Optimization for Microbial Degumming of Ramie with Bacillus subtilis DZ5 in Submerged Fermentation by Orthogonal Array Design and Response Surface Methodology
2023-10-29LIUFenCHENYangdong陈杨栋ZHUPengCAOZhangjun曹张军ZHANGXingqun张兴群
LIU Fen(刘 芬), CHEN Yangdong(陈杨栋), ZHU Peng(朱 鹏), CAO Zhangjun(曹张军), ZHANG Xingqun(张兴群), *
1 College of Biological Science and Medical Engineering, Donghua University, Shanghai 201620, China 2 Key Laboratory of Science &Technology of Eco-Textile, Ministry of Education, Donghua University, Shanghai 201620, China 3 College of Textile and Clothing, Xinjiang University, Urumqi 830046, China
Abstract:As a kind of natural fiber, ramie fiber has distinctive advantages in textile application, but the application is limited due to the traditional degumming mode. Compared with the traditional degumming process, the microbial degumming process has many advantages. To obtain the optimal conditions for degumming ramie with Bacillus subtilis DZ5(BSDZ5), a combined statistical approach of orthogonal array design (OAD) and response surface methodology (RSM) was used. The influences of initial pH of the bacteria medium, culture temperature, shaking speed, degumming time and inoculum size on submerged fermentation degumming were evaluated by using fractional factorial design. The main factors in the analysis were culture temperature, shaking speed and initial pH. The residual gum mass fraction was used as the optimization index, and the optimal conditions for degumming were determined by central composite design and RSM. Thus with only a limited number of experiments, an optimal ramie microbial degumming condition was found as the culture temperature of 40 ℃, the initial pH in the culture medium of 8.5, the shaking speed of 205 r/min, the degumming time of 96 h and the inoculum size of 5%. After microbial degumming of ramie under the optimal conditions, there was only 10.6% residual gum by mass in the fiber. In addition, the effective degumming of BSDZ5 was also confirmed by a scanning electron microscope (SEM).
Key words:microbial degumming; optimization; ramie; orthogonal array design (OAD); response surface methodology(RSM)
0 Introduction
Ramie (Boehmerianivea) is a member of the nettle family (Uricaceae) and grows mainly in temperate and tropical areas. It can be used to produce the strongest and longest plant fibers which are lustrous with an almost silky appearance. The ramie fibers possess excellent mechanical properties and are resistant to bacteria, mildew and insect attack[1-3]. The ramie fibers obtained from the outer part of the ramie stem can be used for the production of clothing fabrics, industrial packaging, twines, cordages, canvas, car outfits, and so on. In order to obtain high-quality ramie fibers, appropriate degumming is essential.
Decorticated ramie fibers contain 20%-35% gum that mainly consists of hemicellulose, wax, pectin, lignin and hydrotope. This gum should be removed as much as possible to fulfill the textile industrial requirement (1.5%-2.5% residual gum by mass)[4-5]. As the conventional degumming process with a solution of NaOH not only consumes a high amount of chemicals and energy, but also results in serious environmental pollution, it is imperative to develop substitutive techniques. Degumming of ramie fibers by microorganisms or their enzymes has attracted wide attention[6-11]. The present study aims to determine the optimal conditions of microbial degumming of ramie and develop an environmentally friendly and efficient degumming process.
1 Materials and Methods
1.1 Source of raw materials and microorganisms
The ramie was collected from Hunan province, China, and its constituents were measured according to the Chinese national standard GB/T 5889—1986[12]. To avoid bias, all the samples taken from the middle section of the ramie bast were tested thrice, and the average values were taken. All the chemicals were purchased from SINOPHARM, China, and used without further treatment.
The bacterial strain was isolated from putrid ramie and maintained on nutrient agar slants at 4 ℃ and stored in glycerol at -78 ℃. The strain was identified asBacillussubtilisDZ5(BSDZ5) based on 16S rDNA sequence analysis. Its physiological and biochemical characteristics were proved to be effective in degumming.
1.2 Inoculum preparation
The inoculum was prepared by liquid shake cultivation in Erlenmeyer flasks (250 mL) containing 100 mL beef extract-peptone broth (0.3% beef extract, 1% meat peptone, 0.5% NaCl, and 0.2% agar by mass; pH 7.4-7.6) at 40 ℃ and 200 r/min for 24 h.
1.3 Treatment of ramie fibers
All the ramie fiber samples (each 5 g) were prepared, and the liquor ratio of fibers to water was 1∶20. The samples were sterilized at 121 ℃ for 20 min and placed in a shaker after the addition of a liquid culture medium comprisingBSDZ5. After degumming, the fibers were washed under running water and put in a drier. The evaluation of degumming was based on the residual gum mass fraction which was assayed according to Ref.[12]. Thus, the optimal conditions employed in microbial degumming of ramie could be determined.
1.4 Experimental design and data analysis
1.4.1Orthogonalarraydesign(OAD)
The experimental design is based on Taguchi’s method[13-14], which is a unique and powerful optimization technique that allows optimization with a minimum number of experiments. The effects of initial pH, culture temperature, inoculum size, degumming time, and shaking speed were investigated (Table 1).
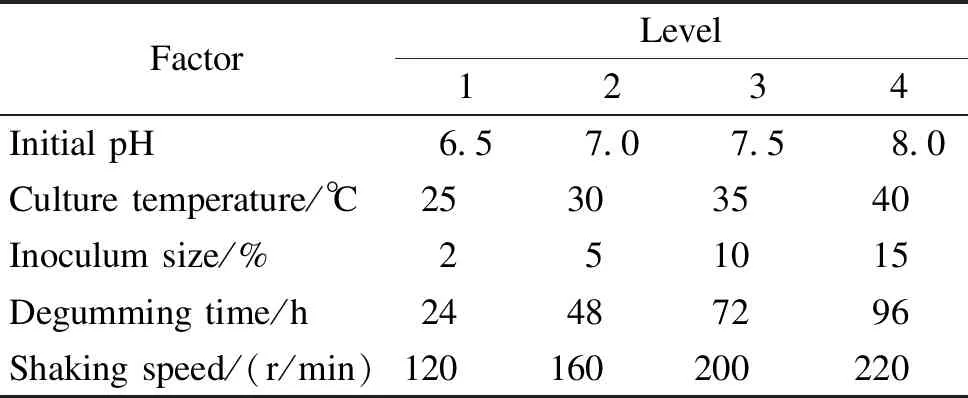
Table 1 Factors and levels of degumming by BSDZ5 based on OAD
1.4.2Centralcompositedesign(CCD)
The CCD approach was used for determining the optimal levels of critical factors identified by the orthogonal test and was also employed as an experimental design for the analysis of the treatment combinations. Three independent factors potentially affecting the degumming effect of the ramie fiber, namely, culture temperature, shaking speed and initial pH, were investigated. The CCD comprised five levels for each factor. The upper and the lower limits of each factor were chosen based on the results of the preliminary experimental responses in the L16(45) matrix. The levels corresponding to the factors are shown in Table 2.

Table 2 Process parameters used in CCD with actual factor levels corresponding to coded factor levels
1.5 Observation of microstructure and surface morphology
A scanning electron microscope(SEM, S-4800, HITACHI, Japan) was used to observe the microstructure and the surface morphology of the treated and the untreated ramie fibers as well as the morphology of the microorganisms used for degumming the ramie fibers.
1.6 Tenacity of a single ramie fiber
The constant rate of elongation (CRE) method was applied to measure the tenacity of the ramie fiber. The tenacity of the fiber was measured with a single-fiber strength tester(YG(B)001A, Wenzhou Darong Textile Instrument Co., Ltd., China), which determined the max load of clean and dry fiber strands (5 cm long). The tenacity of the samples was calculated from the ratio of the max load to its linear density.The test speed was 10 mm/min. A paper frame was used to support the fibers to avoid fibers bending or breaking during the sample loading, and the frame was cut prior to the stress-strain scan. To obtain reliable results, 10 samples of each type were employed for single-fiber tenacity testing.
2 Results and Discussion
2.1 Analysis of OAD
In this study, the effects of five important factors, including initial pH, culture temperature, inoculum size, degumming time, and shaking speed, at different levels were studied by using Taguchi’s method. The L16(45) matrix employed to assign the considered factors is shown in Table 3. The mean values of the responses (i.e.the residual gum mass fractions for each factor at different levels), denoted byr1,r2,r3,andr4, are shown in Table 4. In the L16(45) OAD, 16 experiments are required to complete the optimization process. All the trials were carried out in triplicate to obtain suitable precision. The sequence of the experiments was randomized to avoid any personal or subjective bias. In the proposed method, no interaction among the factors was found in the matrix, and the focus was placed on the main effects of the five most important factors.
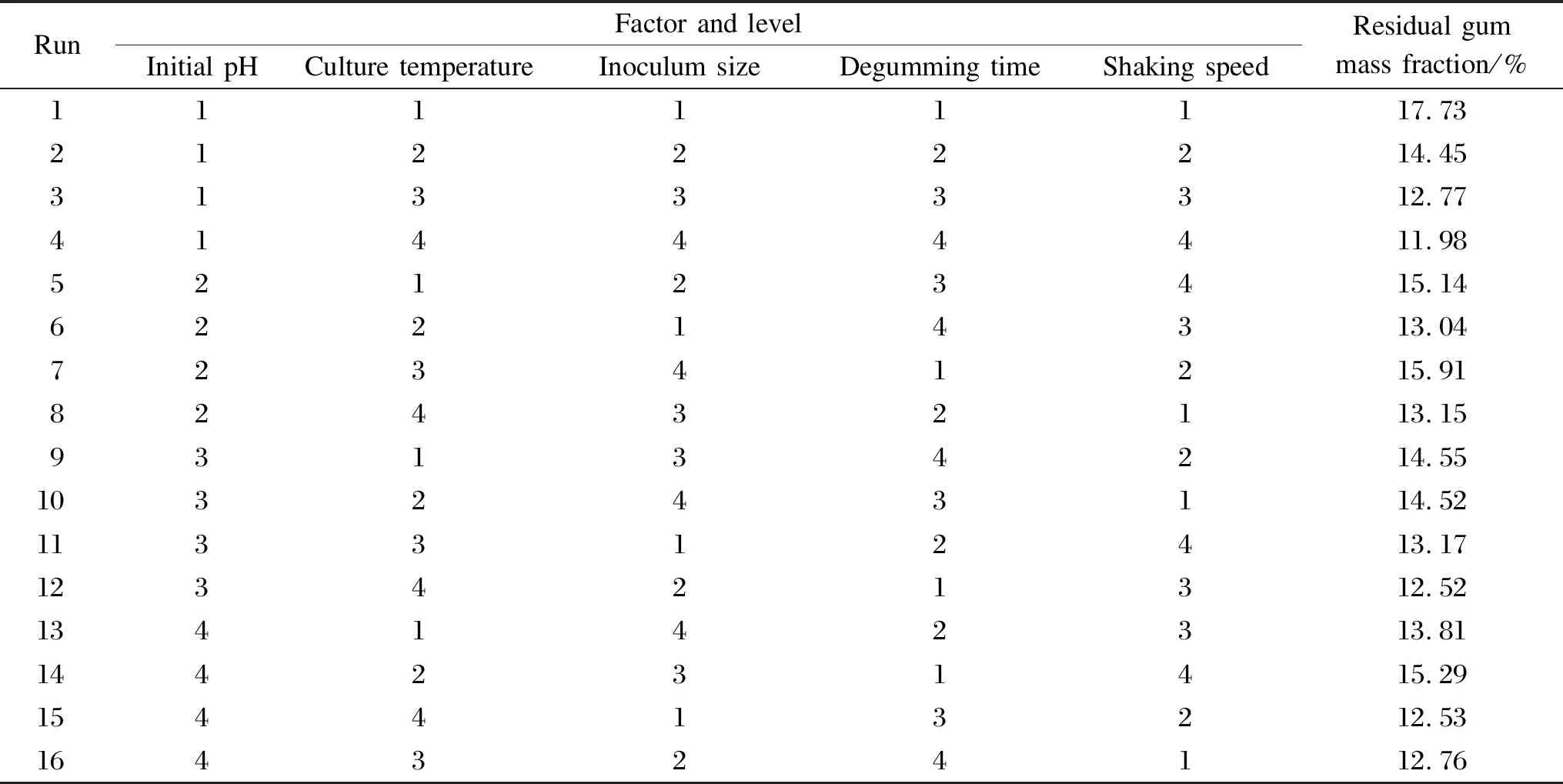
Table 3 L16 (45) matrix optimization of the degumming conditions
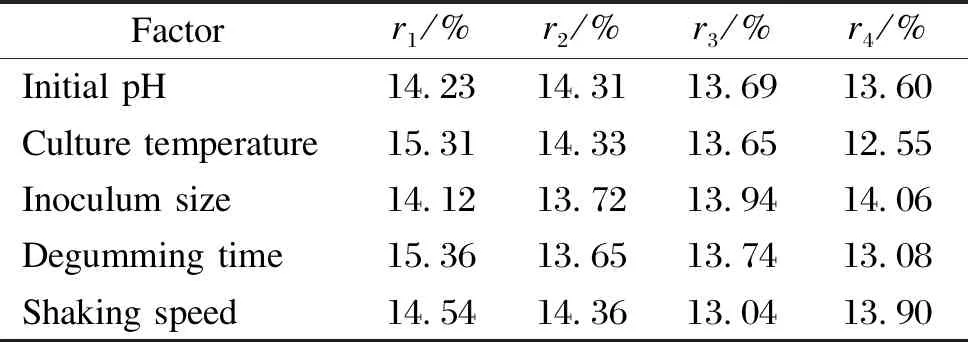
Table 4 Analysis results of L16(45) experiment data
According to the results in Table 4 and Fig.1, the optimal level of each factor is as follows: the initial pH of 8.0, the temperature of 40 ℃, the inoculum size of 5%, the degumming time of 96 h, and the shaking speed of 200 r/min.
Analysis of variance (ANOVA) was used to assess the OAD experiment results, and the sums of squares (SS) for different variables were calculated (Table 5). The error estimation of the experiments was calculated and used in ANOVA because no dummy column was assigned in the L16(45) matrix. The SS of the error was obtained by subtracting all the SS of the items from the total SS. The ANOVA results showed that all the factors contributed to the degumming efficiency (p<0.001), with the most important factor being culture temperature, followed by degumming time and inoculum size. The residual gum mass fraction under the optimal conditions (initial pH of 8.0, culture temperature of 40 ℃, inoculum size of 5%, degumming time of 96 h, and shaking speed of 200 r/min) evaluated from the L16(45) matrix, was 10.8%.
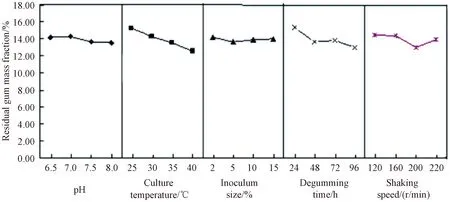
Fig.1 Graphical analysis of relationship between factors and residual gum mass fractions
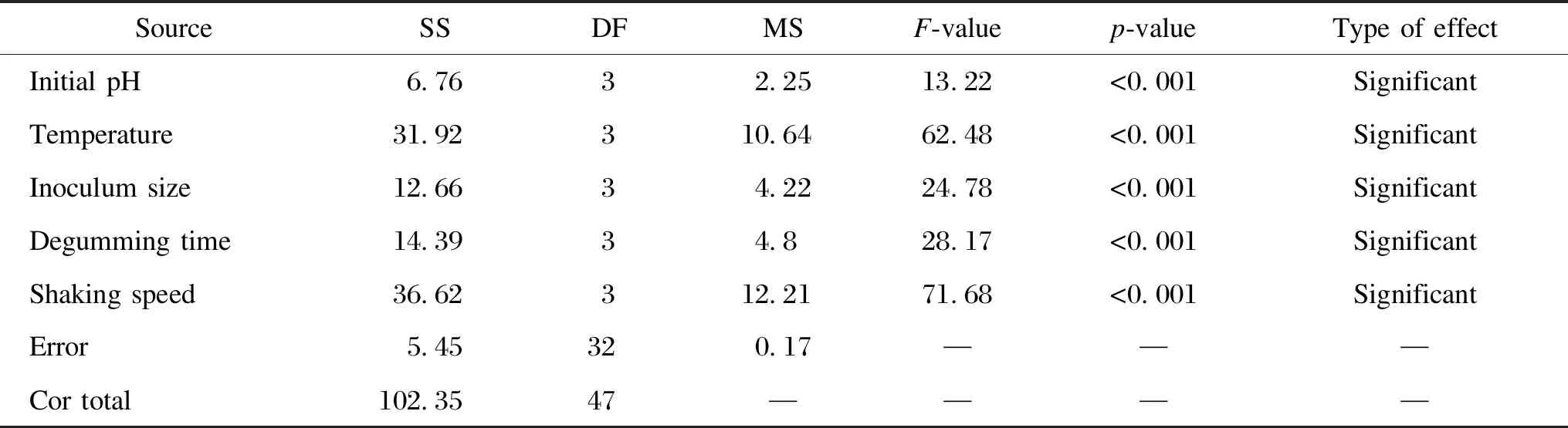
Table 5 ANOVA results for experimental responses in L16(45) matrix
2.2 Analysis of CCD
The optimal degumming time and the optimal inoculum size, identified by the OAD experiment, were chosen, and then the optimal levels of the other three critical factors, namely, initial pH, culture temperature and shaking speed were determined by the CCD approach. The total number of the experiments was 20, including 16 at factorial points and 4 replications at the center point. The response was the residual gum mass fraction of the treated ramie. The pattern of the levels of three factors is shown in Table 6, along with the response values obtained from individual experiments. The data acquired from CCD were analyzed by using the design expert system to yield ANOVA, regression coefficients, and regression equations.
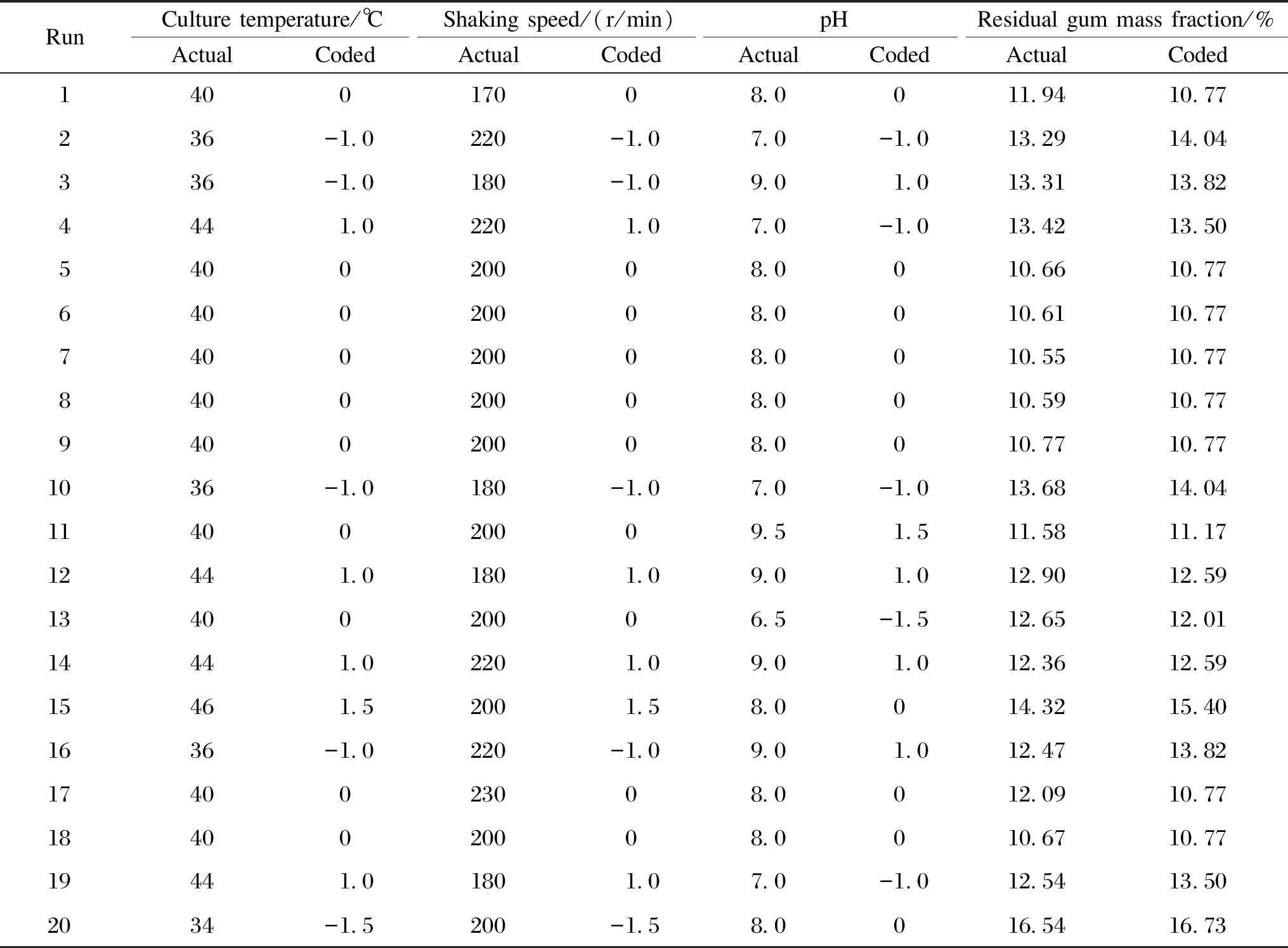
Table 6 Results of optimization of residual gum mass fraction by RSM
Table 7 shows the ANOVA analysis of the results obtained by using CCD after 96 h of fermentation, which demonstrated that the model was highly significant (p<0.050). The measure of goodness of fit of the model (R2) showed that 92.03% of the model variation could be explained by the model. Furthermore, no abnormality was observed from the analyses of the residues. Thus, it was concluded that the model was statistically valid. In addition, the cross-product terms were also not significant, indicating that there were no interactions between the factors studied.
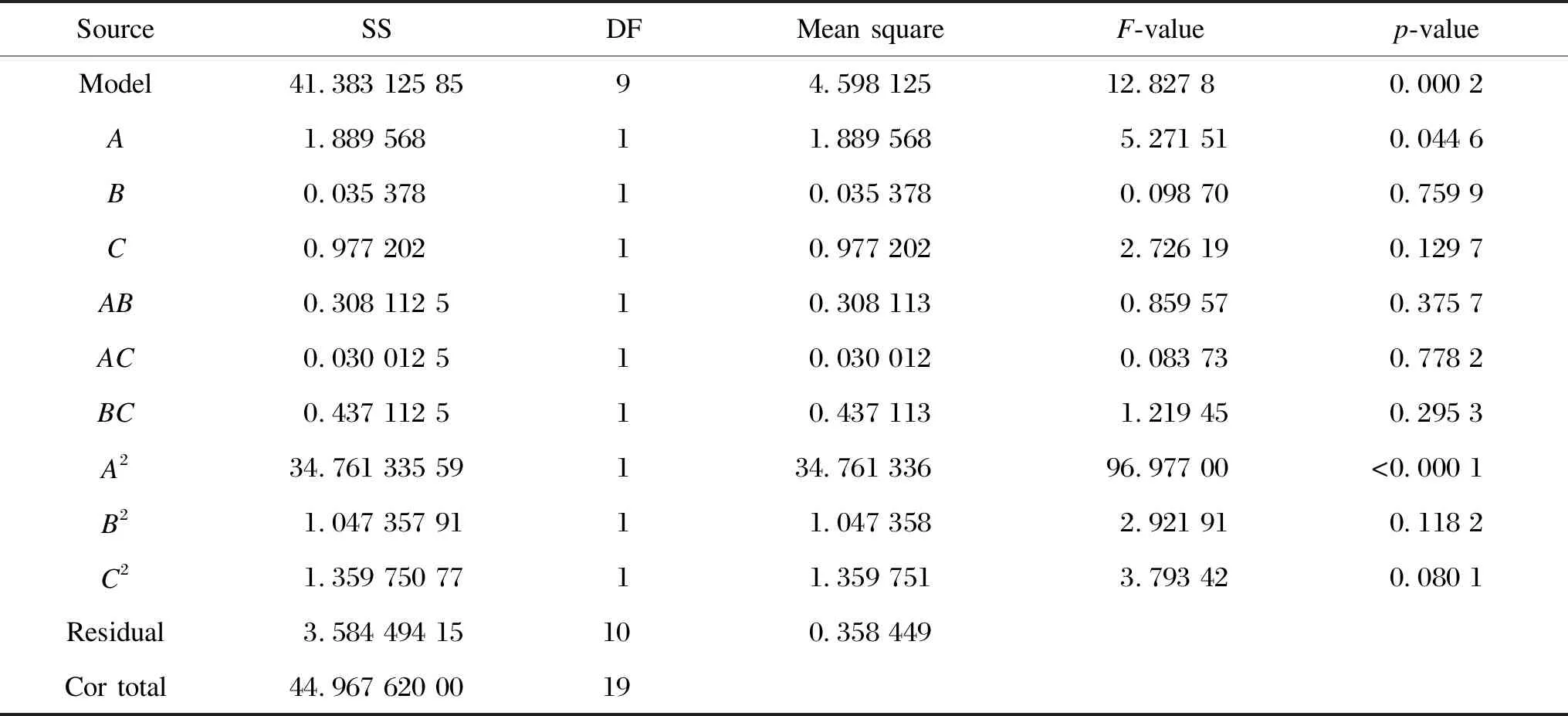
Table 7 ANOVA for experimental responses in CCD
The final equation in terms of coded factors is
R1=10.772 9-0.388 8A-0.053 2B-0.279 6C+
0.196 3AB+0.061 3AC-0.233 8BC+1.836 6A2+
0.318 8B2+0.363 2C2,
whereR1denotes the residual gum mass fraction multiplied by 100.
The final equation in terms of actual factors is
R1=262.037 8-9.893 2A-0.326 1B-
4.366 4C+0.002 5AB+0.015 3AC-0.011 7BC+
0.114 8A2+0.000 8B2+0.356 2C2.
To determine the most adequate operating conditions and analyze the process for the low residual gum mass fraction, contour diagrams were plotted by using the polynomial equation for all possible combinations. According to the regression model, the low residual gum mass fraction of 10.7% was obtained, which corresponded to the code 0.086, 0.220, and 0.448 and an actual values of 40(temperature,℃), 205 (shaking speed, r/min), and 8.5 (initial pH), respectively. To confirm the optimal conditions (culture temperature of 40 ℃, initial pH of 8.5, cultivation time of 24 h, degumming time of 96 h, inoculum size of 5%), a set of five replicate experiments was carried out to affirm the predicted residual gum mass fraction. The confirmatory experiments revealed a residual gum mass fraction of 10.6% which was close to the estimated result (10.7%). Figure 2 illustrates the three-dimensional (3D) curve and contour plot of the calculated response surface obtained from the interaction between culture temperature and shaking speed at an actual initial pH of 8.5.
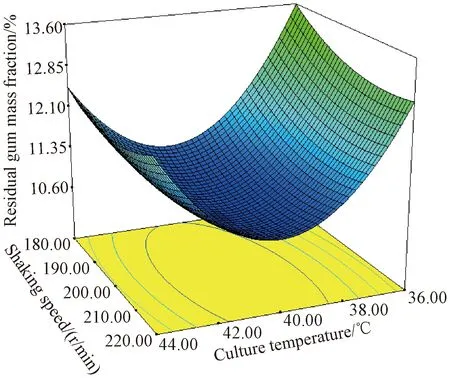
Fig.2 3D curve and contour plot of calculated response surface from interaction between culture temperature and shaking speed at an actual initial pH 8.5
Although the residual gum mass fraction of the degummed ramie fibers did not reach the textile industrial requirements, the pectinase and xylanase produced byBSDZ5caused significant changes in the molecular structure of the gum and disrupted the stability of the colloidal complex, thereby activating the chemical reaction properties of the macromolecules and enhancing the sensitivity to alkali. AfterBSDZ5treated ramie was treated by 5 g/L NaOH solution for 30 min at atmospheric pressure, the residual gum mass fraction decreased to less than 2% which met the textile industrial requirement.
2.3 Evaluation of degumming
The constituents in ramie were measured according to Ref. [12]. The results are listed in Table 8. The ramie fibers were degummed under optimal conditions for 96 h. The tensile properties of the degummed ramie fibers were tested. The results are shown in Table 9. It is demonstrated that after degumming, owing to their high intrinsic strength, the fibers retain enough tenacity which meets the textile requirements.
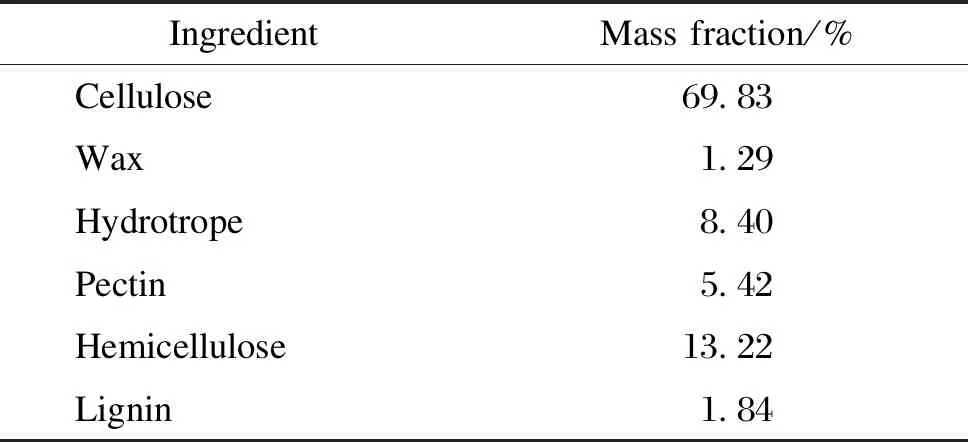
Table 8 Ingredients of ramie

Table 9 Tensile properties of degummed ramie fiber
2.4 Observation of fibers
The surface structure of ramie fibers was analyzed by SEM as shown in Fig.3.BSDZ5was fixed in glutaraldehyde (2.5%, volume fraction) and dehydrated in an acetone gradient, and all samples were sputter-coated with gold particles.
During the degumming process of ramie fibers byBSDZ5, both pectinase and xylanase were active. Treatment of ramie fibers withBSDZ5alone resulted in fibers with almost smooth surfaces and a few residual gum materials (Fig.3(b)). However, the fibers treated byBSDZ5and NaOH had a smoother surface and almost no gum (Fig.3(c)), which revealed that the bio-chemical-degummed sample exhibited a better degumming effect. Furthermore, the separation of the fibers was also improved by the bio-chemical treatment.
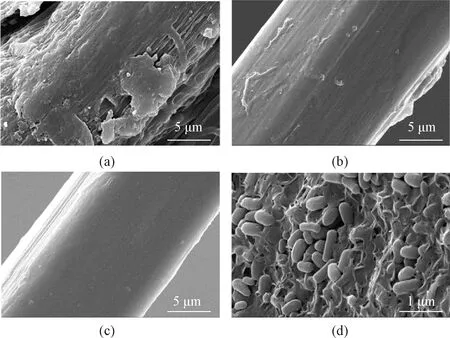
Fig.3 SEM images of ramie fibers: (a) raw; (b) bio-degummed; (c) bio-chemical-degummed; (d) BSDZ5 on fibers
3 Conclusions
By employing OAD and CCD, with only a limited number of experiments, the optimal conditions for microbial degumming of ramie, including the culture temperature of 40 ℃, the initial pH of 8.5 in culture medium, the shaking speed of 205 r/min, the degumming time of 96 h, and the inoculum size of 5%, were obtained. AfterBSDZ5degumming under the optimal conditions, the resultant ramie fibers contained only 10.6% of the original gum.
杂志排行
Journal of Donghua University(English Edition)的其它文章
- Oscillation Reduction of Breast for Cup-Pad Choice
- Effect of Weft Binding Structure on Compression Properties of Three-Dimensional Woven Spacer Fabrics and Composites
- pth-Moment Stabilization of Hybrid Stochastic Differential Equations by Discrete-Time Feedback Control
- Semantic Path Attention Network Based on Heterogeneous Graphs for Natural Language to SQL Task
- Device Anomaly Detection Algorithm Based on Enhanced Long Short-Term Memory Network
- Image Retrieval Based on Vision Transformer and Masked Learning
