‘Whansan’ pears. Horticulture, Environment, and Biotechnology, 2020, 61(1): 51-59.
[19] 杨磊, 冯贝贝, 靳娟, 樊丁宇, 刘晶, 克里木∙伊明, 郝庆. 新疆6个大果红枣裂果性差异及其内在原因分析. 西北植物学报, 2021, 41(8)1364-1370.
YANG L, FENG B B, JIN J, FAN D Y, LIU J, KELIMU∙Yimin, HAO Q. Differences in fruit cracking of six big fruit type jujube cultivars from Xinjiang and its internal causes. Acta Botanica Boreali- Occidentalia Sinica, 2021, 41(8): 1364-1370. (in Chinese)
[20] CHOI D, KIM J H, LEE Y. Expansins in plant development// Advances in Botanical Research. Amsterdam: Elsevier, 2008: 47-97.
[21] SEKI Y, KIKUCHI Y, YOSHIMOTO R, ABURAI K, KANAI Y, RUIKE T, IWABATA K, GOITSUKA R, SUGAWARA F, ABE M, SAKAGUCHI K. Promotion of crystalline cellulose degradation by expansins from. Planta, 2015, 241(1): 83-93.
[22] MINOIA S, BOUALEM A, MARCEL F, TROADEC C, QUEMENERB, CELLINI F, PETROZZA A, VIGOUROUX J, LAHAYE M, CARRIERO F, BENDAHMANE A. Induced mutations in tomatoalter cell wall metabolism and delay fruit softening. Plant Science, 2016, 242: 195-202.
[23] KASAI S, HAYAMA H, KASHIMURA Y, KUDO S, OSANAI Y. Relationship between fruit cracking and expression of the expansin geneBorkh.). Scientia Horticulturae, 2008, 116(2): 194-198.
[24] WANG Y, LU W J, LI J G, JIANG Y M. Differential expression of two expansin genes in developing fruit of cracking-susceptible and-resistantcultivars. Journal of the American Society for Horticultural Science, 2006, 131(1): 118-121.
[25] 辛海青, 周军永, 孙耀星, 穆文磊, 杨健, 马福利, 孙俊, 薛峥嵘, 陆丽娟, 孙其宝. 枣易裂与抗裂品种灌水后果皮结构和扩张蛋白基因表达差异研究. 园艺学报, 2021, 48(9): 1785-1793.
XIN H Q, ZHOU J Y, SUN Y X, MU W L, YANG J, MA F L, SUN J, XUE Z R, LU L J, SUN Q B. Differences in the pericarp structure and the expression of expansin genes after irrigation between easily cracked and resistant jujube. Acta Horticulturae Sinica, 2021, 48(9): 1785-1793. (in Chinese)
[26] GRABHERR M G, HAAS B J, YASSOUR M, LEVIN J Z, THOMPSON D A, AMIT I, ADICONIS X, FAN L, RAYCHOWDHURY R, ZENG Q D, CHEN Z H, MAUCELI E, HACOHEN N, GNIRKE A, RHIND N, DI PALMA F, BIRREN B W, NUSBAUM C, LINDBLAD-TOH K, FRIEDMAN N, REGEV A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 2011, 29(7): 644-652.
[27] BUCHFINK B, XIE C, HUSON D H. Fast and sensitive protein alignment using DIAMOND. Nature Methods, 2015, 12(1): 59-60.
[28] 邓泱泱, 荔建琦, 吴松锋, 朱云平, 陈耀文, 贺福初. nr数据库分析及其本地化. 计算机工程, 2006, 32(5): 71-73, 76.
DENG Y Y, LI J Q, WU S F, ZHU Y P, CHEN Y W, HE F C. Integrated nr database in protein annotation system and its localization. Computer Engineering, 2006, 32(5): 71-73, 76. (in Chinese)
[29] APWEILER R, BAIROCH A, WU C H, BARKER W C, BOECKMANN B, FERRO S, GASTEIGER E, HUANG H Z, LOPEZ R, MAGRANE M, MARTIN M J, NATALE D A, O'DONOVAN C, REDASCHI N, YEH L S L. UniProt: The universal protein knowledgebase. Nucleic Acids Research, 2004, 32: D115-D119.
[30] TATUSOV R L, GALPERIN M Y, NATALE D A, KOONIN E V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research, 2000, 28(1): 33-36.
[31] KOONIN E V, FEDOROVA N D, JACKSON J D, JACOBS A R, KRYLOV D M, MAKAROVA K S, MAZUMDER R, MEKHEDOV S L, NIKOLSKAYA A N, RAO B S, ROGOZIN I B, SMIRNOV S, SOROKIN A V, SVERDLOV A V, VASUDEVAN S, WOLF Y I, YIN J J, NATALE D A. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biology, 2004, 5(2): R7.
[32] HUERTA-CEPAS J, SZKLARCZYK D, FORSLUND K, COOK H, HELLER D, WALTER M C, RATTEI T, MENDE D R, SUNAGAWA S, KUHN M, JENSEN L J, VON MERING C, BORK P. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Research, 2016, 44: D286-D293.
[33] KANEHISA M, GOTO S, KAWASHIMA S, OKUNO Y, HATTORI M. The KEGG resource for deciphering the genome. Nucleic Acids Research, 2004, 32(suppl_1): D277-D280.
[34] XIE C, MAO X Z, HUANG J J, DING Y, WU J M, DONG S, KONG L, GAO G, LI C Y, WEI L P. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 2011, 39(suppl_2): W316-W322.
[35] JONES P, BINNS D, CHANG H Y, FRASER M, LI W Z, MCANULLA C, MCWILLIAM H, MASLEN J, MITCHELL A, NUKA G, PESSEAT S, QUINN A F, SANGRADOR-VEGAS A, SCHEREMETJEW M, YONG S Y, LOPEZ R, HUNTER S. InterProScan 5: Genome-scale protein function classification. Bioinformatics, 2014, 30(9): 1236-1240.
[36] EDDY S R. Profile hidden Markov models. Bioinformatics, 1998, 14(9): 755-763.
[37] FINN R D, TATE J, MISTRY J, COGGILL P C, SAMMUT S J, HOTZ H R, CERIC G, FORSLUND K, EDDY S R, SONNHAMMER E L L, BATEMAN A. The Pfam protein families database. Nucleic Acids Research, 2008, 36: D281-D288.
[38] LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 2014, 15(12): 550.
[39] 张锐, 周开兵. 荔枝果实膨大期果皮总RNA提取方法的筛选. 中国南方果树, 2013, 42(5): 11-14.
ZHANG R, ZHOU K B. Screening of the methods for extraction of total RNA frompericarp at the swollen stage. South China Fruits, 2013, 42(5): 11-14. (in Chinese)
[40] SCHMITTGEN T D, LIVAK K J. Analyzing real-time PCR data by the comparative CTmethod. Nature Protocols, 2008, 3(6): 1101-1108.
[41] WEI W, YANG C, LUO J, LU C M, WU Y J, YUAN S. Synergism between cucumber α-expansin, fungal endoglucanase and pectin lyase. Journal of Plant Physiology, 2010, 167(14): 1204-1210.
[42] COSGROVE D J. Loosening of plant cell walls by expansins. Nature, 2000, 407(6802): 321-326.
[43] WANG X Y, HUANG B R. Lipid- and calcium-signaling regulation of-mediated heat tolerance in tall fescue. Environmental and Experimental Botany, 2017, 136: 59-67.
[44] GAI W X, MA X, LI Y, XIAO J J, KHAN A, LI Q H, GONG Z H. CaHsfA1d improves plant thermotolerance via regulating the expression of stress- and antioxidant-related genes. International Journal of Molecular Sciences, 2020, 21(21): 8374.
[45] ZHAO Y, YU W G, HU X Y, SHI Y H, LIU Y, ZHONG Y F, WANG P, DENG S Y, NIU J, YU X D. Physiological and transcriptomic analysis revealed the involvement of crucial factors in heat stress response of. Gene, 2018, 660: 109-119.
[46] CHO J H, CHOI M N, YOON K H, KIM K N. Ectopic expression of SjCBL1, calcineurin B-like 1 gene from, rescues the salt and osmotic stress hypersensitivity incbl1 mutant. Frontiers in Plant Science, 2018, 9: 1188.
[47] XU G Y, LI M J, ZHANG H, CHEN Q S, JIN L F, ZHENG Q X, LIU P P, CAO P J, CHEN X, ZHAI N, ZHOU H N., a novel RLK-like protein kinase from, positively regulates drought tolerance in transgenic. Biochemical and Biophysical Research Communications, 2018, 503(3): 1235-1240.
[48] XU M, LI H, LIU Z N, WANG X H, XU P, DAI S J, CAO X, CUI X Y. The soybean CBL-interacting protein kinase, GmCIPK2, positively regulates drought tolerance and ABA signaling. Plant Physiology and Biochemistry, 2021, 167: 980-989.
[49] LI C X, ZHANG W L, YUAN M, JIANG L N, SUN B, ZHANG D J, SHAO Y, LIU A Q, LIU X Q, MA J H. Transcriptome analysis of osmotic-responsive genes in ABA-dependent and-independent pathways in wheat (L.) roots. PeerJ, 2019, 7: e6519.
[50] REDILLAS M C F R, JEONG J S, KIM Y S, JUNG H, BANG S W, CHOI Y D, HA S H, REUZEAU C, KIM J K. The overexpression ofalters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnology Journal, 2012, 10(7): 792-805.
[51] SUN X L, SUN M Z, LUO X, DING X D, JI W, CAI H, BAI X, LIU X F, ZHU Y M. AABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses.Planta, 2013, 237(6): 1527-1545.
[52] DAS A, BASU P, KUMAR M, ANSARI J, SHUKLA A, THAKUR S, SINGH P, DATTA S, CHATURVEDI S, SHESHSHAYEE M, BANSAL K, SINGH N. Transgenic chickpea (L.) harbouring AtDREB1a are physiologically better adapted to water deficit. BMC Plant Biology, 2021, 21: 39.
[53] HU W, LV Y Y, LEI W R, LI X, CHEN Y H, ZHENG L Q, XIA Y, SHEN Z G. Cloning and characterization of thewall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant and Soil, 2014, 384(1/2): 335-346.
[54] ZANDALINAS S I, RIVERO R M, MARTÍNEZ V, GÓMEZ- CADENAS A, ARBONA V. Tolerance of citrus plants to the combination of high temperatures and drought is associated to the increase in transpiration modulated by a reduction in abscisic acid levels.BMC Plant Biology, 2016, 16: 105.
[55] HASANUZZAMAN M, NAHAR K, ALAM M M, FUJITA M. Modulation of antioxidant machinery and the methylglyoxal detoxification system in selenium-supplementedseedlings confers tolerance to high temperature stress. Biological Trace Element Research, 2014, 161(3): 297-307.
[56] BATCHO A A, SARWAR M B, RASHID B, HASSAN S, HUSNAIN T. Heat shock protein gene identified from(AsHSP70) confers heat stress tolerance in transgenic cotton (). Theoretical and Experimental Plant Physiology, 2021, 33(2): 141-156.
[57] CONTRAN N, TONELLI M, CROSTI P, CERANA R, MALERBA M. Antioxidant system in programmed cell death of sycamore (L.) cultured cells. Acta Physiologiae Plantarum, 2012, 34(2): 617-629.
A Transcriptome Analysis Identifies Candidate Genes Related to Fruit Cracking in Pomelo Fruits
LU YanQing, LIN YanJin, WANG XianDa, LU XinKun
Institute of Pomology, Fujian Academy of Agricultural Sciences, Fuzhou 350013
【Objective】Fruit cracking is a universal physiological disorder that occurs during growth in citrus fruits. However, the molecular mechanisms that regulate cracking in citrus fruits remain unclear. The aim of this study was to screen genes that were related to resistance to fruit cracking. 【Method】 Normal fruits from a pomelo ((L). Osbeck) cultivar (Duxin 1) resistant to cracking, as well as normal and cracked fruits from Duwei, a cultivar sensitive to cracking, were collected on August 3, 2021 andAugust 20, 2021, respectively. The pericarp surrounding blossom ends of the fruits (the blossom end was considered the center, approximate 30 millimeters radius) were sampled for RNA-seq. 【Result】 The differentially expressed genes (DEGs) in each stage were screened based on the comparisons of a transcriptome between cracked fruits from the cracking-sensitive cultivar and normal fruits from both cultivars. In the stage A, 1 660 DEGs were obtained, and 104 DEGs were common between the comparison. A total of 1 972 DEGs were screened in stage B, and 82 were common in the comparison. All the DEGs screened at both stages were used for a Gene Ontology enrichment analysis. In the classification of biological process, the major common sub-classifications, including ‘metabolic process’ , ‘cellular process’ , ‘single-organism process’ , ‘biological regulation’ , ‘response to stimulus’ , and ‘signaling’ were identified in both stages. All the screened DEGs were also analyzed using Kyoto Encyclopedia of Genes and Genomes enrichment. Many genes were enriched in several metabolic pathways, including ‘carbon metabolism’, ‘MAPK signaling pathway-plant’, ‘plant hormone signal transduction’ and ‘protein processing in endoplasmic reticulum’. In addition, these pathways were identified in both stages. Several genes related to resistance to fruit cracking were identified in this study. The levels of transcription ofwere significantly higher in the pericarp of normal fruits from the two cultivars than that in the pericarp of cracked fruits from the sensitive cultivar.gene was highly expressed in the pericarp of normal fruits from both cultivars when compared with the pericarp of cracked fruits from the sensitive cultivar. However, this difference disappeared at the stage B. The genes for,,, andwere upregulated in the pericarp of cracked fruits from the sensitive cultivar compared with the pericarp of normal fruits from the two cultivars in both stages. 【Conclusion】These findings suggested that the genes related to strength of pericarp, water movement, and responsing to high temperature and water deficiency stresses were critical to regulating resistance to fruit cracking.
pummelo (L. Osbeck); fruit cracking; pericarp; transcriptome; cracking-resistant gene

10.3864/j.issn.0578-1752.2023.20.013
2023-04-18;
2023-08-15
福建省属公益类科研院所基本科研专项(2021R1028001,2020R10280014,2022R1028006)、福建省自然科学基金(2022J01472)、科技部、财政部国家科技资源共享服务平台项目(NHGRC2023-NH18-2)、福建省农业科学院农业科技创新联盟专项(CXLM2021003)
卢艳清,E-mail:lluyqing2006@126.com。通信作者卢新坤,E-mail:gsskg@126.com
(责任编辑 赵伶俐)
